Hyperspectral Imaging as a Tool to Study Optical Anisotropy in Lanthanide-Based Molecular Single Crystals
Summary
Here, we present a protocol to obtain luminescent hyperspectral imaging data and to analyze optical anisotropy features of lanthanide-based single crystals using a Hyperspectral Imaging System.
Abstract
In this work, we describe a protocol for a novel application of hyperspectral imaging (HSI) in the analysis of luminescent lanthanide (Ln3+)-based molecular single crystals. As representative example, we chose a single crystal of the heterodinuclear Ln-based complex [TbEu(bpm)(tfaa)6] (bpm=2,2’-bipyrimidine, tfaa– =1,1,1-trifluoroacetylacetonate) exhibiting bright visible emission under UV excitation. HSI is an emerging technique that combines 2-dimensional spatial imaging of a luminescent structure with spectral information from each pixel of the obtained image. Specifically, HSI on single crystals of the [Tb-Eu] complex provided local spectral information unveiling variation of the luminescence intensity at different points along the studied crystals. These changes were attributed to the optical anisotropy present in the crystal, which results from the different molecular packing of Ln3+ ions in each one of the directions of the crystal structure. The HSI herein described is an example of the suitability of such technique for spectro-spatial investigations of molecular materials. Yet, importantly, this protocol can be easily extended for other types of luminescent materials (such as micron-sized molecular crystals, inorganic microparticles, nanoparticles in biological tissues, or labelled cells, among others), opening many possibilities for deeper investigation of structure-property relationships. Ultimately, such investigations will provide knowledge to be leveraged into the engineering of advanced materials for a wide range of applications from bioimaging to technological applications, such as waveguides or optoelectronic devices.
Introduction
Hyperspectral Imaging (HSI) is a technique that generates a spatial map where each x-y coordinate contains a spectral information that could be based on any kind of spectroscopy, namely photoluminescence, absorption and scattering spectroscopies1,2,3. As a result, a 3-dimensional set of data (also called “hyperspectral cube”) is obtained, where the x-y coordinates are the spatial axes and the z coordinate is the spectral information from the analyzed sample. Therefore, the hyperspectral cube contains both spatial and spectral information, providing a more detailed spectroscopic investigation of the sample than traditional spectroscopy. While HSI has been known for years in the field of remote sensing (e.g., geology, food industries4), it recently emerged as an innovative technique for the characterization of nanomaterials2,5 or probes for biomedical applications3,6,7,8. Generally speaking, it is not limited to the UV/visible/near-infrared (NIR) domain, but can also be extended using other radiation sources, such as X-rays – for instance in order to characterize elemental distribution in different materials9 – or Terahertz radiation, where HSI was used to perform thermal sensing in biological tissues8. Further, photoluminescence mapping has been combined with Raman mapping to probe the optical properties of monolayer MoS210. Yet, amongst the reported applications of optical HSI, there are still only a few examples on HSI of lanthanide-based materials11,12,13,14,15,16,17. For instance, we can cite: detection of cancer in tissues6, analysis of the light penetration depth in biological tissues7, multiplexed biological imaging3, analysis of multicomponent energy transfer in hybrid systems11, and investigation of aggregation-induced changes in spectroscopic properties of upconverting nanoparticles12. Clearly, the attractiveness of HSI arises from its suitability for generating knowledge about environment-specific luminescence, providing simultaneous spatial and spectral information about the probe.
Taking advantage of this powerful technique we herein describe a protocol to investigate the optical anisotropy of the heterodinuclear Tb3+-Eu3+ single crystal [TbEu(bpm)(tfaa)6] (Figure 1a)13. The optical anisotropy observed resulted from the different molecular packing of the Ln3+ ions in the different crystallographic directions (Figure 1b), resulting in some crystal faces showing brighter, others showing dimmer photoluminescence. It was suggested that the increased luminescence intensity at specific faces of the crystal was correlated with more efficient energy transfer along those crystallographic directions where the Ln3+···Ln3+ ion distances were the shortest13.
Motivated by these results, we propose the establishment of a detailed methodology to analyze optical anisotropy through HSI, opening the path for better understanding of ion-ion energy transfer processes and tunable luminescent properties stemming from specific molecular arrangement18,19. These structure-properties relationships have been recognized as important aspects for innovative optical materials design including, but not limited to waveguide systems and opto-magnetic storage devices at nano and microscale – addressing the demand for more efficient and miniaturized optic systems20.
Protocol
CAUTION: It is recommended to use safety goggles specific for the excitation wavelength being used at all times when operating the imager.
1. Configuration of the hyperspectral microscope
NOTE: An overview of the hyperspectral imaging system is given in Figure 2a, with the main components of the imager being described. The imaging system can be used for the detection of the visible or the near-infrared (NIR) emission from a sample. Depending on which detection is desired (visible or NIR), the light goes through two different light paths (Figure 2e). A combination of different beam turning cubes and dichroic filter cubes (optical cubes) must be positioned at specific positions in the instrument to select the respective path.
- Power on the computer which is connected to the imaging system. Turn on the computer’s monitor.
- Set the appropriate optical cube configuration (Figure 2b,c).
NOTE: Here, the imager configuration (optical cube configuration) for HSI mapping using UV excitation and visible emission detection is described. However, it is also possible to change it for NIR excitation and visible or NIR emission detection, depending on the sample analyzed. Refer to the section Representative Results for an example.- Starting from the microscope stage (1 in Figure 2a) and following the emission beam pathway towards the detectors (3 in Figure 2a), leave the first position for an optical cube (4 in Figure 2b) vacant and place the confocal microscope optical cube (DFM1-P01) in the position indicated as 5 in Figure 2b, so that the emission from the sample is directed through the visible light path.
- Looking along the optical path towards the detector, place the visible optical cube (CM1-P01), which contains the dichroic mirror and the filters to direct the visible emission to the detection paths, in the position indicated as 6 in Figure 2b.
- Continuing the path towards the detector, place the confocal pinhole optical cube (DFM1-P01) in the position indicated as 7 in the Figure 2b to direct the light through the visible light detection path. Then, following the path, place the confocal spectrometer optical cube (DFM1-P01) in position 8 in Figure 2c so that the emitted light reaches the detector.
- For the HSI mapping, manually control the detector slit opening (9 in Figure 2c) in order to match with the size of the pinholes that are used (around 50 mm is optimal).
- In the PHySpec software, choose the aperture of the pinhole (5 in Figure 3).
NOTE: The smaller the pinhole aperture, the better is the HSI resolution, at the cost of signal intensity.
- Turn on the broadband lamp (Figure 2d, inset) by positioning the switch (10 in Figure 2d) into the ON position. To control the intensity of the excitation light, turn the knob indicated by 11 (Figure 2d) to higher (32 – lowest intensity) or lower values (1 – highest intensity). Keep the broadband lamp shutter (12 in Figure 2d) closed during the set-up.
NOTE: The higher values correspond to higher attenuation of the power density emitted by the lamp, while lower values correspond to lower attenuation. - Turn on the following hardware in the order given below by setting their switches into the ON position:
- Turn on the ThorLabs motion controller.
- Turn on the Nikon power source.
- Turn on the ASI controller.
- Turn on the Galvo controller.
- Turn on the ProEm detector.
- Turn on the Bayspec detector.
- At the computer, open the PHySpec software by double clicking on its icon.
- Press the F8 key on the keyboard in order to initialize the IMA Upconversion system and click the OK button on the Connect to System window.
NOTE: Step 1.5.1. can also be performed by clicking on the System tab and then clicking on Connect to arrive to the Connect to System window. Then, the OK button can be clicked to connect the imaging system to the software. - Ensure that all menus appear on the interface (Color camera, ProEm and Bayspec) as well as the instrument control panel, on the left side of the screen, as shown in Figure 3.
- Press the F8 key on the keyboard in order to initialize the IMA Upconversion system and click the OK button on the Connect to System window.
2. Hyperspectral imaging of a [TbEu(bpm)(tfaa)6] single crystal
- In order to prepare the sample, place the crystal on a microscopy glass slide. In case it is needed to use higher magnification, cover the crystal with a thin cover glass and secure it with tape, so that the sample can be placed with the thin cover glass facing toward the objective lens.
- Place the glass slide on which the sample has been prepared on the microscope stage and secure it using the metal arms (Figure 4a,b).
- Move the sample using the joystick (Figure 4c) of the ASI controller in order to position the sample over the objectives in use.
- Manually position the right filter cube in the wheel underneath the objectives (3 in Figure 5) to select the UV excitation of the lamp and to let pass the visible emission towards the detector.
NOTE: Additional filter cubes are available to use either green or blue light excitation, thus, having the right filter cube in place is important for the appropriate excitation wavelength. - Manually position the 20X objective (indicated by 5 in Figure 5) under the sample and press the white button (6 in Figure 5) on the left side of the microscope to turn on the white light.
- Adjust the brightness by turning the knob underneath the white light power button (7 in Figure 5).
- In the PHySpec software, press the Play (video) button on the color camera window, which will cause the acquisition of a live scan.
- If the color camera window shows a black image, increase the Exposure Time (2 in Figure 3) and/or the Gain Value (3 in Figure 3) found in the instrument control panel, under the Color Camera tab. If the image viewed is too bright, decrease the exposure time and/or gain value.
- Ensure the forward knob on the right side of the microscope (2 in Figure 5) is set to R in order to send 20% of the signal to the camera/binoculars and 80% of the signal to the detector.
- Focus on the sample by adjusting the distance between the objective and the stage (Figure 4b). This is done by turning the knobs shown in Figure 4d on the right side of the microscope.
NOTE: The larger knob is used for coarse adjustments, while the smaller knob is for more delicate and small focus changes. - Ensure that the manually chosen objective is also selected at the software. First, click on the View button in the top menu bar and then click one Show/hide scale bar to display the scale bar on the image (1 in Figure 3). Then, go to the Galvanometer tab in the instruction control panel and select the Objective used (4 in Figure 3). Ensure that the displayed scale bar is correct by selecting the proper objective in the software.
- In the software, select the appropriate detector by going to the tab Diverter (ProEM – 6 in Figure 3) and gratings by going to the tab Filter (1200 gr/mm in case of ProEM – 7 in Figure 3) under the SpectraPro SP-2300 tab.
- Open the broadband lamp shutter (12 in Figure 2) to allow the UV excitation of the sample to take place. Turn the intensity knob (11 in Figure 2d) to the desired position (e.g., 8 – intermediate intensity) to control the intensity of the broadband lamp (UV) excitation.
- To choose between wide field illumination (open aperture) or a smaller spot illumination (more closed aperture), control the size of the UV lamp field aperture using the stick and knobs shown in 4 in Figure 5.
- Under the SpectraPro SP-2300 tab, select a wavelength to observe the sample emission.
- If the emission wavelengths of the sample are unknown, acquire an emission spectrum.
- In the sequencer, click on the + sign to add a new sequence (“node”) for the acquisition of an emission spectrum.
- Click on Spectrometer and then Spectrum Acquisition (with spectral scan).
- Input a Minimum Wavelength (i.e., 400 nm) and a Maximum Wavelength (i.e., 700 nm) and click OK to set the spectral range in which the spectrum will be recorded.
- Choose the adequate Exposure Time in the left side menu of the software. Choose shorter times (e.g., 0.1 s) for very bright samples and longer times (e.g., 2 s) for dim emitters.
- Adjust the excitation power in case of the broadband lamp (UV) excitation (see step 1.3 above).
NOTE: In case of NIR diode excitation, it can be adjusted from the Neutral Density drop-down menu at the left side of the PHySpec software.
- On the sequencer, click on the double play button to run the entire sequence. Once the spectrum is shown, note the regions of interest for the detection of the sample emission (e.g. 580 to 640 nm in case of Tb3+– and Eu3+-based samples).
- As needed, optimize signal detection by either changing the focus of the sample or by adjusting the Exposure Time in the PHySpec software. Achieve further optimization of signal detection through the increase of the sample emission intensity by changing the power of the excitation source (broadband lamp), as described above.
- In the sequencer, click on the + sign to add a new sequence (“node”) for the acquisition of an emission spectrum.
- Adjust the Exposure Time (e.g., 0.5 s – 2 in Figure 3) and Gain (3 in Figure 3) of the Color Camera accordingly, to obtain a good quality image. If needed, add the scale bar to the image by clicking at the button Show/hide scale bar at the second row of the menu in the top of the PHySpec software window.
- Recommendation: Prior to acquiring the hyperspectral cube, record a bright field optical microscopy image of the crystal under white light (Figure 6a) and/or UV full (Figure 6b) or confined (Figure 6c) illumination (UV illumination controlled by the shutter aperture, shown as 4 in Figure 5). To do so, with the sample in focus, click on the play button of the color camera.
- Click on File and then Export Window View, choose the desired format to export the obtained image, and save the file with the desired extension (.h5, .JPEG).
- Before acquiring the hyperspectral image, turn off the white light illumination as well as the room light.
- To obtain the hyperspectral cube, write a new sequence. Therefore, in the sequencer, click on the + sign to add a new node.
- Click on Confocal Imager.
- Click on Multi-Spectrum Acquisition. Here, the desired field of view is defined by the number of points to acquire in the x and y directions and the step size. For example, use 100 points in x and 100 points in y with a 5 µm step size to obtain an image of 500 by 500 µm.
Note: The total number of acquisition points and the integration time at each point will directly affect the total acquisition time of the hyperspectral cube.- Input the desired X Position (e.g., 100) and Y Position (e.g., 100) counts as well as the desired Step Size (e.g., 5 µm). Select the Hardware option for the camera sync, for visible emission mapping (and Software option in case of NIR detection). Click OK.
- Click on Multi-Spectrum Acquisition. Here, the desired field of view is defined by the number of points to acquire in the x and y directions and the step size. For example, use 100 points in x and 100 points in y with a 5 µm step size to obtain an image of 500 by 500 µm.
- Click on Confocal Imager.
- In the sequencer, click on the newly added Multi-Spectrum Acquisition line to highlight the node.
- Click the Play button to run the selected node.
NOTE: The remaining time that the acquisition will take will appear next to the node (in minutes, e.g., 28 min). - Once the acquisition is complete, save the hyperspectral cube in the appropriate file format (.h5).
3. Hyperspectral data analysis
- Right after the acquisition, if the saved hyperspectral cube does not open automatically in the software, recover the hyperspectral cube, which was saved as .h5 file, by clicking on File in the top menu bar and then hovering the cursor and click at Open File…. When the window titled Select data(s) to open pops up, choose the folder where the .h5 file is saved and double click on the file to open it.
- Once the hyperspectral cube file is imported, modify the displayed hyperspectral cube image to show the intensity of a specific spectral wavelength by moving the bar on the top of the cube image to the left (lower wavelength, e.g., 580 nm) or right (higher wavelength, e.g., 638 nm).
NOTE: The selected wavelength is displayed in the left side of this upper bar (1 in Figure 7). - After choosing the wavelength of interest for the analysis (e.g., the maximum intensity, which in the case of [TbEu(bpm)(tfaa)6] is 613.26 nm), make one (or all) of the three possible types of spectral analysis: (A) the spectral distribution in form of an image (2 in Figure 7); (B) an emission intensity profile across a region of interest (3 in Figure 7); (C) extraction of a spectrum at a specific point or region of interest (4 in Figure 7).
- In case of the spectral distribution from an image, use the Crop and Bin function to increase the signal-to-noise ratio in the image. In order to do that, click in the top menu Processing then choose Data and then the option Crop and Bin.
- For an emission intensity profile, on the cube image, right click and select the Create Target or Create X profile or Create Y profile depending if only one point (Target – 5 and 6 in Figure 7) or a line (Profile – 7 and 8 in Figure 7) needs to be analyzed. Select the area of analysis by dragging the target, the horizontal or the vertical line profile with the cursor and move it across the cube.
- Once the profile has been properly selected, right click on the region and select the Add Target to Graph. Chose the option to create a new graph to display the emission intensity (y axis) as a function of the physical position of the target (x axis). The spectrum will appear on the new graph which was inserted (6 and 7 in Figure 7).
NOTE: Multiple targets can be created, and these will show up as different colored emission profiles (5 and 6 in Figure 7).
- Once the profile has been properly selected, right click on the region and select the Add Target to Graph. Chose the option to create a new graph to display the emission intensity (y axis) as a function of the physical position of the target (x axis). The spectrum will appear on the new graph which was inserted (6 and 7 in Figure 7).
- Alternatively, obtain an emission spectrum of a specific area of the sample (9 in Figure 7). To begin with, hover the cursor over the cube image and right click. Click on the Rectangle Selection or Ellipse Selection options on the tab that pops-up.
- Draw the selection shape (e.g., a rectangle) over the desired region by clicking and dragging the cursor across the cube. Once the area has been properly selected, right click on the region and select the Add Selection to Graph.
- At the appearing window Add to Graph, select Create a New Graph to display the emission spectra of the target and click OK.
NOTE: A new colored line (8 in Figure 7) will appear on the graph in which the target emission is being shown, with the emission intensity as the y axis and the wavelength at the x axis. This spectrum corresponds to the averaged intensity of the selected area for each wavelength.
- Once the spectrum is obtained, save it before selecting a new region because only one region can be selected at a time. To do so, select the window in which contains the graph. In the File menu, select Save as and choose to save the graph in the folder of choice, using the name of choice, either in .h5 format, which can be opened in the PHySpec software, or in .csv format, which can be imported in Excel.
Representative Results
To illustrate the configuration of the hyperspectral microscope for the data acquisition on a Ln-based, molecular single crystal (i.e., [TbEu(bpm)(tfaa)6], Figure 1a), Figure 2 shows an overview of the system as well as the right placement of the optical cubes in the setup. Figure 3 shows a screen shot of the PHySpec software containing the menus used during the HSI acquisition. Figure 4 and Figure 5 show the microscope stage in greater detail, including the placement of the glass slide containing the sample to be analyzed. The selected UV illumination was turned on to show the visible red luminescence of the crystal (Figure 4a and 1 in Figure 5). Figure 6a shows a bright field image of the crystal recorded after adjusting the sample in the proper focus. The needle-like morphology of the crystal can be clearly seen. Figure 6b,c show the image of the same crystal under UV excitation with either full view (Figure 6b) or locally confined (Figure 6c) illumination. Under the wide UV illumination, the differences of emission brightness from the different faces of the crystal are immediately visible. The confined illumination can be used as an option, mainly to investigate any effects of energy or light transfer in the crystal, which may trigger waveguide-like behavior. In this case, a strong emission is detected in a point not directly under excitation. This suggests that efficient energy migration takes place through the crystal13 (5 and 6 in Figure 7).
From the acquired hyperspectral cube, it is further possible to obtain the spectral distribution in form of an image representing a specific wavelength, the intensity profile of a specific emission wavelength, as well as the emission spectra at any pixel or area of the acquired hyperspectral cube. As an example, the emission spectra given in Figure 7 (panel 4) show the most characteristic emission bands of the Eu3+ ion: the band observed at 590 nm is assigned to the magnetic dipole (MD) 5D0→7F1 transition of Eu3+, while the emission peaks in the region from 610 to 630 nm stem from the hypersensitive forced electric dipole (ED) 5D0→7F2 Eu3+ transition. The ratio between the integrated intensity of these two transitions is well known to be an excellent probe of the chemical environment around the Ln3+ ion in the structure of the single crystal21: the lower the symmetry around the Ln3+ ion, the larger is the ED/MD ratio. This allows to draw conclusions about the symmetry character of the chemical environment of the Ln3+ ion. Moreover, the Stark splitting of the 5D0→7F2 transition can also be correlated with the symmetry around the Ln3+ in its crystallographic environment – the lower the symmetry, the higher is the number of Stark sub-levels. In case of the needle-like polymorph crystallized in the low symmetric triclinic crystal system, the 5D0→7F2 transition splits into four sub-peaks (spectra shown in Figure 7, panel 4). Such analysis is particularly appealing when comparing the optical properties of several polymorphs of a luminescent crystal. We previously demonstrated that the information about the chemical environment deduced from the optical analysis correlated well with the molecular crystal structure obtained by single crystal X-ray analysis13. Moreover, the spectral profile along the different crystal faces shown in Figure 7 (panel 3), indicating brighter emission at the tip and side faces, can also be correlated with the Ln3+···Ln3+ ion distances in the three spatial directions (Figure 1b): the denser Ln3+ packing along the axes perpendicular to the tip and side faces, respectively, favor ion-ion energy transfer. Hence, emission enhancement is observed at the respective faces, thus, optical anisotropy.
Overall, the various options of data analysis, shown in Figure 7 and Figure 8, constitute the most important features of the combined spectroscopic and spatial information, which is possible to be explored by HSI analysis of luminescent samples.
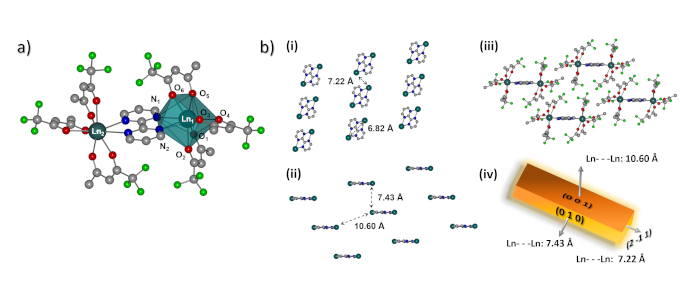
Figure 1: Molecular structure and crystallographic arrangement. (a) Structure of the heterodinuclear Ln-based complex [TbEu(bpm)(tfaa)6], where Ln1 and Ln2 are Tb3+ and Eu3+ ions. Disordered groups and hydrogen atoms are omitted for clarity. Color code: Eu: dark cyan; C: grey; O: red; N: blue; F: lime green. (b) Representation of the molecular packing in the crystal: (i) top view and (ii) tip view of the needle-like single crystal structure with selected intermolecular and intramolecular Ln···Ln distances (tfaa subunits and hydrogen atoms are omitted for clarity). (iii) Crystal packing arrangement of the [TbEu(bmp)(tfaa)6] dimers (hydrogen atoms are omitted for clarity). (iv) Diagram of the crystal growth faces of the dimer revealing the shortest Ln···Ln distances in the (0 1 0) and (2 -1 1) crystallographic directions. The figure has been modified from reference 13. Please click here to view a larger version of this figure.
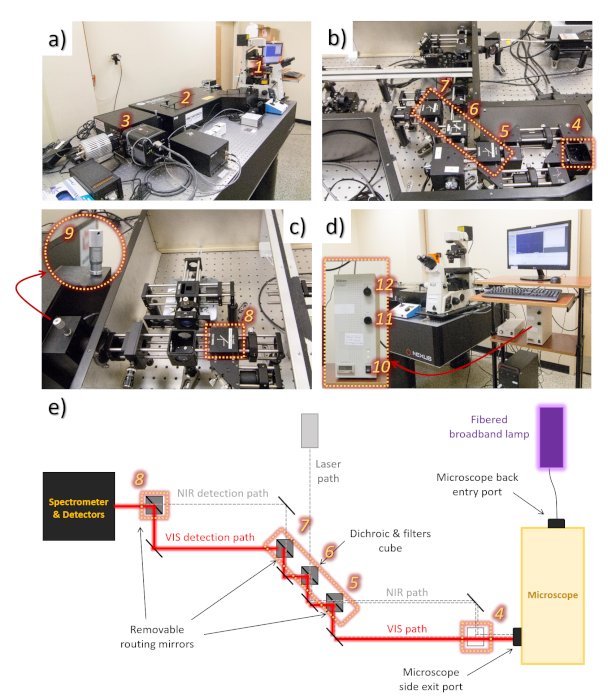
Figure 2: Overview of the Hyperspectral Imaging System. Shown is the configuration required for luminescence mapping at the visible spectral region using UV excitation. (a) General view of the system, where 1 is the microscope stage, 2 is the section containing the optical configuration, and 3 is the spectrometer with the visible and NIR detectors. (b) Open view of the optical set-up close to the microscope stage (right side of a) showing the optical configuration for the experiment: optical cube position 4 remains empty and the confocal microscope cube is placed in position 5 in order to route the light through the visible path, the visible cube is placed in position 6 in order to direct the visible light to the detection path, and the confocal pinhole cube is placed in position 7 in order to route light to the visible detection path. (c) Open view of the optical set-up closer to the detectors (left side of a), showing position 8, where the confocal spectrometer cube is placed to reflect light to the spectrometer and visible camera. The inset 9 shows the screw to adjust the opening width of the spectrometer slit. (d) View of the microscope stage, computer and broadband lamp (used for UV excitation) controller. In the inset, the broadband lamp controller is shown with more detail: 10 is the on/off button, 11 is the knob to control the intensity of the lamp, and 12 is the shutter button. (e) Scheme showing the visible/NIR optical path from the microscope stage to the detectors, including the optical cube positions from 4 auf 8. Please click here to view a larger version of this figure.
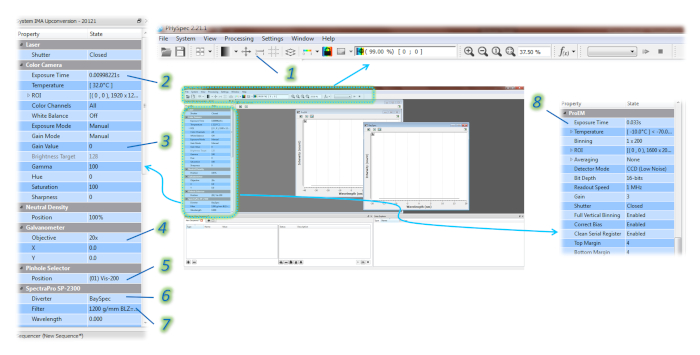
Figure 3: Screenshot of the PHySpec software showing the menu with the parameters to be adjusted for the HSI. 1 allows to insert the scale bar in the color camera image; 2 and 3 allow to control exposure time and gain value of the color camera, respectively; the proper objective lens must be selected in 4; 5 allows the selection of the aperture of the pinhole ; 6 (Diverter) and 7 (Filter) allow to choose the detector and grating, respectively; the exposure time for the visible detector is set in 8. Please click here to view a larger version of this figure.
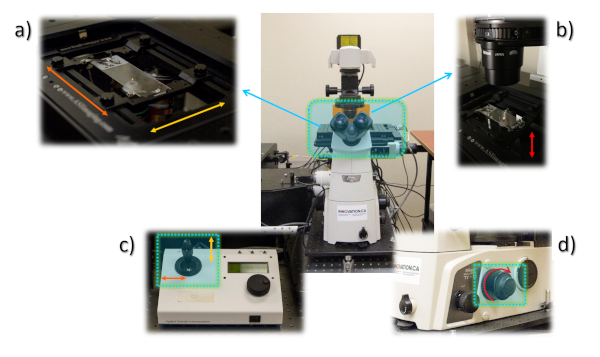
Figure 4: General view of the microscope stage. (a) Placement of the glass slide containing the sample at the stage, with the UV illumination ON showing the sample’s red luminescence (small red dot in the center of the glass slide). (b) View of the microscope stage with the white light illumination condenser on top. (c) Stage controller showing the joystick that controls the movement of the stage in the directions indicated by orange and yellow arrows (also shown in (a). (d) Detailed view of the focus button, which moves the stage in the directions indicated by the red arrow (also shown in (b)). Please click here to view a larger version of this figure.

Figure 5: The components of the microscope stage. 1 microscope stage with the sample on the glass slide placed on the sample stage on top of the objective lenses; 2 wheels to adjust the focus (large wheel) and to direct the captured emission (small wheel) either only to the detector (L), partially to the detector and partially to the camera (R), or solely to the binocular lenses (eye); 3 excitation/emission filters wheel used to choose the excitation wavelength range. The detail on the right shows the filter cube holding the UV filter and long-pass filter used in this experiment; 4 in the top/bottom are show the knobs to move the excitation beam through the sample, while in between, the circular field aperture control; 5 objective lenses; 6 ON/OFF button of the white light illumination; 7 knob to adjust the brightness of the white light lamp. Please click here to view a larger version of this figure.
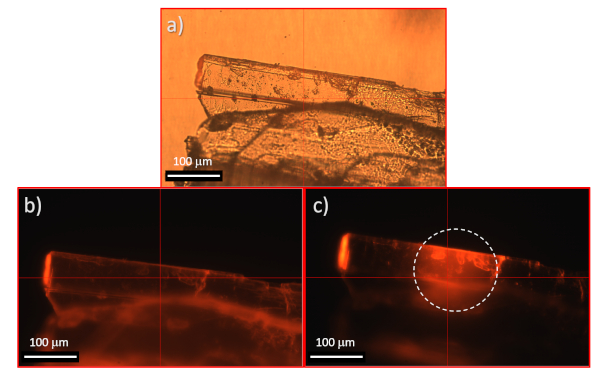
Figure 6: Optical microscopy images of the analyzed single crystal. These images were obtained under (a) white light illumination, (b) full-view UV illumination, using the excitation circular aperture completely open, and (c) locally confined UV illumination (marked by the white circle), using a closer excitation circular aperture. Please click here to view a larger version of this figure.
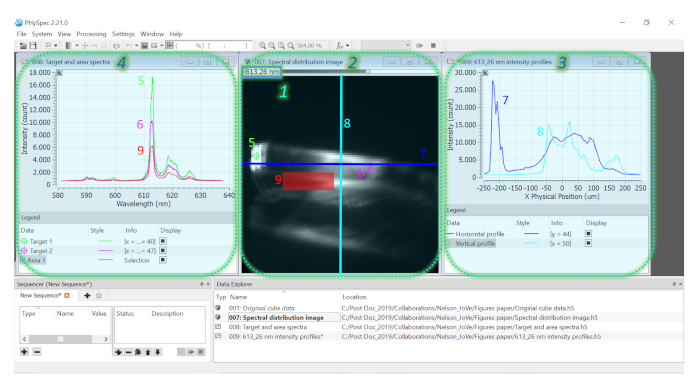
Figure 7: Screenshot of the PHySpec software showing the hyperspectral cube data analysis process. Diverse spectral analysis methods can be applied on the acquired hyperspectral cube: 1 shows the wavelength which was chosen for the spectral image distribution shown in 2; 3 shows the 613.26 nm horizontal (7) and vertical (8) intensity profiles; 4 shows the emission spectra extracted from the targets 5 and 6 as well as from the area highlighted in 9. Please click here to view a larger version of this figure.
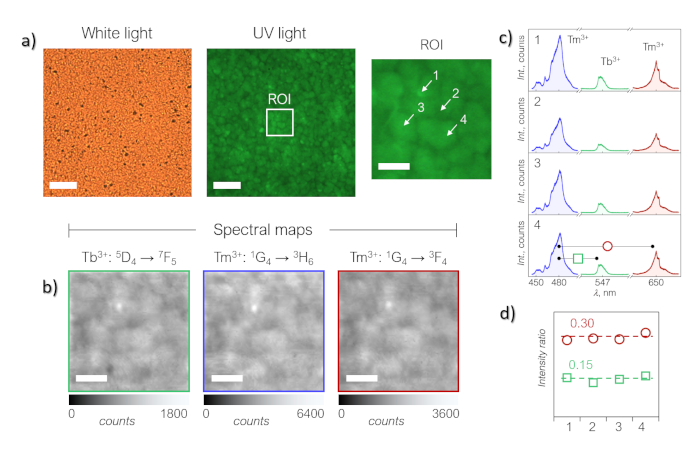
Figure 8: Alternative application of HSI probing the synergy between upconverting nanoparticles and lanthanide complexes. This example shows the hyperspectral analysis of a hybrid system comprised of molecular crystals ([Tb2(bpm)(tfaa)6]) combined with upconverting nanoparticles (NaGdF4:Tm3+,Yb3+). (a) Photomicrographs under white and UV light illumination along with the region of interest (ROI) used for hyperspectral imaging under 980 nm light irradiation. (b) Tm3+ and indirect Tb3+ emissions monitored over an area of 20 x 20 µm2. (c) Variation of the absolute intensity of the emission bands fluctuated throughout the hybrid system indicating some variability in the total amount of material distributed over the surface. (d) The constancy of the ratio between the integrated emission of the complex vs. Tm3+: 1G4→3H6 (squares) and Tm3+: 1G4→3F4 (circles) confirmed the simultaneous presence of the two moieties throughout the hybrid system and the homogeneous interaction between them. Scale bars are 20 µm in the photomicrographs and 5 µm in ROIs and spectral maps. Photomicrographs are presented in real colors. The Figure has been modified from reference 11. Please click here to view a larger version of this figure.
Discussion
The hyperspectral imaging protocol here described provides a straightforward approach that allows to obtain spectroscopic information at precise locations of the sample. Using the described setup, the spatial resolution (x and y mapping) can reach down to 0.5 µm while the spectral resolution can be of 0.2 nm for the mapping at the visible range and 0.6 nm for the NIR range.
In order to conduct hyperspectral mapping on a single crystal, sample preparation follows an easy procedure: the crystal can simply be placed on a glass microscopy slide, covered by a cover glass as needed. Focusing the sample using the proper objective lenses and the bright field image at the Color Camera set-up in the software is a very important step during the pre-analysis stage to obtain best resolved hyperspectral images. Typically, higher emission intensities are obtained when the sample is well focused. Once this is done, the choice of the parameters of the analysis such as the x and y counts and the step size will dictate the field of view and spatial sampling, respectively of the obtained hyperspectral cube. However, the objective´s numerical aperture and the excitation/emission wavelengths dictate the real volume of the sample that is probed at each acquisition point. For example, for the objective used in this study, with numerical aperture (NA) of 0.4, and using the excitation in the UV spectral range (390 nm), the focused laser spot has a size of approximately 0.6 x 0.6 µm in the x and y direction. The size of the laser spot was calculated using the website (https://www.microscopyu.com/tutorials/imageformation-airyna, accessed on Sep. 26, 2019). If the chosen step size is larger than the spatial sampling, one may actually be sampling an area smaller than the one given by the step size. If the sample is homogeneous, undersampling may not be an issue. Yet, if spatial variations in the sample are important to detect, optimal sampling is obtained with a step size set at half the size of the laser beam on the sample. The chosen exposure time and the intensity of the UV selected illumination will control the intensity of the obtained spectra. Such parameters vary from sample to sample, depending on the emission intensity and the sensitivity towards the UV excitation.
At this point, the critical steps within the protocol can be listed as: the alignment of the optical system, correct placement of the optical cubes, regulation of the circular aperture of the excitation and detector slit openings, choice of the pinhole, focus of the sample in the color camera using the proper objective lenses, the intensity of the broadband lamp and proper choice of the long pass filter cube (allowing UV excitation), as well as choice of the proper step size and exposure time as mentioned above. Lastly, the room lights must be off during all time of the hyperspectral cube acquisition.
In case of poor signal detection either with the color camera or the spectrometer, the troubleshooting of the technique should include checking carefully each one of the critical steps given above, before starting the hyperspectral cube acquisition. The configuration of the output signal at the microscope stage is also important. The three possible configurations are: eye (100 % of the output signal is sent to the microscope binocular mount), L (100 % of the output signal is sent to the detectors) and R (80 % of the output signal is sent to the detectors and 20 % to the microscope binocular mount). During the hyperspectral cube acquisition, the R or L configuration must be used. If all the above listed parameters are properly chosen, high resolution spatial and spectral information of the sample can be obtained.
Some possible modifications of the technique herein described can be exemplified by the hyperspectral imaging of other systems, such as luminescent microparticles14 or optical hybrid systems comprised of molecular crystals combined with upconverting nanoparticles (Figure 8)11. In these examples, the NIR laser diode (980 nm) was used as the excitation source, replacing UV excitation, while detecting the generated visible emission. In the latter example of the hybrid system, HSI revealed the homogeneity of hybrid films that combine upconverting nanoparticles (NaGdF4:Tm3+,Yb3+) and [Tb2(bpm)(tfaa)6] crystals into a multiwavelength-responsive isotropic system, exhibiting energy transfer between materials and molecules (Figure 8)11. Furthermore, by using the InGaAs detector of the system, detection of emissions in the NIR spectral region (1000 to 1700 nm) becomes possible. This is of particular interest when seeking the investigation of NIR-based optical probes for biomedical applications3. In this case, the optical cubes configuration of the system (Figure 2e) has to be set for the NIR path. In case of NIR excitation-NIR emission, one of the limitations of the hyperspectral technique here described becomes evident: as being wavelength-dependent, the spectral resolution in the NIR region is lower than that for the visible detection, i.e. approximately 0.6 nm (vs. 0.2 nm). Moreover, for sub-1 µm features, in e.g. smaller molecular crystals, nanoparticles or hybrid systems, the spatial resolution, which is dictated by the system configuration (objectives used and excitation/emission wavelengths), becomes another potential limitation.
Finally, (irrespective of the chosen HSI configuration and wavelength regime) the data manipulation can be done either with the instrument’s software or, as shown for the case of the spectral profiles, can be exported to be analyzed in other software packages such as Origin® or Microsoft Excel. In our example, the optical anisotropy of the crystal was also promptly revealed at the color camera image, namely by the strong intensity variation along the different crystal faces. Moreover, under the wide UV excitation, different emission intensities are obtained depending on which face is analyzed (Figure 7). The possibility to obtain the emission intensity profile in different points of interest at the crystal (targets in Figure 7) further allows to study variation in emission intensity and, if present, also in spectral shape. The block-like polymorph of the [TbEu(bpm)(tfaa)6] constitutes an example where two crystal faces exhibited equally high emission intensities and a lower emission intensity for the third one13. Expending on this, in case of a system without any anisotropy, the emission intensity would be the same for all crystal faces.
Complementary methods to probe optical anisotropy are for instance related to the presence of polarized emission from a sample. These include polarization memory or spectroscopic ellipsometry. The first consists in the correlation between the polarization state of the light emitted by the material with the polarization state of the incident excitation light,22,23 while the latter measures the change in the polarization state of the light after being reflected obliquely by a thin sample film24. However, an advantage of using hyperspectral imaging as a tool for probing optical anisotropy, as shown here, comes with the fact that the presence of polarization is not a requirement for sample analysis. Moreover, the sample preparation does not require the fabrication of thin films, neither a very careful orientation of the crystal with respect to the incident and collected light. These aspects make the technique of HSI potentially more widely applicable. Moreover, the anisotropic features are promptly visualized upon image acquisition, and data analysis is straightforward (as exemplified above).
Considering a broader scope, the significance of the HSI technique can be attributed to its unique characteristic to corelate the optical signal with environment-dependent features. For instance, such connection is essential to improve the understanding of nano-bio interactions3,15,16 in the growing field of nanomedicine or even to understand structure-properties relationship in materials science10,11,12,13. As such, future potential applications of the herein described technique can be named as, but not being limited to: analysis of biological samples in vitro, ex vivo and in vivo performing the mapping of molecules of biological interest4,6, adaptation of the microscope stage to study environment-specific opto-electronic properties (e.g. samples embedded in electrical circuits), optical temperature sensing8,15 (by adding a temperature controller to the microscope stage) or gas sensing (by adapting a gas chamber to the microscope stage). HSI has further been demonstrated as suitable for fluorescence excitation techniques instead of fluorescence emission25. A good example of the use of this particular hyperspectral technique adaptation is the detection of cancer cells in biological tissues6. Consequently, the protocol here described has the potential to be largely extended to the study of spectroscopic features of many different types of luminescent structures.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors thank Mr. Dylan Errulat and Prof. Muralee Murugesu from the Department of Chemistry and Biomolecular Sciences of the University of Ottawa for the provision of [TbEu(bpm)(tfaa)6] single crystals. E.M.R, N.R., and E.H. gratefully acknowledge the financial support provided by the University of Ottawa, the Canadian Foundation for Innovation (CFI), and the Natural Sciences and Engineering Research Council Canada (NSERC).
Materials
| Microscope glass slides | FisherBrand | 12-550-15 | Glass slides used for sample preparation |
| Visible and Near Infrared Hyperspectral Confocal Imager | PhotonETC | Microscope used for the analysis, builted according to the user needs, therefore it is no catalog number |
Referenzen
- ElMasry, G., Sun, D. W. Principles of Hyperspectral Imaging Technology. Hyperspectral Imaging for Food Quality Analysis and Control. , 3-43 (2010).
- Dong, X., Jakobi, M., Wang, S., Köhler, M. H., Zhang, X., Koch, A. W. A review of hyperspectral imaging for nanoscale materials research. Applied Spectroscopy Reviews. 54 (4), 285-305 (2019).
- Yakovliev, A., et al. Hyperspectral Multiplexed Biological Imaging of Nanoprobes Emitting in the Short-Wave Infrared Region. Nanoscale Research Letters. 14 (243), 1-11 (2019).
- Cheng, W., Sun, D. W., Pu, H., Wei, Q. Heterospectral two-dimensional correlation analysis with near-infrared hyperspectral imaging for monitoring oxidative damage of pork myofibrils during frozen storage. Food Chemistry. 248, 119-127 (2018).
- Liu, Y., Liu, L., He, Y., Zhu, L., Ma, H. Decoding of quantum dots encoded microbeads using a hyperspectral fluorescence imaging method. Analytical Chemistry. 87 (10), 5286-5293 (2015).
- Leavesley, S. J., et al. Colorectal cancer detection by hyperspectral imaging using fluorescence excitation scanning. Optical Biopsy XVI: Toward Real-Time Spectroscopic Imaging and Diagnosis. 10489, (2018).
- Zhang, H., Salo, D., Kim, D. M., Komarov, S., Tai, Y. -. C., Berezin, M. Y. Penetration depth of photons in biological tissues from hyperspectral imaging in shortwave infrared in transmission and reflection geometries. Journal of Biomedical Optics. 21 (12), 126006 (2016).
- Naccache, R., et al. Terahertz Thermometry: Combining Hyperspectral Imaging and Temperature Mapping at Terahertz Frequencies. Laser and Photonics Reviews. 11 (5), 1-9 (2017).
- Jacques, S. D. M., Egan, C. K., Wilson, M. D., Veale, M. C., Seller, P., Cernik, R. J. A laboratory system for element specific hyperspectral X-ray imaging. Analyst. 138 (3), 755-759 (2013).
- Birmingham, B., et al. Probing the Effect of Chemical Dopant Phase on Photoluminescence of Monolayer MoS2 Using in Situ Raman Microspectroscopy. Journal of Physical Chemistry C. 123 (25), 15738-15743 (2019).
- Marin, R., et al. Harnessing the Synergy between Upconverting Nanoparticles and Lanthanide Complexes in a Multiwavelength-Responsive Hybrid System. ACS Photonics. 6 (2), 436-445 (2019).
- Gonell, F., et al. Aggregation-induced heterogeneities in the emission of upconverting nanoparticles at the submicron scale unfolded by hyperspectral microscopy. Nanoscale Advances. 1, 2537-2545 (2019).
- Errulat, D., Gabidullin, B., Murugesu, M., Hemmer, E. Probing Optical Anisotropy and Polymorph-Dependent Photoluminescence in [Ln2] Complexes by Hyperspectral Imaging on Single Crystals. Chemistry – A European Journal. 24 (40), 10146-10155 (2018).
- Panov, N., Marin, R., Hemmer, E. Microwave-Assisted Solvothermal Synthesis of Upconverting and Downshifting Rare-Earth-Doped LiYF4 Microparticles. Inorganic Chemistry. 57 (23), 14920-14929 (2018).
- Debasu, M. L., Brites, C. D. S., Balabhadra, S., Oliveira, H., Rocha, J., Carlos, L. D. Nanoplatforms for Plasmon-Induced Heating and Thermometry. ChemNanoMat. 2 (6), 520-527 (2016).
- Nadort, A., et al. Quantitative Imaging of Single Upconversion Nanoparticles in Biological Tissue. PLoS ONE. 8 (5), 1-13 (2013).
- Sava Gallis, D. F., et al. Tunable Metal-Organic Framework Materials Platform for Bioimaging Applications. ACS Applied Materials and Interfaces. 9 (27), 22268-22277 (2017).
- Varghese, S., Das, S. Role of molecular packing in determining solid-state optical properties of π-conjugated materials. Journal of Physical Chemistry Letters. 2 (8), 863-873 (2011).
- Yan, D., Evans, D. G. Molecular crystalline materials with tunable luminescent properties: From polymorphs to multi-component solids. Materials Horizons. 1 (1), 46-57 (2014).
- Mu, S., Oniwa, K., Jin, T., Asao, N., Yamashita, M., Takaishi, S. A highly emissive distyrylthieno[3,2-b]thiophene based red luminescent organic single crystal: Aggregation induced emission, optical waveguide edge emission, and balanced ambipolar carrier transport. Organic Electronics: Physics, Materials, Applications. 34, 23-27 (2016).
- Binnemans, K. Interpretation of europium(III) spectra. Coordination Chemistry Reviews. 295, 1-45 (2015).
- Koyama, H., Fauchet, P. M. Anisotropic polarization memory in thermally oxidized porous silicon. Applied Physics Letters. 77 (15), 2316-2318 (2000).
- Kushida, T., Takushi, E., Oka, Y. Memories of photon energy, polarization and phase in luminescence of rare earth ions under resonant light excitation. Journal of Luminescence. 12-13, 723-727 (1976).
- Onuma, T., et al. Spectroscopic ellipsometry studies on β-Ga2O3 films and single crystal. Japanese Journal of Applied Physics. 55 (12), (2016).
- Favreau, P. F., et al. Excitation-scanning hyperspectral imaging microscope. Journal of Biomedical Optics. 19 (4), 046010 (2014).

