Phage Therapy Application to Counteract Pseudomonas aeruginosa Infection in Cystic Fibrosis Zebrafish Embryos
Summary
Presented here is a protocol for Pseudomonas aeruginosa infection and phage therapy application in cystic fibrosis (CF) zebrafish embryos.
Abstract
Antimicrobial resistance, a major consequence of diagnostic uncertainty and antimicrobial overprescription, is an increasingly recognized cause of severe infections, complications, and mortality worldwide with a huge impact on our society and on the health system. In particular, patients with compromised immune systems or pre-existing and chronic pathologies, such as cystic fibrosis (CF), are subjected to frequent antibiotic treatments to control the infections with the appearance and diffusion of multidrug resistant isolates. Therefore, there is an urgent need to address alternative therapies to counteract bacterial infections. Use of bacteriophages, the natural enemies of bacteria, can be a possible solution. The protocol detailed in this work describes the application of phage therapy against Pseudomonas aeruginosa infection in CF zebrafish embryos. Zebrafish embryos were infected with P. aeruginosa to demonstrate that phage therapy is effective against P. aeruginosa infections as it reduces lethality, bacterial burden and pro-inflammatory immune response in CF embryos.
Introduction
Phage therapy, the use of the natural enemies of bacteria to fight bacterial infections, is garnering renewed interest as bacterial resistance to antibiotics becomes widespread1,2. This therapy, used for decades in Eastern Europe, could be considered a complementary treatment to antibiotics in curing lung infections in patients with CF and a possible therapeutic alternative for patients infected with bacteria that are resistant to all the currently in use antibiotics2,3. Advantages of antibiotic therapy are that bacteriophages multiply at the infection site, whereas antibiotics are metabolized and eliminated from the body4,5. Indeed, the administration of cocktails of virulent phages isolated in different laboratories has proven to be effective in treating Pseudomonas aeruginosa infections in animal models as different as insects and mammals6,7,8. Phage therapy was also shown to be able to reduce the bacterial burden in burn wounds infected with P. aeruginosa and Escherichia coli in a randomized clinical trial9.
Zebrafish (Danio rerio) has recently emerged as a valuable model to study infections with several pathogens, including P. aeruginosa10,11, Mycobacterium abscessus and Burkolderia cepacia12,13. By microinjecting bacteria directly into the embryo blood circulation14 it is easy to establish a systemic infection that is counteracted by the zebrafish innate immune system, which is evolutionary conserved with neutrophils and macrophage generation similar to the human counterpart. Moreover, during the first month of life, zebrafish embryos lack the adaptive immune response, making them ideal models for studying the innate immunity, which is the critical defense mechanism in human lung infections15. Zebrafish recently emerged as a powerful genetic model system to better understand the CF onset and to develop new pharmacological treatments10,16,17. The CF zebrafish model of cftr knock-down generated with morpholino injection in zebrafish presented a dampened respiratory burst response and reduced neutrophil migration10, while the cftr knock-out leads to impaired internal organ position and the destruction of the exocrine pancreas, a phenotype that mirrors human disease16,17. Of greatest interest was the finding that the P. aeruginosa bacterial burden was significantly higher in cftr-loss-of-function embryos than in controls at 8 hours post-infection (hpi), which parallels the results obtained with mice and human bronchial epithelial cells2,18.
In this work, we demonstrate that phage therapy is effective against P. aeruginosa infections in zebrafish embryos.
Protocol
Adult zebrafish (Danio rerio) from the AB strain (European Zebrafish Resource Center EZRC) are maintained according to international (EU Directive 2010/63/EU) and national guidelines (Italian decree 4th March 2014, n. 26) on the protection of animals used for scientific purposes. Standard conditions are set in the fish facility with a 14 h of light/10 h dark cycle and tank water temperature at 28° C.
1. Preparation of solutions and tools
- Prepare a 50x stock solution and 1x working solutions for E3 embryo medium for growing zebrafish embryos (see Table 1).
- Make pigmentation blocking solution, to block the embryo pigmentation from 24 hours post fertilization (24 hpf) (see Table 1).
- Prepare 25x stock and 1x working Tricane anesthetic solution as described in Table 1.
NOTE: To avoid the repeated freezing and thawing of the solution that could damage the stock solution, make aliquots of 2 mL of 25x tricaine and store them at -20 °C. - Prepare tracks for embryo microinjection by dissolving 1 g of agarose in 100 mL of distilled H2O in a microwavable flask or bottle. Microwave for 1-3 min until the agarose is completely dissolved.
- Position zebrafish microinjection molds (see Table of Materials) turned upside down in a Petri dish (90 mm x 15 mm) and cover the entire surface by pouring the liquid agarose gel with a width of approximately 10 mm.
- Remove the mold once the agarose has solidified. Fill the Petri dish with 1x E3 embryo medium. This can be stored at 4 °C for approximately one week. Before use, replace with the fresh E3 medium containing Tricaine.
NOTE: Older molds might develop fungi or bacterial contaminations. - Use fire polished 10 cm borosilicate capillaries to prepare needles for microinjection and secure them to the micropipette puller by tightening the handles. Set the puller with the following settings: heat 500, velocity 100, time 150, pull 150.
2. P. aeruginosa (PAO1) inoculum preparation
- Inoculate 1 colony of GFP+ P. aeruginosa strain PAO119 in 5 mL of LB broth (see Table 1) and grow overnight at 37 °C with shaking (200 rpm).
- Dilute the above overnight culture to OD600 = 0.1 in 10 mL of LB broth and grow to OD600 = 0.5 at 37 °C with shaking (200 rpm).
- Centrifuge 2 mL of the culture for 2 min at 4 °C at 16,100 x g and resuspend the cell pellet to OD600 = 1, equivalent to about 1 x 109 cfu/mL, in 1 mL of the physiological solution (see Table 1).
- Dilute the cell suspension to about 5 x 107cfu/mL in physiological solution and store at 4 °C.
3. Phage stock preparation
- Inoculate 1 colony of P. aeruginosa strain PAO1 in 5 mL of LB broth and incubate overnight at 37 °C with shaking. Dilute the overnight culture to OD600 = 0.01 in 500 mL of LB broth and grow at 37 °C with shaking to OD600 = 0.05, equivalent to about 2.5 x 107 cfu/mL.
- To the diluted culture of P. aeruginosa, add about 1.25 x 107 phages, to get the multiplicity of infection (M.O.I.) of 10-3. Incubate at 37 °C with shaking until the OD600 drops to about 0.1-0.3 (in about 3 to 5 h, depending on the phage used).
NOTE: Four phages were selected for this experiment that infect PAO1 strain: Two Podoviridae, GenBank accession numbers vB_PaeP_PYO2, MF490236, and vB_PaeP_DEV, MF490238, and two Myoviridae, vB_PaeM_E215, MF490241, and vB_PaeM_E217, MF490240. Perform the steps below for each phage individually. - Incubate the lysate with 1 mg/mL of DNase and RNase for 30 min at 37 °C. Centrifuge at 5,000 x g for 30 min at 4 °C and carefully recover the supernatant (SN). Filter the SN with a filter having 0.8 μm pore diameter.
- To the SN, add 58 g/L of NaCl and 105 g/L of PEG6000. Dissolve with stirring at RT 20 min and then keep it overnight at 4 °C. Centrifuge at 20, 000 x g for 30 min at 4 °C for phage precipitation. Remove the SN and carefully dissolve the phage pellet in 15 mL of TN buffer (see Table 1).
- Purify the phages with CsCl density gradient as described in steps below.
- In a polyallomer ultracentrifuge tubes for SW41 rotor, prepare a CsCl step density gradient by stratifying 2 mL of the four CsCl solutions described in Table 1. Add 3.5 mL of phage suspension in each of the 4 tubes.
NOTE: CsCl is toxic, adopt proper safety procedures to handle and discard it. - Introduce the tubes into the rotor and ensure that the tubes are balanced (the weight difference between the two facing tubes must be ≤ 0.01g).
- Centrifuge for 2 h at 4 °C and 100, 000 x g (25,000 rpm for SW41 rotor). After centrifugation, slowly remove the tubes and carefully immobilize them on a support. Using a syringe with 19 G needle, draw up the white/opalescent band that is located usually between the d=1.5 and the d=1.4 density region.
- Transfer the suspensions from the syringe into new polyallomer tubes for SW60 rotor and use the d=1.4 solution to precisely balance the tubes.
- Centrifuge the suspension at 4 °C and 150,0000 x g (38,000 rpm for the SW60 rotor) for at least 16 h.
- Collect the visible band as above (see step 3.3.3) and transfer them into dialysis tubes with 6,000 Da cut-off. Dialyze the obtained visible band 2x for 20 min against 500 mL of water and overnight against 500 mL of TN buffer. Filter with a 0.22 µm pore filter and store at 4 °C.
- In a polyallomer ultracentrifuge tubes for SW41 rotor, prepare a CsCl step density gradient by stratifying 2 mL of the four CsCl solutions described in Table 1. Add 3.5 mL of phage suspension in each of the 4 tubes.
4. Phage cocktail preparation
- Make a mix of four phages that infect the strain PAO1, previously isolated and characterized8. Before mixing, estimate the titer of each phage stock by plaque assay20.
- Assemble the phage cocktail, with an overall titer of 5 × 108 pfu/mL, by equally mixing four phage preparations at the same pfu/mL immediately before each experiment, to ensure accurate phage titers.
5. Collection and preparation of zebrafish embryos for cftr morpholinos microinjection
NOTE: Collect 1-2 cell embryos from wild type zebrafish for cftr morpholinos (cftr-MOs) microinjection.
- Two days before the bacterial microinjection experiment, set up breeding pairs and collect embryos as described previously21. Adult zebrafish (Danio rerio) of the AB strain were purchased from the European Zebrafish Resource Center, EZRC.
- The day after, collect the embryos immediately after fertilization with a plastic pipette or using a fine-mesh strainer. Place the collected embryos in a Petri dish containing E3 embryo medium and carefully remove the debris with a plastic pipette to avoid contamination that might damage the embryo development.
- Prepare 5 µL of morpholino injection solution by diluting the two morpholino stock solutions (1 mM) in sterile water to a final concentration of 0.25 pmol/embryo ATG-MO + 0.25 pmol/embryo splice-MO. To trace the embryo injection, add 0.5 µL of phenol red to the morpholino injection solution mix.
- Load a microinjection needle with approximately 5 μL of the morpholino mix solution using a 20 μL micropipette with a fine gel loading tip. Secure the needle to the micromanipulator connected to a stand and position it under a stereomicroscope.
NOTE: It is not necessary that the solution reaches the tip of the needle as the pressure of the microinjector pump will push it. - Clip off the tip of the needle by cutting the needle tip with fine sterile tweezers. Measure the diameter of the drop using a scale bar in the ocular of the microscope. Alternatively, create a drop of mineral oil onto a micrometer slide and set the microinjection volume by injecting the solution into the oil-drop to evaluate the droplet size. Use a drop with a 156 μm diameter to inject a volume of 2 nL.
- Set the microinjector by adjusting the compensation pressure to 15 hPa, injection time to 0.5 s, and injection pressure between 300 and 600 hPa to obtain the correct injection volume depending on the needle used.
- With a plastic pipette arrange the embryos from step 5.2 onto a glass positioned in a 96 mm diameter Petri dish.
NOTE: Too much E3 medium will prevent the proper penetration of the microinjection needle into the chorion of the embryos. - Penetrate the chorion and then the yolk with the needle to inject 2 nL into the embryo as described previously22.
- Place injected embryos into a Petri dish with E3 medium and put them in an incubator at 28 °C to let them develop until the day after.
6. Microinjection of zebrafish embryos with bacteria and phage cocktail
NOTE: To perform a systemic infection, the embryo must have blood circulation that usually starts after 26 hpf.
- After 26 hpf (for zebrafish developmental stages refer to Kimmel21) dechorionate embryos with pronase solution prepared by dissolving pronase powder in E3 medium at a concentration of 2 mg/mL. Gently pipette the embryos to break the chorion. Discard the pronase/E3 medium and rinse the dish several times with fresh E3 to remove all pronase.
- Anesthetize zebrafish embryos in E3 medium containing Tricaine approximately 5 min prior to injections.
- Pipette the anesthesized embryos into the track prepared in step 1. Use a fine gel loading tip to line the embryos in the lateral position.
- Load a microinjection needle with approximately 5 μL of the P. aeruginosa preparation as described in part 5.
- Insert the needle dorsally to the starting point of the duct of Cuvier where it starts spreading over the yolk sac. Inject a volume of 1-3 nL and ensure that the volume expands directly within the duct and enters into the circulation.
- Approximately after every 50 injected-embryo, inject a drop of the bacteria into a 1.5 mL centrifuge tube with 100 μL of sterile PBS to check if the injection volume remains the same. Plate the inoculum on LB agar (see Table 1) at 37 °C overnight to assess the CFU.
- Transfer the microinjected embryos in two clean Petri dishes with fresh E3 medium + PTU and incubate them at 28 °C.
- At two selected time points: 30 min or 3 h after the bacterial injection, take out one of the Petri dishes with bacterial-injected embryos from the incubator and align them in the track as described in 6.3 for the phage cocktail injection.
- Load a microinjection needle with approximately 5 μL of the phage cocktail (prepared in step 4) with a fine gel loading tip and fix it to the microinjector (see step 5).
- Inject the phage cocktail in the duct of Cuvier of the embryos previously injected with bacteria.
- Transfer the microinjected embryos in two clean Petri dishes with fresh E3 medium + PTU and incubate them at 28 °C.
7. Evaluation of the bacterial burden of embryos injected with PAO1 and phages
- At 8 hpi, pipette 15 anesthetized embryos from the Petri dish to a 1.5 mL centrifuge tube.
- Replace the anesthetic solution with 300 μL of 1% Triton X-100 in PBS (PBSTritonX) and homogenize the embryos by passing them at least 15x through an insulin syringe with a sterile 27-G needle.
- Prepare serial dilutions of homogenates in sterile PBS by transferring 100 μL of the diluted homogenate into 900 μL of sterile PBS.
- To select for the naturally amp-resistant PAO1 strain, plate 100 μL of the dilutions on LB agar added with ampicillin (100 μg/mL), and incubate at 37 °C over-night.
- The day after, count the number of colonies, multiply by the dilution factor to determine the total number of CFU, and divide by the number of embryos homogenized to obtain the average number of CFU/infected embryo.
8. Evaluation of the lethality of embryos injected with PAO1 and phages
- To evaluate the lethality of the PAO1 infection, score the injected embryos at 20 hpi under a stereomicroscope and count for the dead embryos (white/not transparent).
- Calculate the half-maximal lethal concentration 50 (LD50) dose of PAO1 that determines the death of 50% of injected embryos at 20 hpf.
9. Embryo preparation for stereomicroscope time-lapse imaging of GFP+ PAO1 infection
- Preparate the mold for live-embryo imaging by dissolving 1.5% low melting agarose in 100 mL of E3 solution.
- Add 1% Tricaine (see Table 1) to anesthetize the embryos.
- At 4 hpi transfer one embryo with a plastic pipette in a glass bottom dish and add the warm low-melting-point agarose solution.
- Position the glass bottom dish under a stereomicroscope and with a tip position the embryo in the desired orientation. Use a plastic pipette to carefully add a drop of agarose on the embryo.
- Let the agarose cool for 5-10 min and gently fill the glass bottom dish with E3 containing the anesthetic solution to keep the embryo moist.
- Place the glass bottom dish with the embryo under the fluorescent stereomicroscope with a fluorescent filter (channel 488 nm) for GFP+ bacteria. Do not remove the Petri dish until further acquisitions with the same parameters at 9, 14 and 18 hpi.
10. Expression analyses of pro-inflammatory cytokines
- At 20 hpi anesthetize the embryos with tricaine 1x solution and transfer them from the Petri dish to a 1.5 mL centrifuge tube.
- Under a fume hood remove the anesthetic solution and replace with 200 μL of guanidium hydrochloride reagent.
- Homogenize embryos by pipetting them repetitively through a 200 μL pipette first and then at least 15x with an insulin syringe with a sterile 27 G needle.
- Extract total RNA from homogenized zebrafish embryos using a commercially available guanidium hydrochloride reagents as per the manufacturer’s instructions.
- Treat 1 μg of each sample of RNA with 2 μL of 1U/μL of DNase I (RNase-free), following the manufacturer’s instructions.
- Use 1 μg of DNase I treated RNA for reverse-transcription reaction (RT) using commercially available kit (see Table of Materials), in a total volume of 20 μL according to manufacturer’s instructions.
- Perform RT qPCR reaction in a total volume of 10 μL containing 1x SYBR Green mastermix using 1.5 μL of a 1:6 dilution of RT reaction. For normalization purposes, use the primers for the house-keeping genes rpl8 and for pro-inflammatory cytokines, use primers for TNF-α and IL-1β (see Table 2). qPCR protocol was: cycle 1 step 1: 95 °C, 2 min, repeat once; cycle 2: step 1 95 °C, 10 s, step 2 55 °C, 30 s, repeat 40x; cycle 3: step 1 72 °C, 15 min.
Representative Results
Results and figures presented here are referred to CF embryos generated through the injection of cftr morpholinos as described previously10 and in step 5. To validate the CF phenotype, the impaired position of internal organs such as heart, liver, and pancreas as previously described17 (Figure 1) were considered. Similar results were obtained in case of the WT embryos as reported in our previous publication19.
Bacterial burden was reduced by phage therapy in CF embryos infected with PAO1. Furthermore, we evaluated the bacterial burden at 8 hpi by homogenizing groups of 15 embryos: the average number of bacteria (cfu/embryo) present in the PAO1 infected embryos was reduced to about 20% after phage administration treatment, thus confirming a less severe infection in the presence of the phage cocktail (Figure 2).
Lethality was reduced by phage therapy in CF embryos infected with GFP+ bacteria PAO1. CF zebrafish embryos at 48 hpf were injected with GFP+ bacteria of the PAO1 strain at a dose that caused 50% lethality after 20 hpi (30 cfu/embryo, Figure 3A). The site of injection was the yolk or the Duct of Cuvier to generate a systemic infection. Phage therapy against PAO1 infection was tested by injecting 2 nL of the equally mixed phage cocktail (300-500 pfu/embryo). The injection was performed at two different time points: 30 min (early) and 7 hours (late) after bacterial injection. In both cases, lethality was reduced at 20 hpi, indicating that phage therapy is effective (Figure 3B).
With live imaging, using a fluorescent stereomicroscope, we also followed the progression of the infection in CF embryos injected with GFP+ PAO1 and showed the efficacy of phage therapy in reducing the spread of fluorescent bacteria over the yolk sac. The CF+PAO1 injected embryo with GFP+ bacteria multiplication at 4, 9, 14 and 18 hpi is shown in the upper side of Figure 4, whereas the CF+PAO1+phages embryo with reduced fluorescence due to phage action against bacteria is shown in the bottom part (Figure 4).
Phage therapy reduced the inflammatory response generated by PAO1 infection in CF embryos. We, also, evaluated the immune response generated by PAO1 and PAO1 + phages injection at 8 hpi. As expected, the expression of the pro-inflammatory cytokines TNF-a and IL-1β analyzed by qPCR techniques was significantly increased following PAO1 injection in comparison to controls, while it was reduced with the co-injection of the phage cocktail (Figure 5A,B).
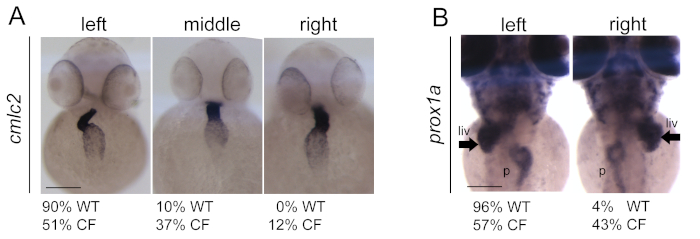
Figure 1: Generation and validation of CF embryos upon cftr morpholinos (cftr-MOs) injection. (A) Impaired position and looping of the heart in CF injected embryos in comparison to wild-type (WT) embryos. Heart is visualized with cmlc2 expression by in situ hybridization technique. (B) Impaired position of the liver (arrows) and pancreas in CF embryos in comparison to WT. Liver and pancreas are visualized with prox1a expression by in situ hybridization techniques. Scale bars indicate 100 μm. liv: liver; p: pancreas. The figure is reprinted from19 Please click here to view a larger version of this figure.
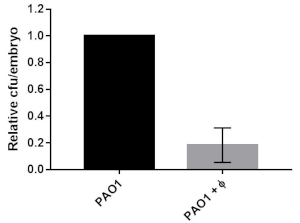
Figure 2: Bacterial burden in CF embryos infected with PAO1 or PAO1+phages. The relative percentage of cfu/embryo in PAO1+phages vs PAO1 embryos are given. The mean and SD of three independent experiments is reported. The figure is reprinted from19. Please click here to view a larger version of this figure.
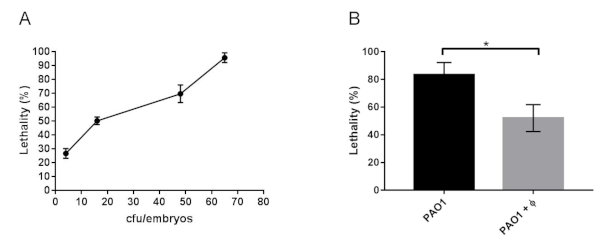
Figure 3: Lethality of CF zebrafish embryos infected with PAO1 and with PAO1+phages. (A) Determination of LD50 in 48 hpf zebrafish embryos microinjected with cftr-MO at 1-cell stage (CF embryos) and infected at 48 hpf with 2 nL of a culture of PAO1 containing increasing number of bacteria (cfu/embryo). Lethality of the embryos was observed at 20 hpi. (B) Lethality at 20 hpi of CF embryos infected with PAO1 at 48 hpf and treated with the phage cocktail (PAO1+ Φ). The mean and SD reported are from six and four experiments, respectively, each with 25-40 embryos. Angular transformation was applied to the percentage of lethality and one-way ANOVA followed by Duncan’s test was used. The figure is reprinted from19. Please click here to view a larger version of this figure.
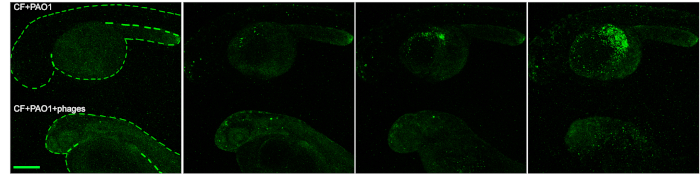
Figure 4: Imaging of the efficacy of phage therapy in zebrafish. Progression of the infection in CF embryos following PAO1 injection (upper embryo) and efficacy of the phage therapy in PAO1+phages injected embryos (bottom embryo) at 4, 9, 14 and 18 hpi. Scale bar indicates 100 microns. The figure is reprinted from19. Please click here to view a larger version of this figure.
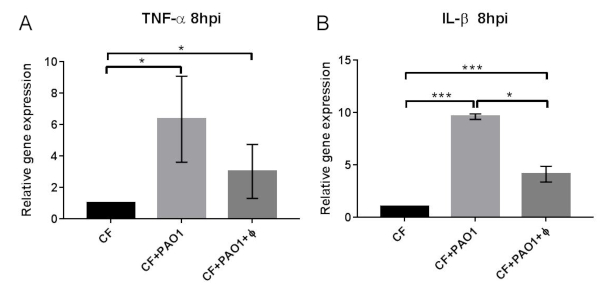
Figure 5: Expression of pro- and anti-inflammatory cytokines following PAO1 and PAO+phage administration. Expression levels of the TNF-a (A) and IL-1β genes measured by RT-qPCR at 8 hpi in CF embryos injected with PAO1 and PAO1+Φ at 48 hpf and normalized using the expression of rpl8. The mean and SD of four experiments are reported. Statistical significance was assessed by ANOVA followed by Duncan’s test: for TNF- a (CF) vs (CF+PAO1) p = 0.015*; (CF) vs (CF+PAO1+Φ) p = 0.019*; (CF+PAO1) vs (CF+PAO1+ Φ) p = 0.77 n.s.; for IL-1β (CF) vs (CF+PAO1) p = 0.00014***; (CF) vs (CF+PAO1+ Φ) p = 0.00068***; (CF+PAO1) vs (CF+PAO1+ Φ) p = 0.031*. The figure is reprinted from19. Please click here to view a larger version of this figure.
| Solutions | Preparation |
| Anaesthetic stock solution 25X | 4 mg/mL of Tricaine in distilled H2O. |
| Anaesthetic working solution 1X | dilute in distilled H2O the Tricaine stock solution 25X preparation to reach the 1X concentration (0.16 mg/mL) Tricaine of distilled H2O. |
| CsCl d=1.3 | 20.49 g in 50 mL TN |
| CsCl d=1.4 | 20.28 g in 50 mL TN |
| CsCl d=1.5 | 34.13 g in 50 mL TN |
| CsCl d=1.6 | 41.2 g in 50 mL of TN |
| E3 embryo medium for zebrafish embryo | 1 L 1of E3 (dilute the 50X stock with distilled H2O) + 200 μl of 0.05% methyl blue . Store at RT. |
| E3 embryo medium stock solution (50X) | 73.0 g NaCl, 3.15 g KCl , 9.15 g CaCl2 , and 9.95 g MgSO4 in 5 L of distilled H2O. Store at RT. |
| LB agar | 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, 10 g/L agar |
| LB broth | for 1L: 950 mL H2O, 10 g Tryptone, 10 g NaCl, 5 g Yeast extract |
| PBST | PBS 1X + Triton X 1% |
| Physiological solution | 0.9% NaCl |
| Pigmentation blocking stock solution 10X | 0.3 mg/mL phenyl thiourea (PTU) powder in E3 embryo medium for zebrafish embryo |
| Pronase stock solution 5X | 5 mg/mL pronase powder in E3 embryo medium for zebrafish embryo |
| TN buffer | 10 mM Tris HCl pH 8.0, 150 mM NaCl |
Table 1: Preparation of solutions.
| Gene name | Primer sequence |
| TNF-alpha Fw | 5’-TGCTTCACGCTCCATAAGACC-3’ |
| TNFalpha Rev | 5’-CAAGCCACCTGAAGAAAAGG-3’ |
| IL1-beta Fw | 5’-TGGACTTCGCAGCACAAAATG-3’ |
| IL1-beta Rev | 5’-CGTTCACTTCACGCTCTTGGATG-3’ |
| rpl8 Fw | 5’-CTCCGTCTTCAAAGCCCAT-3’ |
| rpl8 Rev | 5’-TCCTTCACGATCCCCTTGAT-3’ |
Table 2: Primers used for RT-qPCR.
Discussion
In this manuscript, we described the protocol to perform P. aeruginosa (PAO1) infection in zebrafish embryos and how to apply phage therapy with a cocktail of phages previously identified as able to infect PAO1 to resolve it. The use of bacteriophages as an alternative to antibiotic treatments has been of increasing interest since the last few years. This is mainly due to the diffusion of multi-drug resistant (MDR) bacterial infections, which constitute a serious issue for public health. Of course, the scope of this work is limited to the application of phage therapy to an animal model and not to humans. However, we generated a cystic fibrosis zebrafish model with the injection of a morpholino targeting the cftr gene, demonstrating the efficacy of phage therapy also in a pathogenetic model particularly susceptible to P.aeruginosa infection.
It is to note that we could easily achieve phage therapy, as in a previous work, we isolated and characterized different phages able to infect P. aeruginosa both in vitro and in vivo8. This step is fundamental to obtain the antimicrobial activity of the phages. The isolation and characterization of phages able to infect a specific bacterial strain are critical steps of phage therapy application. Indeed, to avoid adverse effects of phages on the animal/human host, it is necessary to use lytic phages instead of those lysogenic. Most importantly, it is necessary to check the phage genomes for the presence of harmful genes as those for antibiotic resistance, virulence and gene transfer.
Phage therapy has already been successfully used in other animal models. We are conscious that zebrafish is not a mammalian model and some effects of phages might be different. However, since zebrafish possess an innate immune system comparable to the human one with a conserved population of neutrophils and macrophages23, we speculate that the data on the immune response could be reproducible in humans. Moreover, zebrafish is well assessed for bacterial infection studies, its use as a tester for phage therapy efficacy might be promising for therapeutic approaches. Indeed, the infection can be systemic if bacteria are injected into the circulation through the Duct of Cuvier, or localized as reported17. We performed both systemic and localized infections and demonstrated that they similarly generated increased lethality and bacterial burden that were both decreased following phage therapy application. Moreover, since GFP+ fluorescent PAO1 bacteria was injected, it was easy to follow the infection at different time points of embryo development confirming the reduction of fluorescent bacteria when phages were injected.
To our knowledge, this is the first description of phage therapy application in zebrafish, with the added value for the demonstration of phages antimicrobial activity against P. aeruginosa in a CF background, that is particularly susceptible to this bacterial infection.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Italian Cystic Fibrosis Foundation (FFC#22/2017; Associazione “Gli amici della Ritty" Casnigo and FFC#23/2019; Un respiro in più Onlus La Mano tesa Onlus).
Materials
| Bacto Agar | BD | 214010 | |
| Calcium chloride | Sigma-Aldrich | 10043-52-4 | |
| CsCl | Sigma-Aldrich | 289329 | |
| Dulbecco's phospate buffered saline PBS | Sigma-Aldrich | D8537 | |
| Ethyl 3-aminobenzoate methanesulfonate | Sigma-Aldrich | 886-86-2 | common name tricaine |
| Femtojet Micromanipulator | Eppendorf | 5247 | |
| Fleming/brown P-97 | Sutter Instrument Company | P-97 | |
| LE-Agarose | Sigma-Aldrich | 11685660001 | |
| Low Melting Agarose | Sigma-Aldrich | CAS 9012-36-6 | |
| Magnesium sulfate | Sigma-Aldrich | 7487-88-9 | |
| Methyl Blue | Sigma-Aldrich | 28983-56-4 | |
| Microinjection needles | Harvard apparatus | ||
| N-Phenylthiourea >=98% | Aldrich-P7629 | 103-85-5 | |
| Oligo Morpholino | Gene Tools | designed by the researcher | |
| PEG6000 | Calbiochem | 528877 | |
| Phenol Red Solution | Sigma-Aldrich | CAS 143-74-B | |
| Potassium chloride | Sigma-Aldrich | 7447-40-7 | |
| Pronase | Sigma-Aldrich | 9036-06-0 | |
| Sodium chloride ACS reagent, ≥99.0% | Sigma-Aldrich | S9888 | |
| Stereomicroscope | Leica | S9I | |
| Tris HCl | Sigma-Aldrich | T5941 | |
| Triton X | Sigma-Aldrich | T9284 | |
| Tryptone | Oxoid | LP0042B | |
| Yeast extract | Oxoid | LP0021B | |
| Z-MOLDS Microinjection | Word Precision Instruments |
Referenzen
- Cisek, A. A., Dąbrowska, I., Gregorczyk, K. P., Wyżewski, Z. Phage Therapy in Bacterial Infections Treatment: One Hundred Years After the Discovery of Bacteriophages. Current Microbiology. 74 (2), 277-283 (2017).
- Trend, S., Fonceca, A. M., Ditcham, W. G., Kicic, A., Cf, A. The potential of phage therapy in cystic fibrosis: Essential human-bacterial-phage interactions and delivery considerations for use in Pseudomonas aeruginosa-infected airways. Journal of Cystic Fibrosis. 16 (6), 663-667 (2017).
- Pacios, O., et al. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics. 9 (2), 65 (2020).
- Dubos, R. J., Straus, J. H., Pierce, C. The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with shigella dysenteriae. Journal of Experimental Medicine. 78 (3), 161-168 (1943).
- Marza, J. A. S., Soothill, J. S., Boydell, P., Collyns, T. A. Multiplication of therapeutically administered bacteriophages in Pseudomonas aeruginosa infected patients. Burns. 32 (5), 644-656 (2006).
- Heo, Y. J., et al. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrobial Agents and Chemotherapy. , 01646 (2009).
- McVay, C. S., Velásquez, M., Fralick, J. A. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrobial Agents and Chemotherapy. , 01028 (2007).
- Forti, F., et al. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrobial Agents and Chemotherapy. , 02573 (2018).
- Jault, P., et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infectious Diseases. 19 (1), 35-45 (2019).
- Phennicie, R. T., Sullivan, M. J., Singer, J. T., Yoder, J. A., Kim, C. H. Specific resistance to Pseudomonas aeruginosa infection in zebrafish is mediated by the cystic fibrosis transmembrane conductance regulator. Infections and Immunity. 78, 4542-4550 (2010).
- Clatworthy, A. E., et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infections and Immunity. 77, 1293-1303 (2009).
- Bernut, A., et al. CFTR Protects against Mycobacterium abscessus Infection by Fine-Tuning Host Oxidative Defenses. Cell Reports. 26 (7), 1828-1840 (2019).
- Semler, D. D., Goudie, A. D., Finlay, W. H., Dennis, J. J. Aerosol phage therapy efficacy in Burkholderia cepacia complex respiratory infections. Antimicrobial Agents and Chemotherapy. , 02388 (2014).
- Benard, E. L., et al. Infection of zebrafish embryos with intracellular bacterial pathogens. Journal of Visualized Experiments. (61), e3781 (2012).
- Doring, G., Gulbins, E. Cystic fibrosis and innate immunity: how chloride channel mutations provoke lung disease. Cellular Microbiology. 11, 208-216 (2009).
- Navis, A., Bagnat, M. Loss of cftr function leads to pancreatic destruction in larval zebrafish. Entwicklungsbiologie. 399, 237-248 (2015).
- Navis, A., Marjoram, L., Bagnat, M. Cftr controls lumen expansion and function of Kupffer’s vesicle in zebrafish. Development. 140, 1703-1712 (2013).
- Balloy, V., et al. Normal and cystic fibrosis human bronchial epithelial cells infected with Pseudomonas aeruginosa exhibit distinct gene activation patterns. PLoS One. 10, 0140979 (2015).
- Cafora, M., et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Science Reports. 9, 1527 (2019).
- Hershey, A. D., Kalmanson, G. M., Bronfenbrenner, J. Quantitative methods in the study of the phage-antiphage reaction. Journal of Immunology. 46, 267-279 (1943).
- Kimmel, C., Ballard, W., Kimmel, S., Ullmann, B., Schilling, T. Stages of embryonic development of the zebrafish. Developmental Dynamics. 203, 253-310 (1995).
- Rosen, J. N., Sweeney, M. F., Mably, J. D. Microinjection of zebrafish embryos to analyze gene function. Journal of Visualized Experiments. (25), e1115 (2009).
- Traver, D., et al. The Zebrafish as a Model Organism to Study Development of the Immune System. Advances in Immunology. 81, 253-330 (2003).

