Analysis of Hematopoietic Stem Progenitor Cell Metabolism
Summary
Hematopoietic stem progenitor cells (HSPCs) transition from a quiescent state to a differentiation state due to their metabolic plasticity during blood formation. Here, we present an optimized method for measuring mitochondrial respiration and glycolysis of HSPCs.
Abstract
Hematopoietic stem progenitor cells (HSPCs) have distinct metabolic plasticity, which allows them to transition from their quiescent state to a differentiation state to sustain demands of the blood formation. However, it has been difficult to analyze the metabolic status (mitochondrial respiration and glycolysis) of HSPCs due to their limited numbers and lack of optimized protocols for non-adherent, fragile HSPCs. Here, we provide a set of clear, step-by-step instructions to measure metabolic respiration (oxygen consumption rate; OCR) and glycolysis (extracellular acidification rate; ECAR) of murine bone marrow-LineagenegSca1+c-Kit+ (LSK) HSPCs. This protocol provides a higher amount of LSK HSPCs from murine bone marrow, improves the viability of HSPCs during incubation, facilitates extracellular flux analyses of non-adherent HSPCs, and provides optimized injection protocols (concentration and time) for drugs targeting oxidative phosphorylation and glycolytic pathways. This method enables the prediction of the metabolic status and the health of HSPCs during blood development and diseases.
Introduction
Since the lifespan of most mature blood cells is short, the homeostasis of blood relies on the self-renewal and differentiation of a long-lived but rare population of hematopoietic stem cells (HSPCs)1. HSPCs are quiescent, but they are quick to proliferate and undergo differentiation upon stimulation to sustain demands of the blood system. As each HSPC cellular state requires a unique bioenergetic demand, the metabolic changes are key drivers of HSPC fate decisions. Therefore, the loss of metabolic plasticity, by altering the equilibrium between quiescence, self-renewal, and differentiation of HSPCs, often leads to myelo- or lympho-proliferative disorders. Together, the understanding of metabolic regulation of HSPC development is critical to uncover mechanisms underlying hematologic malignancies2,3,4,5.
Mitochondrial respiration and glycolysis generate ATP to drive intracellular reactions and produce the building blocks necessary for macromolecule synthesis. Since HSPCs have low mitochondrial mass compared to differentiated cells6 and they sustain quiescence in hypoxic bone marrow niches, HSPCs primarily rely on glycolysis. Activation of HSPCs enhances their mitochondrial metabolism that leads to the loss of quiescence and their subsequent entry into the cell cycle. Such metabolic plasticity of HSPCs allows the maintenance of the HSPC pool throughout adult life6,7,8,9,10,11,12. Therefore, it is critical to investigate their metabolic activities, such as the oxygen consumption rate (OCR; index of oxidative phosphorylation) and the extracellular acidification rate (ECAR; index of glycolysis) to analyze the HSPC activation and the health status. Both the OCR and the ECAR can be measured simultaneously, in real time, using an extracellular flux analyzer. However, the current method requires large numbers of cells and is optimized for adherent cells13. Since HSPCs cannot be isolated in large quantities from mice14, require sorting to obtain a pure population, are non-adherent cells15, and cannot be cultured overnight without avoiding differentiation16, it has been difficult to measure the OCR and the ECAR of HSPCs. Here, we provide a set of clear, step-by-step instructions to accompany video-based tutorials on how to measure metabolic respiration and glycolysis of few thousands murine bone marrow-LineagenegSca1+c-Kit+ (LSK) HSPCs.
Protocol
This protocol was approved by Nationwide Children’s Hospital Animal Care and Use Committee (IACUC).
NOTE: The protocol is described in chronological order that spans over the period of two days. Use fresh reagents as described in the protocol below.
1. Preparation of Reagents on the Day Prior to the Assay
- Hydrate the sensor cartridge.
- Incubate 5 mL of the calibrant (Table of Materials) in a non-CO2 37 °C incubator overnight.
- Open the flux assay kit (Table of Materials) and separate the sensor cartridge (green; top) from the utility plate (transparent; bottom). Next, place the sensor cartridge upside down and adjacent to the utility plate.
- Using sterile water, fill the wells of the utility plate (200 µL) and the chambers around the wells (400 µL).
- Submerge the sensor cartridge into the utility plate making sure that the sensors are completely covered by water. Tap 3x to avoid the formation of bubbles.
- Place the cartridge submerged in the utility plate in a non-CO2 37 °C incubator overnight. To prevent evaporation of the water, make sure that the incubator is properly humidified. Place an open beaker containing H2O as an extra precaution next to the cartridge-utility plate, particularly if using a regular oven.
- Prepare the assay plate.
- Sterilize the surface of biosafety cabinet class 2 using 70% ethanol. Open the assay plate under the hood.
- Add 40 µL of commercially available 0.01% (w/v) poly-L-lysine (PLL) solution to each well of the assay plate under the hood.
- Cover the assay plate with the lid provided in the kit. Incubate the closed assay plate at room temperature (RT) under the hood for 1 h.
NOTE: The goal of the incubation is to let PLL coat the surface of the assay plate to facilitate adhesion of suspension cells to the surface of the assay plate. - After 1 h incubation of the assay plate, remove the excess solution with a sterile vacuum-based aspirator and air dry the well under the hood.
NOTE: It takes ~30−60 min to air-dry the wells of the assay plate following the excessive PLL removal using the aspirator.
2. Day of the Assay
- Prepare the cartridge.
- Lift the sensor cartridge. Place it upside down in the tissue culture hood and discard the water from the utility plate.
- Fill the utility plate wells with 200 μL of the pre-warmed calibrant.
- Fill the chambers around the wells with 400 μL of the calibrant.
- Submerge the sensor cartridge into the utility plate, making sure that the sensors are completely covered by the calibrant. Tap 3x to avoid the formation of bubbles.
- Equilibrate the sensor cartridge submerged in the utility plate in a non-CO2 37 °C incubator for 45−60 min.
- Harvest murine bone marrow-derived LSK HSPCs.
- To accommodate sufficient biological replicates, plan to use HSPCs derived from one mouse per well of the assay plate.
NOTE: This bone marrow-harvesting method provides ~50,000−80,000 LSK HSPCs from each mouse. This protocol to measure extracellular flux is optimized for ~70,000 LSK HSPCs per well of a 96 well-plate. - Euthanize mice using CO2 overdose and cervical dislocation, following local IACUC approved methods.
- Sterilize the surface of dissecting tools and the bench using 70% ethanol.
- For each mouse, pre-fill one Petri dish with 1x phosphate-buffered saline (PBS) containing 2% heat inactivated fetal bovine serum (FBS) at RT.
NOTE: Do not use pre-chilled PBS as it will create clumps in the following steps. - Spray 70% ethanol (v/v) on the entire euthanized mouse. Isolate all bones, including upper and lower limbs, hip bones, sternum, rib cage, and spine, from the mouse17,18. Pull out white matter from the spinal cord of the mouse as it can contaminate LSK cells. Place all bones in 1x PBS (+ 2% FBS), as they are being collected, in a Petri dish until further use.
- Invert a 50 mL conical tube (new each time) and use it to triturate bones submerged in 1x PBS (+ 2% FBS) in the Petri dish.
- Using a 10 mL serological pipette, pipette up and down (~10x) to uniformly flush out cells from bones, after crushing bones.
- Place a 40 µm cell strainer on a 50 mL conical tube. Pre-wet the surface of the strainer by passing 1 mL of 1x PBS (+ 2% FBS).
- Harvest bone marrow-derived cell suspension from step 2.2.7 and pass it through the pre-wet surface of the cell strainer to remove bone debris.
- Repeat steps 2.2.6 through 2.2.9 until all bone materials turn white as a marker that most of bone marrow cells are collected in the 50 mL conical tube.
NOTE: It often takes two 50 mL conical tubes per mouse to collect all bone marrow-derived cells. - Centrifuge 50 mL conical tubes containing bone marrow-derived cells for 5 min at 500 x g and RT.
- Remove the supernatant. Resuspend bone marrow-derived cells (combine contents of both 50 mL tubes) in 5 mL of 1x PBS (+ 2% FBS) as a final volume and keep cells at RT.
- To accommodate sufficient biological replicates, plan to use HSPCs derived from one mouse per well of the assay plate.
- Harvest mononucleated murine bone marrow cells.
- Add 5 mL of density gradient medium (i.e., Ficoll) to a 15 mL conical tube. Then slowly add 5 mL of the bone marrow cell suspension. Make sure that cells remain as a layer above the density gradient medium.
- Centrifuge for 30 min at 500 x g and RT. Do not use a brake in the centrifuge. Make sure the centrifuge is at the lowest possible acceleration (e.g., 1 acceleration and 0 deceleration).
- Harvest the middle interface of mononucleated cells (white color) following centrifugation into a fresh 15 mL conical tube.
- Wash cells, harvested from density gradient medium, with 5 mL of 1x PBS (+ 2% FBS). Centrifuge for 5 min at 500 x g and 4 °C. Remove the supernatant.
- Repeat step 2.3.4.
- Resuspend cell contents of the tube in 300 µL of 1x PBS (+ 2% FBS). Aliquot 10 µL of cell suspension for unstained or single-color control in a FACS tube.
- Harvest LSK HSPCs from mononucleated murine bone marrow cells.
- Make a cocktail of biotin-antibodies by mixing 3 µL per sample of the following antibodies: Gr1, Cd8a, Cd5, B220, Ter119. Add 15 µL of the biotin-antibody cocktail to 300 µL of mononucleated bone marrow cells.
NOTE: Each antibody is used at 1:100 dilution. - Incubate cells with the biotin-antibody cocktail for 30 min at 4 °C with agitation to avoid cells clumping in the bottom of the tube.
- Add 10 mL of pre-chilled 1x PBS (+ 2% FBS) to cells mixed with the biotin-antibody cocktail.
- Centrifuge the tube for 5 min at 500 x g and 4 °C. Discard the supernatant and resuspend the cell pellet in 400 µL of 1x PBS (+ 2% FBS). Aliquot 10 µL for streptavidin-single color control.
- Briefly vortex anti-biotin microbeads (Table of Materials) before use. Add 80 µL of microbeads to each cell sample (of 400 µL). Mix well and incubate for additional 20 min at 4 °C, with agitation.
- Add 10 mL of pre-chilled 1x PBS (+ 2% FBS) to cells. Centrifuge the tube for 5 min at 500 x g and 4 °C.
- Discard the supernatant and resuspend the cell pellet in 1 mL of 1x PBS (+ 2% FBS). Store at 4 °C while setting up magnetic separation unit.
- Place a column (Table of Materials) in the magnetic field of the magnetic assisted cell sorting (MACS) separator at 4 °C. Prepare the column for magnetic separation by rinsing it with 3 mL of 1x PBS (+ 2% FBS) under the gravity flow at 4 °C.
- Add the cell suspension from step 2.4.7 to the pre-wet column at 4 °C. Allow the cells to pass through the column at 4 °C and collect effluent in a 15 mL conical tube.
NOTE: The fraction with unlabeled cells in such effluent represents the enriched lineage negative cells. - Wash column with 3 mL of 1x PBS (+ 2% FBS) at 4 °C. Repeat 3x. Collect the flow-through and keep it at 4 °C. Count the eluted viable cells by trypan blue exclusion using a hemocytometer.
- Centrifuge the 15 mL conical tube containing the flow-through for 5 min at 500 x g and 4 °C. Discard the supernatant. Resuspend cells in 0.5 mL of 1x PBS (+ 2% FBS) and transfer the contents to a FACS tube.
- Add 24 µL of the LSK antibody cocktail to each 107 cells. The antibody cocktail contains equal concentration of 450-streptavidin antibody, PE-CY7-Sca1 antibody, and APC-c-Kit antibody.
- Incubate for 1 h at 4 °C with agitation under dark (covered with tin foil).
- Add 3 mL of 1x PBS (+ 2% FBS) to the FACS tube. Centrifuge for 5 min at 500 x g and 4 °C. Discard the supernatant.
- Resuspend antibody-labelled cells in 1 mL of 1x PBS (+ 2% FBS). Add 1 µL of 1 mg/mL propidium iodide to cell suspension just before sorting.
- Filter contents of the FACS tube using a 40 µm strainer right before sorting LSK cells.
- Collect LSK cells, via FACS sorting, into 1.5 mL tube containing 0.5 mL of complete media supplemented with 2 mM glutamine, 3 mg/mL glucose, 1 mM pyruvate, 1x thrombopoietin (TPO), 1x stem cell factor (SCF), 0.5x penicillin/streptomycin (P/S), pH 7.4 (Table 1).
- Make a cocktail of biotin-antibodies by mixing 3 µL per sample of the following antibodies: Gr1, Cd8a, Cd5, B220, Ter119. Add 15 µL of the biotin-antibody cocktail to 300 µL of mononucleated bone marrow cells.
3. Mitochondrial Respiration and Glycolysis Assays of LSK HSPCs
- LSK seeding in the assay plate
- Centrifuge LSK cells from step 2.4.17 for 5 min at 500 x g and RT. Discard the supernatant.
- Resuspend cells in complete media to a final concentration of at least 70,000 cells/40 µL.
- Seed contents of 40 µL media (containing 70,000 cells) in the PLL-coated 8-well plate from step 1.2.4. Leave all corner wells empty.
- Centrifuge for 1 min at 450 x g and RT. Do not apply the brake. Make sure that cells are attached to the well bottom using the inverted microscope.
- Add 135 µL of complete media to the cells in each well for a final volume of 175 µL. Add 175 µL of complete media to the 2 corners of the plate as blanks.
- Incubate the cells in the non-CO2 incubator for 2 h at 37 °C.
- Set up a program to add drugs to each well of the well plate in the analyzer using a metric described in Table 2 (mitochondrial stress test) and Table 3 (glycolysis stress test).
NOTE: While cells are incubating, turn on the instrument and make sure it is at 37 °C.
- Mitochondrial stress test
- Prepare 45 µM stock solutions of oligomycin, 50 µM carbonyl cyanide 4-(trifluromethoxy)phenylhydrazone (FCCP) and 25 µM stock solutions of rotenone/antimycin A. To prepare 45 µM oligomycin stock solution, dissolve the contents of the commercially available pouch in 280 µL of the complete media. To prepare 50 µM FCCP stock solution, dissolve the contents of the commercially available pouch in 288 µL of the complete media. To prepare 25 µM rotenone/antimycin A stock solution, dissolve the contents of the commercially available pouch in 216 µL of the complete media.
- Take the sensor cartridge in the utility plate out from the incubator and load its ports (A, B, and C) such that each well would have final concentration of 2 µM oligomycin, 1.5 µM FCCP, and 0.5 µM of rotenone/antimycin A as needed, following dilution metrics described in Table 4 (for oxygen consumption rate).
- Remove the lid from the sensor cartridge assembled in the utility plate and place it on the instrument tray. Start the calibration that will take 20 min.
- After the calibration, remove the utility plate and substitute it with assay plate containing LSK cells, which are now adhered to the bottom of the well.
- Press Weitermachen to start the program described in Table 2. After the completion of the program, retrieve the data and analyze them using the Wave Desktop software.
NOTE: The data generated from the extracellular flux assays can be plotted using the dot plot from the Wave Desktop software or exported to web-based statistics program.
- Glycolysis stress test
- Reconstitute glucose in 300 µL (100 mM), oligomycin in 288 µL (50 µM), 2-D glucose (2-DG) in 300 µL (500 mM) of complete media from the commercially available pouch.
- Gently pipette up and down (~10x) to solubilize the compounds. Vortex the 2-DG for approximately 1 min to ensure proper dissolution into media.
- Remove the sensor cartridge out from the incubator. Load ports A through C following dilution metrics described in the Table 5 to obtain a final concentration of 10 mM glucose, 2 µM oligomycin, and 50 mM 2-DG.
- Repeat steps 3.2.3 and 3.2.4.
- Press Weitermachen to start the program described in Table 3. After the completion of the program, retrieve the data and analyze them using the Wave Desktop software.
Representative Results
Our extraction method allowed us to harvest up to ~80,000 LSK HSPCs per mouse. The viability and numbers of LSK cells were improved with our method, because we: (1) combined bone marrow from upper and lower limbs, hip bones, sternum, rib cage, and spine, (2) avoided using red cell lysis buffer that would have increased cell-death and clumping, (3) used the density gradient medium separation of mono-nucleated cells, and (4) avoided using pre-chilled buffer that would have caused the loss of cells-of-interest in clumps.
Although extracellular flux analysis has been traditionally used for adherent cells, our use of the PLL coating of wells, followed by centrifugation of cells on it, facilitated adherence of LSK HSPCs to the surface of the well. This allowed us to measure the extracellular flux, and thus metabolic health of LSK HSPCs. Considering the limited number of cells that can be harvested from a mouse and the long duration of the protocol for their isolation, our use of the analyzer with its 8 well format has emerged as the most cost-effective and feasible solution (Figure 1).
Cells use glycolysis and mitochondrial respiration to replenish their energy requirements and to produce intermediates needed for their proliferation and growth19. The hexokinase enzyme converts glucose in glucose-6-phosphate and that is subsequently transformed into pyruvate20. Pyruvate can then be processed into lactate and is exported from the cell with protons21. ECAR measures the acidification of the media and is thus an indicator of glycolysis. Pyruvate can also be transported into the mitochondria and transformed in acetyl coenzyme A (CoA). Acetyl CoA enters the TCA cycle, which provides energy intermediates to drive the electron movements of the Electron Transport Chain (ETC) and generates a proton gradient in the mitochondrial inter-membrane space22. Oxygen acts as the final electron acceptor, and protons move back to the mitochondrial matrix through the ATP synthase complex while generating ATP23. OCR measures the oxygen consumption and it is therefore used to quantify the mitochondrial respiration.
In order to analyze the OCR and the ECAR in basal and stressed conditions, we used sequential injection of drugs that interfere with glycolysis and mitochondrial respiration. We used glucose and 2-deoxyglucose (2-DG), a glucose analogue, to initiate and block glycolysis respectively24. We used Rotenone (a complex I-specific inhibitor of the ETC), antimycin A (a complex III-specific inhibitor of the ETC), oligomycin (inhibitor of ATP synthase), and the uncoupling agent carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) to block specific events of the ETC25. We titrated such reagents to find the optimal concentration for LKS HSPCs (Figure 2A,B).
To perform the glycolysis stress test, we cultured the LSK HSPCs in a glucose/pyruvate deprived media (as recommended by the manufacturer), or in glucose/pyruvate containing media. As expected, we found the basal level of the ECAR was higher for LSK HSPCs cultured in glucose/pyruvate+ media compared to LSK HSPCs cultured in glucose/pyruvate- media. The first injection with glucose did not change the basal level of ECAR for LSK HSPCs cultured in glucose/pyruvate+ media while it boosted glycolysis in LSK HSPCs cultured in glucose/pyruvate- media. However, the basal level of the ECAR, after the injection with glucose, remained lower compared to the glucose/pyruvate+ group. The second injection with oligomycin, which could block the production of ATP through oxidative phosphorylation, activated the glycolysis at its maximum level of LSK HSPCs in glucose/pyruvate+ media, but it did not affect the glucose/pyruvate- group. The last injection with the glucose analogue 2-DG returned the ECAR to its non-glycolytic level (Figure 2C).
For the mitochondrial stress test, we measured the basal level of OCR of LSK HSPCs in glucose/pyruvate+ media. We first injected oligomycin that initially hyperpolarized the mitochondrial membrane, prevented more proton pumping through the ETC complexes, and thus reduced the rate of mitochondrial respiration. The second injection of FCCP ionophores pushed ETC and OCR levels to their maximum as cells tried to recover the mitochondrial membrane potential. The final injection with other two of the ETC inhibitors (antimycin A and rotenone) caused the complete stop of mitochondrial respiration and thus the OCR reverted to its minimum level (Figure 2D).
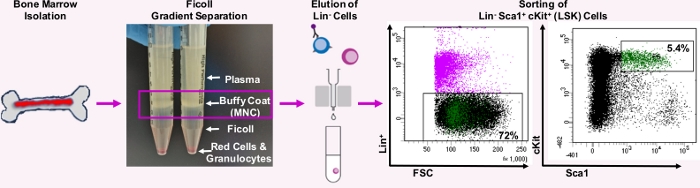
Figure 1: Schema demonstrating isolation of LineagenegSca1+c-Kit+ (LSK) hematopoietic stem progenitor cells from mouse bone marrow. Bone marrow is extracted from bones and mononuclear cells (MNCs) are isolated through density gradient medium gradient separation. Next, cells are incubated with biotinylated Lineage+ antibodies and streptavidin-conjugated magnetic beads to elute Lineage negative (Lin–) cells following their magnetic separation. Lin– cells are subsequently incubated with LSK antibodies and LSK cells isolated by cell sorting. Please click here to view a larger version of this figure.
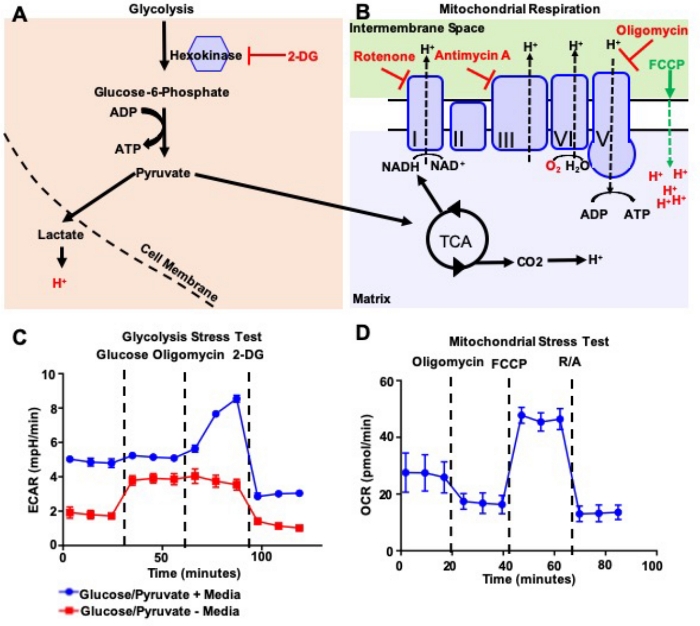
Figure 2: Extracellular flux analyses of murine LineagenegSca1+c-Kit+ (LSK) hematopoietic stem progenitor cells. (A,B) Mechanistic description of drugs utilized for extracellular flux analyses during glycolysis and mitochondrial respiration. (C) Representative results of glycolysis stress test on murine LSK HSPCs in presence or absence of glucose/pyruvate in the media. (D) Representative results of mitochondrial stress test on murine LSK HSPCs. Error bars represent the standard deviation of the mean (S.D.) Please click here to view a larger version of this figure.
| Stock | Final concentration | Volume for 30 mL | |
| Complete media | 28.691 mL | ||
| P/S | 100x | 0.5x | 150 µL |
| L-Glutamine | 200 mM | 2 mM | 300 µL |
| Pyruvate | 100 mM | 1 mM | 300 µL |
| Glucose | 1 M/180.2 mg/mL | 3 mg/mL | 499.4 µL |
| TPO | 100 µg/mL | 100 ng/mL | 30 µL |
| SCF | 100 µg/mL | 100 ng/mL | 30 µL |
Table 1: Contents and preparation of the complete XF media.
| Basal | Oligomycin (2 µM) | FCCP (1.5 µM) | R/A (0.5 µM) | |
| Repetition | 3 times | 3 times | 3 times | 3 times |
| Mix | 3 min | 3 min | 3 min | 3 min |
| Wait | 0 min | 0 min | 0 min | 0 min |
| Measure | 4 min | 4 min | 4 min | 4 min |
Table 2: Injection protocol for the mitochondrial stress test.
| Basal | Glucose (10 mM) | Oligomycin (2 µM) | 2-DG (50 mM) | |
| Repetition | 3 times | 3 times | 3 times | 3 times |
| Mix | 3 min | 3 min | 3 min | 3 min |
| Wait | 0 min | 0 min | 0 min | 0 min |
| Measure | 7 min | 7 min | 7 min | 7 min |
Table 3: Injection protocol for the glycolysis stress test.
| Port | Drug | Final well concentration (µM) | Stock solution volume (µL) | Media volume (µL) | Port solution (µM) | Volume added to port (µL) |
| Port A | Oligomycin | 2 | 100 | 181.25 | 16 | 25 |
| Port B | FCCP | 1.5 | 100 | 270.4 | 13.5 | 25 |
| Port C | Rotenone/antimycin A | 0.5 | 60 | 240 | 5 | 25 |
Table 4: Dilution metrics to obtain optimum drug concentration for mitochondrial stress test in each well of the analyzer.
| Port | Drug | Final well concentration | Stock solution volume (µL) | Media volume (µL) | Port solution | Volume added to port (µL) |
| Port A | Glucose | 10 mM | 300 | 75 | 80 mM | 25 |
| Port B | Oligomycin | 2 µM | 108 | 192 | 18 µM | 25 |
| Port C | 2-DG | 50 mM | 300 | 0 | 500 mM | 25 |
Table 5: Dilution metrics to obtain optimum drug concentration for glycolysis stress test in each well of the analyzer.
Discussion
Here, we demonstrate the isolation of a maximum amount of pure and viable murine LSK HSPCs population as well as the measurement of their glycolysis and mitochondrial respiration with an extracellular flux analyzer. Specifically, the protocol overcomes the following technical issues for the use of LSK HSPCs : i) the low frequency of LSK HSPCs in murine bone marrow14, ii) low basal metabolic activity of LSK HSPCs26, iii) the fragility of LSK HSPCs27, and iv) the non-adherence of LSK HSPCs to culture vessels15. In addition, we have optimized the drug concentrations and media composition for the optimum performance of the extracellular flux assays.
Bone marrow-derived HSPCs reside in the hypoxic niche and they display a glycolytic phenotype, which is crucial for the maintenance of their stemness28. Conversely, respiration is essential for HSPC differentiation6. As metabolic dysfunction of HSPCs leads to blood diseases; here, we have described the protocol and video-based tutorials to measure the hallmarks of metabolic functions, such as the OCR and the ECAR, for murine bone marrow-derived LSK HSPCs.
Contrary to manufacturer recommendations for adherent cells, we found that the glycolysis stress test results on LSK HSPCs are optimum when cells are cultured in glucose/pyruvate+ media compared to glucose/pyruvate- media. We realized that the lack of glucose in media for LSK HSPCs results in cell death during the ~2 h equilibration period in a non-CO2 incubator. As the glucose injection did not change the basal level of ECAR for LSK HSPCs cultured in glucose/pyruvate+ media, our modification of the protocol not only preserves the essence of the glycolytic stress test, but it also allows us to distinguish between the basal glycolysis (before any injections), glycolytic capacity (after oligomycin injection) and non-glycolytic acidification (after injection with 2-DG) of LSK HSPCs.
Our mitochondrial stress test of LSK HSPCs showed that the ATP produced by oxidative phosphorylation in LSK HSPCs is minimal as seen with the small reduction in the OCR after the oligomycin injection. Conversely, the elevation in the OCR upon the FCCP injection affirms that LSK HSPCs are able to respond to higher energy demand.
Together, the goal of this protocol was to provide a set of clear, concise instructions to accompany video-based tutorials on how to measure the metabolic functions, such as the OCR and the ECAR, of HSPCs. With key modifications and additional recommendations for harvesting higher numbers of healthy LSK HSPCs, making them adherent to the surface of the well, as well as the optimization of the incubation time and drug concentration, this protocol will empower investigators to analyze glycolysis and mitochondrial respiration status of HSPCs as well as non-adherent hematopoietic cells during blood development and diseases.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work is in part supported by the funding support from the National Institutes of Health (HL131645, CA016058), the St. Baldrick’s Foundation, and the Pelotonia Foundation.
Materials
| 0.01% (w/v) poly-L-lysine solution | Sigma | P8920 | Used for LSK attachment |
| 40 µm cell strainer | Fisher Scientific | 22-363-547 | Used for cell filtration after bone crushing |
| Anti-Biotin MicroBeads | Miltenyi | 130-090-485 | Used for Lin- separation |
| Biotin Rat Anti-Mouse CD45R/B220 Clone RA3-6B2 | BD Biosciences | 553086 | Used for Lin- separation |
| Biotin Rat Anti-Mouse CD5 Clone 53-7.3 | BD Biosciences | 553019 | Used for Lin- separation |
| Biotin Rat Anti-Mouse CD8a Clone 53-6.7 | BD Biosciences | 553029 | Used for Lin- separation |
| Biotin Rat Anti-Mouse Ly-6G and Ly-6C Clone RB6-8C5 | BD Biosciences | 553125 | Used for Lin- separation |
| Biotin Rat Anti-Mouse TER-119/Erythroid Cells Clone TER-119 | BD Biosciences | 553672 | Used for Lin- separation |
| CD117 (c-Kit) Monoclonal Antibody (2B8), APC | eBioscience | 17-1171-83 | Used for LSK sorting |
| Falcon 15 ml Conical Centrifuge Tubes | Falcon-Fischer Scientific | 14-959-53A | Used in cell isolation |
| Falcon 50 ml Conical Centrifuge Tubes | Falcon-Fischer Scientific | 14-432-22 | Used in cell isolation |
| Falcon Round-Bottom Polypropylene Tubes | Falcon-Fischer Scientific | 14-959-11A | Used for LSK sorting |
| Fetal Bovine Serum | Neuromics | FBS001-HI | Used in FACS buffer |
| Histopaque-1083 | Sigma | 10831 | Used for ficoll gradient separation |
| L-glutamine 100x | Fisher Scientific | 25-030-081 | Used for the assay media |
| LS Column | Miltenyi | 130-042-401 | Used for Lin- separation |
| Ly-6A/E (Sca-1) Monoclonal Antibody (D7), PE-Cyanine7 | eBioscience | 25-5981-82 | Used for LSK sorting |
| Murine Stem Cell Factor (SCF) | PeproTech | 250-03-100UG | Used for the assay media |
| Murine Thrombopoietin (TPO) | PeproTech | 315-14-100UG | Used for the assay media |
| PBS 1% | Fisher Scientific | SH3002802 | Used for FACS buffer |
| Penicillin-Streptomycin (10,000 U/mL) | Fisher Scientific | 15140122 | Used for the assay media |
| Propidium Iodide | Fisher Scientific | P1304MP | Used for LSK sorting |
| Seahorse XFp Cell Culture Miniplate | Agilent Technologies | 103025-100 | Used for LSK seeding |
| Sodium Pyruvate (100 mM) | ThermoFisher | 11360070 | Used for the assay media |
| Streptavidin eFluor 450 Conjugate | eBioscience | 48-4317-82 | Used for LSK sorting |
| XF Calibrant | Agilent Technologies | 100840-000 | Used for cartridge equilibration |
| XF media | Agilent Technologies | 103575-100 | Used for the assay media |
| XFp Glycolysis Stress Test Kit | Agilent Technologies | 103017100 | Drugs for glycolysis stress test |
| XFp Mitochondrial Stress Test Kit | Agilent Technologies | 103010100 | Drugs for mitochondrial stress test |
| XFp Sensor Cartridge | Agilent Technologies | 103022-100 | Used for glycolysis and mitochondrial stress test |
Referenzen
- Dharampuriya, P. R., et al. Tracking the origin, development, and differentiation of hematopoietic stem cells. Current Opinion in Cell Biology. 49, 108-115 (2017).
- Wilkinson, A. C., Yamazaki, S. The hematopoietic stem cell diet. International Journal of Hematology. 107, 634-641 (2018).
- Kohli, L., Passegue, E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends in Cell Biology. 24, 479-487 (2014).
- Abdel-Wahab, O., Levine, R. L. Metabolism and the leukemic stem cell. The Journal of Experimental Medicine. 207, 677-680 (2010).
- Papa, L., Djedaini, M., Hoffman, R. Mitochondrial Role in Stemness and Differentiation of Hematopoietic Stem Cells. Stem Cells International. 2019, 4067162 (2019).
- Vannini, N., et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nature Communication. 7, 13125 (2016).
- Anso, E., et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nature Cell Biology. 19, 614-625 (2017).
- Wanet, A., Arnould, T., Najimi, M., Renard, P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells and Development. 24, 1957-1971 (2015).
- Maryanovich, M., et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nature Communication. 6, 7901 (2015).
- Suda, T., Takubo, K., Semenza, G. L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 9, 298-310 (2011).
- Zhang, C. C., Sadek, H. A. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxidants & Redox Signaling. 20, 1891-1901 (2014).
- Snoeck, H. W. Mitochondrial regulation of hematopoietic stem cells. Current Opinion in Cell Biology. 49, 91-98 (2017).
- Wu, M., et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American Journal of Physiology-Cell Physiology. 292, 125-136 (2007).
- Chen, J., et al. Enrichment of hematopoietic stem cells with SLAM and LSK markers for the detection of hematopoietic stem cell function in normal and Trp53 null mice. Experimental Hematology. 36, 1236-1243 (2008).
- Jung, Y., et al. Hematopoietic stem cells regulate mesenchymal stromal cell induction into osteoblasts thereby participating in the formation of the stem cell niche. Stem Cells. 26, 2042-2051 (2008).
- Masuda, S., Li, M., Izpisua Belmonte, J. C. Niche-less maintenance of HSCs by 2i. Cell Research. 23, 458-459 (2013).
- Anderson, H., et al. Hematopoietic stem cells develop in the absence of endothelial cadherin 5 expression. Blood. 126, 2811-2820 (2015).
- Lo Celso, C., Scadden, D. Isolation and transplantation of hematopoietic stem cells (HSCs). Journal of Visualized Experiment. (157), (2007).
- Van Wyngene, L., Vandewalle, J., Libert, C. Reprogramming of basic metabolic pathways in microbial sepsis: therapeutic targets at last. EMBO Molecular Medicine. 10, (2018).
- Olson, K. A., Schell, J. C., Rutter, J. Pyruvate and Metabolic Flexibility: Illuminating a Path Toward Selective Cancer Therapies. Trends in Biochemical Sciences. 41, 219-230 (2016).
- Halestrap, A. P., Wilson, M. C. The monocarboxylate transporter family–role and regulation. IUBMB Life. 64, 109-119 (2012).
- Kim, A. Mitochondria in Cancer Energy Metabolism: Culprits or Bystanders. Toxicological Research. 31, 323-330 (2015).
- Fosslien, E. Mitochondrial medicine–molecular pathology of defective oxidative phosphorylation. Annals of Clinical & Laboratory Science. 31 (1), 25-67 (2001).
- Wick, A. N., Nakada, H. I., Wolfe, J. B. Localization of the primary metabolic block produced by 2-deoxyglucose. The Journal of Biological Chemistry. 224 (2), 963-969 (1957).
- Linnett, P. E., Beechey, R. B. Inhibitors of the ATP synthetase systems. Methods in Enzymology. 55, 472-518 (1979).
- Rimmele, P., et al. Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO3. EMBO Reports. 16, 1164-1176 (2015).
- Riviere, I., Dunbar, C. E., Sadelain, M. Hematopoietic stem cell engineering at a crossroads. Blood. 119, 1107-1116 (2012).
- Ito, K., Suda, T. Metabolic requirements for the maintenance of self-renewing stem cells. Nature Reviews Molecular Cell Biology. 15 (4), 243-256 (2014).

