Semi-Quantitative Analysis of Peptidoglycan by Liquid Chromatography Mass Spectrometry and Bioinformatics
Summary
This protocol covers a detailed analysis of peptidoglycan composition using liquid chromatography mass spectrometry coupled with advanced feature extraction and bioinformatic analysis software.
Abstract
Peptidoglycan is an important component of bacterial cell walls and a common cellular target for antimicrobials. Although aspects of peptidoglycan structure are fairly conserved across all bacteria, there is also considerable variation between Gram-positives/negatives and between species. In addition, there are numerous known variations, modifications, or adaptations to the peptidoglycan that can occur within a bacterial species in response to growth phase and/or environmental stimuli. These variations produce a highly dynamic structure that is known to participate in many cellular functions, including growth/division, antibiotic resistance, and host defense avoidance. To understand the variation within peptidoglycan, the overall structure must be broken down into its constitutive parts (known as muropeptides) and assessed for overall cellular composition. Peptidoglycomics uses advanced mass spectrometry combined with high-powered bioinformatic data analysis to examine peptidoglycan composition in fine detail. The following protocol describes the purification of peptidoglycan from bacterial cultures, the acquisition of muropeptide intensity data through a liquid chromatograph—mass spectrometer, and the differential analysis of peptidoglycan composition using bioinformatics.
Introduction
Peptidoglycan (PG) is a defining characteristic of bacteria that serves to maintain cell morphology, while providing structural support for proteins and other cellular components1,2. The backbone of PG is composed of alternating β-1,4-linked N-acetyl muramic acid (MurNAc) and N-acetyl glucosamine (GlcNAc)1,2. Each MurNAc possesses a short peptide bound at the ᴅ-lactyl residue that can be crosslinked to adjacent disaccharide-linked peptides (Figure 1A,B). This crosslinking produces a mesh-like structure that encompasses the entire cell and is often referred to as a sacculus (Figure 1C). During PG synthesis, precursors are generated in the cytoplasm, and transported across the cytoplasmic membrane by flippases. Precursors are subsequently incorporated into the mature PG by transglycosylase and transpeptidase enzymes, which produce the glycosidic and peptide bonds, respectively3. However, once assembled, there are numerous enzymes produced by the bacteria that modify and/or degrade the PG to carry out a number of cellular processes, including growth and division. In addition, various modifications of the PG have been shown to confer adaptations specific to the strain, growth conditions, and environmental stress, which have been implicated in cell signalling, antimicrobial resistance, and host immune evasion4. As examples, a common modification is the addition of a C6 acetyl group on the MurNAc that confers resistance by limiting access to the glycan β-1,4 linkages to host-produced lysozyme enzymes which degrade PG4,5,6. In Enterococci, substitution of the terminal ᴅ-Ala of the peptide sidechain with ᴅ-Lac confers a greater resistance to the antimicrobial, vancomycin7,8.
The general procedure for PG isolation and purification has remained relatively unchanged since it was described in the 1960s9. Bacterial membranes are dissolved through heat treatment with SDS, followed by enzymatic removal of bound proteins, glycolipids, and remaining DNA. The purified intact sacculus can be subsequently digested into the individual components by hydrolysis of the β-1,4 linkage between GlcNAc and MurNAc. This digestion produces GlcNAc-MurNAc disaccharides with any structural modifications and/or crosslinks intact and are called muropeptides (Figure 1B).
Compositional analysis of PG was initially conducted through high pressure liquid chromatographic separation (HPLC) to purify each muropeptide followed by manual identification of muropeptides10,11. This has since been superseded by liquid chromatography tandem mass spectrometry (LC-MS), which increases detection sensitivity and decreases the manual workload of purifying each individual muropeptide. However, the time consuming and complex nature of the manual identification of muropeptides has remained a limiting factor, reducing the number of studies conducted. In recent years with the emergence of “omic” technologies, automated LC-MS feature extraction has become a powerful tool, allowing for rapid detection and identification of individual compounds in complex samples from very large datasets. Once the features are identified, bioinformatic software statistically compares the variation between samples using differential analysis isolating even minimal differences among the complex dataset and displaying them graphically to the user. The application of feature extraction software for the analysis of PG composition has only just begun to be explored12,13,14 and coupled to bioinformatic analysis12. Unlike proteomic analysis which benefits from the readily available protein databases that predict peptide fragmentation allowing for fully automated identification, no fragmentation library currently exists for peptidoglycomics. However, feature extraction can be coupled with known and predicted structural databases to predict muropeptide identification12. Here we present a detailed protocol for the use of LC-MS-based feature extraction combined with a muropeptide library for automated identification and bioinformatic differential analysis of PG composition (Figure 2).
Protocol
1. Peptidoglycan sample preparation
- Growth of bacterial cultures
NOTE: The growth of the bacterial cultures will vary depending on the bacterial species and the growth conditions being examined. The experimental parameters to be tested will define the growth conditions.- Grow bacterial cultures under growth conditions required for the bacterial strain and experimental design. Grow bacteria as triplicate cultures (biological replicates) i.e., three separate colonies per strain or growth condition.
NOTE: Growth conditions and growth phase are known to have significant effects on PG composition1,2,10. Great care must be taken to maintain consistency between cultures and replicates to ensure compositional changes are due to the experimental parameters and not experimental error. - Rapidly cool the culture to 4 °C, collect by centrifugation (11,000 x g, 10 min, 4 °C) and freeze the cell pellet at -20 °C. Wash the cell pellet with pre-cooled 4 °C, 20 mM sodium phosphate pH 6.5 prior to freezing. Production of the frozen cell pellet should be done as quickly and as consistently between samples as possible to limit the activity of enzymes which could modify and/or degrade the PG during the collection process. Samples can be processed directly through the extraction process (section 1.2) without freezing; however, ensure that all samples are processed in a similar manner.
NOTE: To ensure sufficient product for later steps, a significant sized wet cell pellet is used. This produces a large enough sacculi pellet, which is easily visualized and maintained during the repetitive washing steps (section 1.2.5 and 1.2.14) without significant loss of product. Depending on bacterium and growth conditions, this yield will likely vary. For the Gram-negative bacterium Pseudomonas aeruginosa, PAO1, 4 L of a 0.5 OD600 culture produced a 3–4 g cell pellet and was sufficient to produce ~10 mg of purified sacculi (section 1.2.17)12. This is a large excess of sacculi than is required for the LC-MS (section 2); however, it will aid in measurement accuracy (section 1.2.17) and normalization (section 1.3).
- Grow bacterial cultures under growth conditions required for the bacterial strain and experimental design. Grow bacteria as triplicate cultures (biological replicates) i.e., three separate colonies per strain or growth condition.
- Extraction of peptidoglycan sacculi
NOTE: The protocol for the extraction of PG is adapted from ref.9,11,15. This protocol will extract the PG from individual bacterial cells as a whole sacculi, free of other cellular components. The protocol can be used with either Gram-negative or Gram-positive bacteria. However, for Gram-positive cells, adjustments may be necessary to isolate the thicker PG structure and to remove cell-wall associated polymers; such as, teichoic acids.- Resuspend frozen cell pellets at approximately 1:10 of the original culture volume of 20 mM sodium phosphate pH 6.5. Perform this step at 4 °C (can be 1–8 mg wet cell pellet weight per mL of buffer11,12).
NOTE: To maintain the acetylation state of the PG, a pH of 6.5 or lower is required15,16. - Add cell suspension dropwise to boiling 8% sodium dodecyl sulfate (SDS) 20 mM sodium phosphate pH 6.5 for a final 1:1 volume (i.e., final concentration is 4% SDS), gently stirring in a round bottom flask outfitted with a water-cooled condenser. (Figure 2, step 2)
- Maintain a gentle boil for 30 min to 3 h with stirring to ensure complete membrane dissociation. Ensure that the resulting mixture is completely clear with no remaining cell clumps or viscosity. Longer boiling is preferred to guarantee complete dissociation.
- Allow it to cool to room temperature. The sample can be left at room temperature overnight.
NOTE: When SDS is present, maintain samples at room temperature to keep SDS in the solution. - Collect sacculi as a pellet through ultracentrifugation at 70,000 x g for 40 min (or the time required to completely sediment sacculi) at 25 °C.
- Repetitively wash sacculi by successive ultracentrifugation (section 1.2.5) and suspension in ~50 mL of room temperature 20 mM sodium phosphate pH 6.5 until the wash buffer has a SDS concentration ~0.001%. Typically, 5 to 7 washes are sufficient.
NOTE: To test the concentration of the remaining SDS in the wash buffer, use the colorimetric dye, Stains-all17. - Resuspend sacculi in 5–10 mL of room temperature 20 mM sodium phosphate pH 6.5.
- Sonicate the sample briefly (~40%, 50 W, 20 kHz, 20 s) at room temperature to disperse clumps.
NOTE: Extended sonication will mechanically cause shearing of the PG structure18. - Supplement the sample with 50 μg/mL each amylase, DNase, and RNase, 10 mM magnesium sulfate, and digest at 37 °C for 1 h with agitation or nutation.
NOTE: Amylase digestion removes any remaining glycogen trapped within the sacculi11. - Add 100 μg/mL pronase and digest at 37 °C overnight with agitation or nutation and ~0.02% sodium azide.
NOTE: Pronase digestion removes the enzymes added (from section 1.2.9) and removes lipoproteins that are covalently linked to the PG.
CAUTION: Sodium azide is highly toxic and requires proper use/disposal methods. - Ultracentrifuge at 70,000 x g for 40 min (or the time required to completely sediment sacculi) at 25 °C to remove sodium azide.
- Resuspend the pellet in 25 mL of 2% SDS 20 mM sodium phosphate pH 6.5.
- Boil for 1 h in a steamer or the round bottom flask with water-cooled condenser (section 1.2.2).
NOTE: The second SDS boiling step removes all the remaining proteins and contaminants from the sacculi. - Repeat the sacculi wash (section 1.2.6) with ~50 mL room temperature double distilled water (ddH2O) until the SDS concentration is ~0.001%.
- Resuspend the pellet in a sufficient quantity of ddH2O to suspend sacculi, as well as wash the container (e.g., 25 mL) and freeze overnight at -80 °C. The volume can vary as the sample will be lyophilized in the next step, although, smaller volumes require less time to lyophilize.
- The next day, lyophilize the suspension and store at room temperature.
- Measure the quantity of lyophilized sacculi obtained on an analytical balance.
- Dilute sacculi in ddH2O to 10 mg/mL and briefly sonicate to break up clumps prior to further analysis.
- Resuspend frozen cell pellets at approximately 1:10 of the original culture volume of 20 mM sodium phosphate pH 6.5. Perform this step at 4 °C (can be 1–8 mg wet cell pellet weight per mL of buffer11,12).
- Quantification of purified peptidoglycan
- Quantify the amount of purified sacculi from section 1.2.18 to ensure mass spec data is equalized during differential analysis (section 3.2). Follow the detailed methodology outlined in Reference15.
NOTE: Purified sacculi (section 1.2.18) are broken down into individual sugar and amino acid components by acid hydrolysis. Individual components are separated and quantified by anion-exchange chromatography using pulsed-amperometric detection. Given the structure of PG (Figure 1), individual muropeptides are composed of a single MurNAc and a single GlcNAc residue. Therefore, quantifying the concentration of either residue represents the quantity of muropeptides 1:1 within the sample. MurNAc is preferred due to the clean peak separation from other PG components during chromatography16.
- Quantify the amount of purified sacculi from section 1.2.18 to ensure mass spec data is equalized during differential analysis (section 3.2). Follow the detailed methodology outlined in Reference15.
2. Mass spectrometry data acquisition
- Preparation of muropeptides for mass spectrometry
- Supplement 800 μg of purified sacculi with 100 μg/mL mutanolysin, 100 mM ammonium acetate pH 5.5, and 50 mM magnesium chloride in a 100 μL reaction.
- Digest at 37 °C overnight.
- Add 1:1 volume 0.5 M borate buffer pH 9.0 and supplement with ~10 mg/mL sodium borohydride (NaBH4).
NOTE: Mutarotation of cyclic sugars between α and β anomeric forms will cause multiple peak formations during the liquid chromatography separation of the muropeptides. The treatment with NaBH4 eliminates interconversion between the two forms by reducing MurNAc into muramitol11. The treatment does not reduce 1,6-anhydro MurNAc or 1,4-linked sugar residues.
CAUTION: The reaction of NaBH4 produces small quantities of hydrogen gas. The NaBH4 reaction will create bubbles and microfuge tubes should be kept open to allow gas to escape. - Incubate the sample at room temperature for ~20–30 min.
- Centrifuge briefly to settle the sample in the microfuge tube and remove bubbles.
- Adjust pH to <4.0 using 1:5 phosphoric acid, added in 5 μL increments. Test pH using litmus pH test strips.
- Centrifuge at ~17,000 x g for 1 min to sediment any remaining insoluble material.
- Filter using 0.2 µm microcentrifuge filters.
- Samples are centrifuged for 10 min at 30,000 x g prior to injection into LC-MS to ensure any particulates are not injected into MS.
- Setup of LC-MS
- Attach a C18 superficially porous particle column (100 mm x 2.1 mm, pore size <3 µm) to a Quadrupole-Time of Flight (QTOF) mass spectrometer with a minimal four decimal point m/z detection accuracy.
- Perform liquid chromatography separation of muropeptides
NOTE: Each biological triplicate (section 1.1.1) should be run through the LC-MS (section 2.2.2 through 2.2.3) three times (technical triplicate). Therefore, each tested condition will have a total of nine LC-MS data files. Data acquisition was performed using commercially available software (see Table of Materials). However, acquisition software should be chosen based on the MS machinery. The following represents a guide for setting up the MS with parameters specific for running this protocol. For a detailed description, please refer to the manufacturer’s manual.- For chromatographic separation, prepare the following solvents 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B).
- Set up a method for chromatographic separation using the following parameters.
- Set the flowrate to 0.4 mL/min.
- Condition the column for 10 min at 2% B (~24 column volumes).
- Using an autosampler, inject 10 µL of sample from section 2.1.9.
NOTE: Run one initial sample through the LC-MS protocol prior to beginning data collection. This run is not used for data but to increase retention time reproducibility for all subsequent runs. The reproducibility of the retention time is required during spectral processing (section 3.1) for the accurate identification and grouping of mass-to-charge ratio (m/z) peaks. - Separate muropeptides using 2% B for 5 min (~12 column volumes), then increasing it to 15% B over 13 min (~30 column volumes), further increasing it to 50% B over 10 min (~24 column volumes), and finally increasing it to 98% B over 2 min (~5 column volumes).
NOTE: Discard (send to waste) the first 2 min and last 5 min of the gradient. - Finish with a column wash at 98% B for 6 min (~14 column volumes) and 20 min (~47 column volumes) re-equilibration.
- Perform mass spectrometry detection of muropeptides
- Calibrate the mass axis in positive mode using a tuning mix in acetonitrile containing LC-MS reference mass standards following MS manufacturer’s instructions.
NOTE: The MS is tuned prior to the beginning of the chromatographic runs (section 2.2.2.1). - Set up a method for MS data acquisition using the following parameters.
NOTE: MS and MS/MS data are collected (sections 2.2.3.2 to 2.2.3.6) simultaneously with the chromatographic separation of the muropeptides (sections 2.2.2.4 and 2.2.2.5). Both chromatographic and MS parameters (sections 2.2.2.2 and 2.2.3.2) are a single method that is added during the setup of a worklist for running multiple samples in sequence. - Set the electrospray capillary voltage at 4.0 kV and the drying gas temperature at 350° C with a flow rate of 13 L/min.
NOTE: Nitrogen (purity >99%) should be used as nebulizing, drying and collision gas during all mass spectrometry data collection. - Set nebulizer pressure to 40 psi and set the fragmentor to 150 V.
- Set the nozzle, skimmer, and octapole RF voltages to 1000 V, 65 V, and 750 V, respectively.
- Set the scan range to 300–2,000 m/z in 4 GHz (extended dynamic range) positive ion mode.
- Set data collection using data dependent MS/MS acquisition with an MS and MS/MS scan rate of 1 spectra/second. Select five precursor mass per cycle, in the order of singly, doubly, and triply charged.
- Set MS/MS fragmentation collision energies to 15, 20 and 30 eV.
- Calibrate the mass axis in positive mode using a tuning mix in acetonitrile containing LC-MS reference mass standards following MS manufacturer’s instructions.
3. Differential analysis of muropeptide abundance
- LC-MS chromatogram spectral processing
NOTE: Recursive feature extraction was performed using commercially available software (see Table of Materials). Other feature extraction software can be used. However, other software may require additional manual data processing to accomplish the highly robust recursive extraction. Various software uses the terminology feature, entity, and compound almost interchangeably. For PG analysis, all refer to the identification of the LC-MS ion chromatogram representative of an individual muropeptide (e.g., Figure 3). During spectral processing (section 3.1), a feature represents the multiple m/z peaks that encompass the multiple possible ion species of a single muropeptide that are grouped together under a single compound label.- Under File, start a new project.
- Add the LC-MS QTOF data files and assign individual data files to an experimental condition / group, e.g., two different growth conditions.
- Run the data processing wizard Batch recursive feature extraction (small molecules/peptides) and set the data processing filters to match the parameters of the LC-MS conditions and instrumentation to accurately identify, group and verify m/z peaks representing individual muropeptides. From the chromatogram, examine the retention time drift and variation of m/z of known similar peaks to set initial filter parameters.
NOTE: Recursive feature extraction uses an initial untargeted molecular feature extraction algorithm to identify and align chromatogram features (m/z peaks) across all the data files. Once created, these features are used to reassess the original data files (recursive) with a targeted molecular feature extraction algorithm to improve the reliability and accuracy of the identified features. It is best to set the initial untargeted extraction with a narrow detection window and use broader detection parameters for the recursive extraction to identify peaks in all data files that may be missing in the initial extraction. Running the recursive feature extraction can take hours to complete depending on the number of samples, the complexity of the data, and the computer hardware present - Review the feature extraction results. If a significant number of features failed to align in a group, adjust the recursive filtering parameters to broaden/restrict the detection window as required. To accomplish this, inspect the chromatogram and isotopic profile of each extracted feature to ensure feature detection was accomplished similarly across all data files. Also, for each identified feature within a data file, examine the score, any warnings and whether the feature passed the chosen filter parameters.
NOTE: Care must be taken to keep detection windows narrow enough that distinct features are not mistakenly grouped together. This is often noted by disparate isotopic profiles between data files, or significant m/z / retention time variations. Conversely, if there are multiple features with similar m/z and retention times, it is possible that the filter parameters were too stringent resulting in one MS peak being split into two features. Therefore, run feature extraction (section 3.1.3) again with adjusted retention time filter parameters to allow better grouping of these features. Visualizing the lowest abundance peaks will indicate whether the filters used (section 3.1.3) accurately identified peaks above background noise. If the lowest abundance peaks appear similar to the background, rerun the feature extraction with new background filter parameters. - Export data as a compound exchange file (.cef) (a format compatible with the statistical software program), or a column separated file (.csv) which contains the mass, retention time, and abundance of each feature for each sample.
- Differential analysis of spectral features
NOTE: Differential analysis was performed using commercially available software (see Table of Materials). Other bioinformatic software can be used.- Open the program and when instructed start a new project.
- Follow the instructions for data import and data analysis. During data import, upload the feature extracted files (section 3.1). During data analysis, choose significance and fold change for differential analysis and select to baseline data to the median intensity across all the data files. Do not set any data filters (if this was done in the previous spectra processing step (section 3.1)) as applying filters again would negate the robust recursive feature extraction. However, similar to feature extraction, differential analysis must be aligned based on mass (m/z) and retention time due to drift in the LC-MS data collection. Use the parameters determined in feature extraction (section 3.1).
NOTE: Normalize between data files using the muropeptide quantitation (section 1.3) to ensure variations are due to experimental parameters and not due to variations in sacculi purification (section 1.2). Use the external scaler option to adjust each data file for differences in sample MurNAc concentration. - Once the analysis is complete, examine the resulting graphical and statistical analyses to identify muropeptides that demonstrate a significant abundance change between the tested experimental conditions.
- Under the project navigator, right-click on the various analyses and choose an export option to save feature details as a column separated (.csv) data file that contains the m/z, retention time, raw and normalized intensity values, p-value, FDR, and fold changes for each feature. Multiple analyses must be saved to obtain all the relevant data.
- Export a second .csv file containing only the muropeptides that have surpassed the statistical analyses including p-value <0.05 and fold change >2.
- Annotating muropeptide identity to spectral features
NOTE: Each identified feature must be assigned a predicted muropeptide structure based on the m/z and this annotation confirmed by examining the MS/MS fragmentation. After confirming the annotations, it may be necessary to perform and refine the differential analysis (section 3.2).- Within the differential analysis software, under results interpretation, select ID browser. Add a library of expected muropeptide structures and select similar parameters as used previously (section 3.1.3). This will produce a predicted muropeptide annotation for each identified feature. A library of muropeptide structures can be produced using the m/z for predicted muropeptide structures and MS database software (see Table of Materials). However, a library of the m/z of >6,000 possible muropeptides can be found in Reference12.
- Select the predicted muropeptide annotation based on the matching score and the biological relevance of the predicted muropeptide, i.e., choose the most likely muropeptide to be present in the biological sample.
- Manually confirm the predicted muropeptide annotation by comparing the m/z peaks of the MS/MS chromatogram to the predicted m/z of all possible fragmentations of a known muropeptide structure (e.g., Figure 4).
- View the MS and MS/MS data using a chromatogram-viewing program (see Table of Materials, Figure 4).
- Draw the predicted muropeptide structure using a molecular editor (chemical structure drawing program) (see Table of Materials, Figure 4, gray inset). Use the mass fragmentation tool to show the m/z of MS fragments when each bond is broken either individually or in combination.
NOTE: Depending on the fragmentation energy used for MS/MS, fragmentation can happen at any bond in the muropeptide structure. However, some bonds are more easily / frequently fragmented at lower energy levels. For example, fragmentation in the peptide sidechain occurs most often at the amide bond between amino acids. While assessing the fragmentation, it is important to note that GlcNAc residues are very easily fragmented from the muropeptide. Therefore, fragmentation of the known muropeptide structure should be assessed with and without GlcNAc. Due to the in-source fragmentation of the GlcNAc, several features extracted in spectral processing (section 3.1) may represent a single muropeptide structure. If found, these features should be merged and the differential analysis reassessed. - Compare all the possible fragmentations of the muropeptide structure that was determined (section 3.3.3.2) to the MS/MS chromatogram (section 3.3.3.1). To confirm the muropeptide annotation, the m/z peaks of multiple fragments should be found in the MS/MS chromatogram with a very minimal m/z alignment window (Figure 4).
- In order to elucidate muropeptide identity in the case of ambiguous MS/MS fragmentation, repeat sections 2.2.2 through 2.2.3 with samples (section 2.1.9) for additional MS/MS data acquisition incorporating a preferred precursor list of m/z and retention time with additional MS/MS fragmentation collision energies.
- For co-eluting entities that were annotated as the same muropeptide, run the differential analysis (section 3.2.2) again and merge the extracted features.
- Assessing global changes in muropeptide modifications
- Edit the .csv file (section 3.2.4) of the statistically significant high fold change muropeptides to include a single column for each muropeptide modification. Populate this column with a designation for each muropeptide annotated (section 3.3) (e.g., acetylated versus de-N-acetylated GlcNAc or MurNAc).
- Upload the modified .csv file into Perseus22,23. Import the normalized intensity values into the Main import box and import the modification designation into the Categorical import box.
- Under Annotate Rows, click on Categorically Annotate Rows and add datafiles to each experimental parameter.
- Under test, click on two sample tests to perform a student’s t-test (p-value < 0.05, FDR < 0.05, s0 = 1).
- Click on 1D to perform 1D annotation22,23. A 1D annotation FDR < 0.05 indicates a significant abundance change for the muropeptide modification between the tested experimental parameters. Setting the Threshold value (s0) = 1 will display the 1D annotation FDR scores for all the muropeptides.
- Within a graphing software (see Table of Materials), produce a heat map of the abundance fold change for each muropeptide modification and show the 1D annotation score to demonstrate significance (Figure 5B). The fold change of each muropeptide modification can be produced in Microsoft Excel using the raw intensities of all individual muropeptides that contain the modification.
Representative Results
Increased detection sensitivity of MS machinery coupled with high-powered peak recognition software has improved the ability to isolate, monitor, and analyze substance compositions of complex samples in very minute detail. Using these technological advancements, recent studies on peptidoglycan composition have begun to use automated LC-MS feature extraction techniques12,13,14,24 over older HPLC-based methodology11,25,26,27,28,29,30,31. Although there are numerous generic feature extraction software packages available, commercial software using recursive feature extraction is rapid and highly robust by automatically identifying and combining all the charges, isotopes, and adduct versions of each muropeptide found within the LC-MS dataset (Figure 3). In addition, initial retention times, m/z and isotopic patterns of extracted features are used to reassess (recursive) the dataset to ensure accurate identification of each feature in all data files. Therefore, the recursive algorithm aids in validating and increasing confidence in peak identification. Most generic feature extraction programs do not group charges/isotopes, etc. and will require this as an additional manual step. In addition, generic programs will be less robust as features are extracted separately within each data file and not as an entire dataset, which is part of the recursive algorithm.
The peptidoglycomic protocol presented here was recently used to examine the compositional changes of PG between two physiological growth conditions, namely, free-swimming planktonic and stationary communal biofilm12. Using a highly sensitive QTOF MS coupled with the recursive feature extraction, 160 distinct muropeptides were recognized and tracked. This represented eight times the number of muropeptides identified in this organism previously29,32, and greater than double the muropeptides identified using other methodologies in other organisms10,14,24.
Associating each m/z peak extracted from the MS data with a particular muropeptide is facilitated by cross-referencing with a database of known and predicted muropeptide structures. The fragmentation MS/MS chromatogram (Figure 4) for each extracted feature is compared to the fragmentation profile (Figure 4, gray inset) of the muropeptide proposed using the database.
Peptidoglycomic data can be viewed in a number of different ways depending on the experimental setup and the questions being asked. Such graphical analysis can include principal components analysis (PCA), scatterplots, volcano plots, heat maps, and hierarchical clustering analysis. For example, volcano plots highlight muropeptides that demonstrate a statistically significant high magnitude of abundance change between the tested conditions (Figure 5A). These selected muropeptides which represent significant abundance changes between the tested conditions can be further examined for muropeptide modifications. These modifications can include the presence of amino acid substitutions, acetylation changes, or the presence of amidase activity. When examined together, multiple muropeptides possessing the same modification can be examined for a trend toward one experimental condition (Figure 5A—highlighted points green) and the entire group assessed for significance (Figure 5B). Tracking a muropeptide modification in this way, can indicate a particular enzymatic activity that is affected by the experimental parameter. In addition, outliers from this trend may indicate enzymatic activity with a particular specificity or biological function (Figure 5A—highlighted points orange).
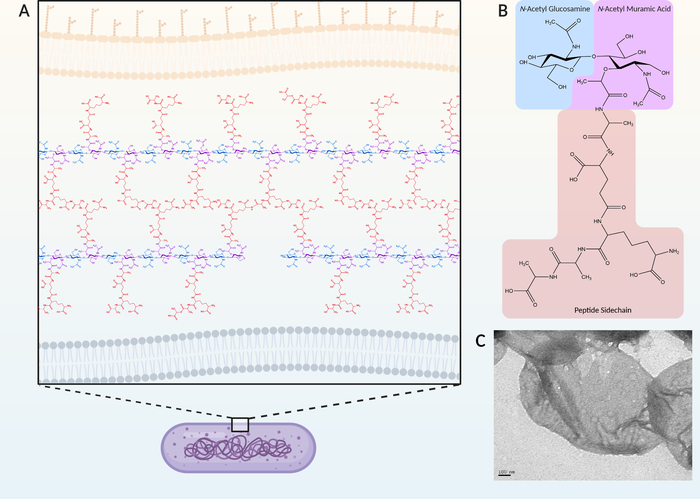
Figure 1: Example of a typical Gram-negative peptidoglycan structure. (A) In Gram-negative bacteria, peptidoglycan is located in the periplasm between the inner and outer membranes. (B) A single muropeptide consists of a β-1,4-linked N-acetyl glucosamine (GlcNAc) (blue) and a N-acetyl muramic acid (MurNAc) (purple) with an appended peptide sidechain (orange). The peptide sidechain can be crosslinked to the sidechain of adjacent muropeptide producing the mature mesh-like peptidoglycan (A). Purification involves the isolation of the peptidoglycan from the entire cell as a sacculus where all other cellular material has been stripped away. (C) Transmission electron micrograph of a peptidoglycan sacculi. In comparison, Gram-positive PG can consist of a greater array of variations in structure and is part of Gram-positive taxonomic classification33. Please click here to view a larger version of this figure.
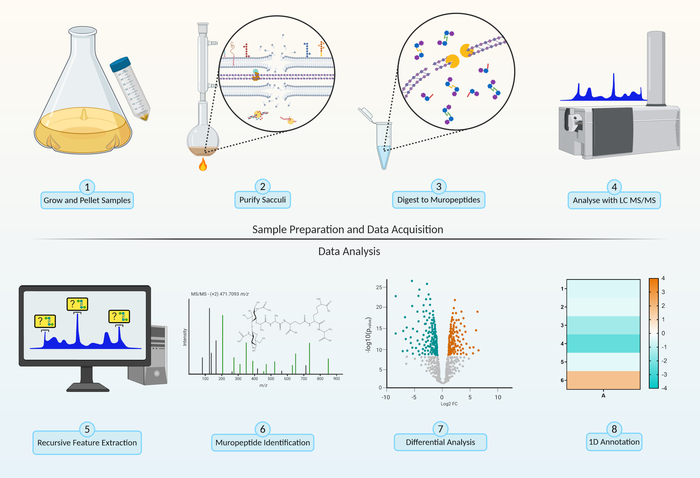
Figure 2: Peptidoglycomics workflow. Sample Preparation. Step 1, grow and pellet bacterial cells (section 1.1). Step 2, purify peptidoglycan sacculi by 4% SDS boil (section 1.2). Data Acquisition. Step 3, enzymatic digestion of sacculi to produce muropeptides by breakage of the β-1,4-linkage between the N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) of the peptidoglycan backbone (section 2.1). Step 4, analysis of muropeptide intensity through LC-MS/MS (section 2.2). Data Analysis. Step 5, recursive feature extraction identifies and collect all charges, adducts and isotopes associated with a single muropeptide (section 3.1). Step 6, identification of muropeptides by comparing predicted fragmentation with MS/MS chromatograms (section 3.3). Step 7, bioinformatic differential analysis (section 3.2) comparing peptidoglycan compositional changes between different experimental parameters. Step 8, examine the global change in muropeptide modifications within the different experimental parameters using 1D annotation (section 3.4). Please click here to view a larger version of this figure.
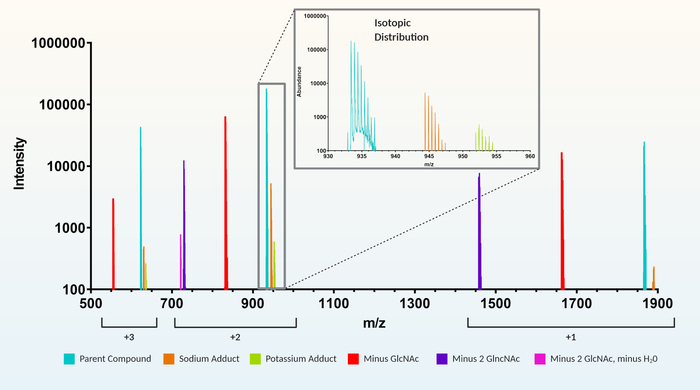
Figure 3: Example of a recursive feature extraction. For a muropeptide representing a peptide sidechain of alanine (A), iso-ᴅ-glutamate (E), meso-diaminopimelic acid (m), alanine (A) crosslinked to the AEmA of the adjacent muropeptide sidechain (1864.8 m/z). Included in the extracted feature for 1864.8 m/z are charges (+1, +2, and +3), adducts (e.g., sodium and potassium), loss of GlcNAc (1 or 2 GlcNAc), and multiple isotopic peaks for each variation (e.g., zoomed inset). Please click here to view a larger version of this figure.
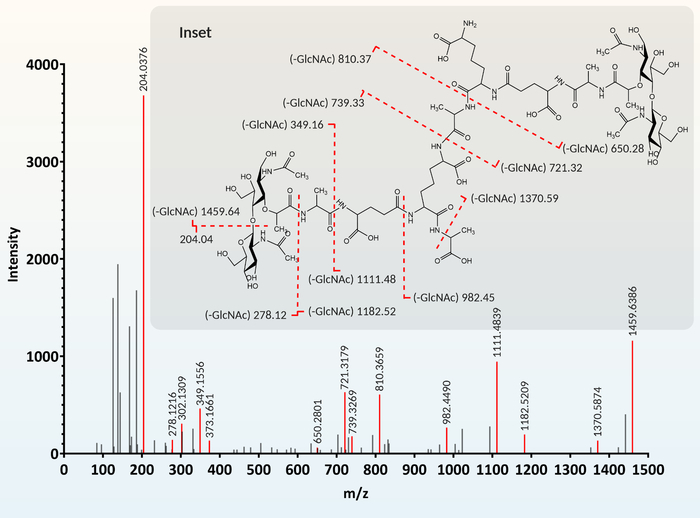
Figure 4: Muropeptide fragmentation and identification. For annotation, each m/z peak (feature) extracted from the MS chromatogram is given a proposed muropeptide structure based on similarity to a muropeptide library. To confirm this proposed structure, predicted MS/MS fragments are generated using a chemical drawing program (gray inset). This predicted fragmentation is compared to the MS/MS chromatogram. When predicted fragments (gray inset) match the MS/MS chromatogram, the proposed muropeptide structure is confirmed. The figure was modified from Reference12. Please click here to view a larger version of this figure.
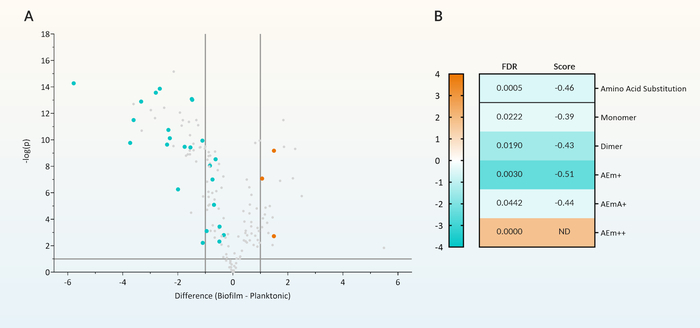
Figure 5: Differential analysis of peptidoglycan composition. (A) Volcano plot of the fold change and statistical significance of changes in muropeptide intensity between peptidoglycan purified from P. aeruginosa grown as either free-swimming planktonic or stationary biofilm culture. All muropeptides that have a modification that represented a change in the typical amino acid arrangement within the peptide sidechain are highlighted. Amino acid substituted muropeptides that showed a trend towards decreased abundance in biofilm-derived peptidoglycan are highlighted in green. Amino acid substituted muropeptides that were outliers to this trend and showed increased abundance in biofilm-derived peptidoglycan are highlighted in orange. (B) Heat map of the global fold change in abundance of all the amino acid substituted muropeptides with increased abundance (orange) and decreased abundance (green) in biofilms. These muropeptides were regrouped and assessed for whether amino acid substitution occurred on monomers, crosslinked dimers, or whether the fourth (AEm+), fifth (AEmA+) or both amino acids (AEm++) were substituted. The significance of each group of muropeptides were assessed by 1D annotation with FDR < 0.05 for significance and the associated 1D score is displayed. 1D annotation can only be performed on more than 2 muropeptides (e.g., AEm++ substitution was only found on two muropeptides). Therefore, in this case, significance must be examined for the individual muropeptides and not on the group. The figure was modified from Reference12. Please click here to view a larger version of this figure.
Discussion
This protocol describes a method to purify peptidoglycan from bacterial cultures, process for LC-MS detection and analyze composition using bioinformatic techniques. Here, we focus on Gram-negative bacteria and some slight modification will be required to enable analysis of Gram-positive bacteria.
The preparation of muropeptides has remained virtually the same since it was first produced in the 1960s9,11,15. Once purified, sacculi (section 1.2.18) are digested into individual muropeptides using the muramidase enzyme mutanolysin from Streptomyces globisporus. Mutanolysin digests the PG structure by breaking the β-1,4-glycosidic linkage releasing individual muropeptides consisting of a GlcNAc-MurNAc disaccharide with appended peptide sidechain and includes any modifications or crosslinkages (Figure 1).
A limitation of previous methodology used to study PG composition has been the time-consuming manual identification of muropeptides. Due to the complexity and difficulty, adducts, charges, and/or isotopes may or may not have been included in the analysis. In addition, most studies restricted analysis to the most abundant, hence easiest to purify, muropeptides. Therefore, because of the complicated nature of the methodology, relatively few high-level detailed PG compositional analyses have been performed. The “omic”-type analyses have used recent technological improvements for the production and statistical analysis of relatively large and complex LC-MS datasets for the high-level overview of biological systems. The application of peptidoglycomics will enable the analysis of PG composition in very fine detail.
Within peptidoglycomics, recursive feature extraction reduces manual workload and increases accuracy by examining all data files at once. A recursive feature extraction algorithm is used to identify, align, and group unique spectral features (m/z peaks) across multiple LC-MS chromatographic data files making identification of muropeptide m/z peaks automated. This algorithm uses isotopic pattern matching which takes the numerous potential isotopes, ion adducts, and charge states and condenses the multiple m/z peaks into its representative single compound (or feature), which in this case would represent a single muropeptide (Figure 3). Verification of the spectral feature group is accomplished by comparing retention time, m/z, and isotopic pattern matching within each chromatographic data file to ensure robust extraction of the feature in the entire dataset. Generic feature finding algorithms may not include isotope matching or align, group, or verify m/z peaks across multiple samples and will require additional manual data processing to accomplish this feature extraction.
Once features are identified, bioinformatic differential analysis algorithms handle the very large dataset as a whole, thus allowing for useful comparisons and interpretations from the complex data. Using these bioinformatic graphical analyses is a powerful way to visualize and interpret large datasets to examine trends which may indicate biologic processes. It was only recently that these high-powered graphical analyses were used to examine peptidoglycan in very fine detail12. Differential analysis (section 3.2) assesses the changes in abundance of individual muropeptides between different experimental conditions. However, within the context of whole bacterial cells, the activity of PG modifying enzymes could result in multiple distinct muropeptide structures depending on the specificity of the catalytic activity (i.e., the addition of an acetyl group on the disaccharide could be with or without a modification of the peptide sidechain). Therefore, assessing the global abundance changes of a particular modification across all individual annotated muropeptides will give insight on the enzymatic activity acting on the PG (Figure 5) Therefore, differential analysis is used to investigate the abundance changes of individual muropeptides; whereas, 1D annotation examines abundance changes of a particular PG modification. Coupling differential analysis with 1D annotation allows PG composition to be assessed both on an individual muropeptide level and also as an indicator of overall PG enzymatic activity.
During differential analysis, it is important to note that PG is composed of a few highly abundant muropeptides and numerous low abundance muropeptides12. Therefore, baselining is very important in order to remove any bias from the high abundance muropeptides during the later steps of the analysis. Also, due to the multiple t-tests performed, a statistical correction to decrease false positives must be applied. The default is often the Benjamini-Hochberg false-discovery rate (FDR)19. Other corrections such as the more conservative Bonferroni familywise error rate (FWER)20,21 are possible.
Within the bioinformatic software, the m/z peak identified in feature extraction is also assigned a predicted structure. Other “omic”-type (e.g., proteomic) analyses benefit from the availability of large compound databases, which allow for compound identification through predictive fragmentation spectra matching. Currently, no muropeptide predicted fragmentation library exists and the confirmation of muropeptide identification remains a manual step. However, as peptidoglycomic fragmentation databases develop and become publicly available, this manual identification step will become more automated and accessible by eliminating or highly reducing sections 3.3.3 and 3.3.4.
In Escherichia coli, PG consists of ~3.5 x 106 muropeptides per cell34. Within the detection limits of the QTOF MS, even the lowest abundant muropeptides can still represent hundreds of copies of a single muropeptide within a cell12. Therefore, understanding the changes to even the lowest abundant muropeptides may provide useful insights into the biological activity of PG-targeted enzymes within the cell.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Dr. Jennifer Geddes-McAlister and Dr. Anthony Clarke for their contributions in refining this protocol. This work was supported by operating grants from CIHR awarded to C.M.K (PJT 156111) and a NSERC Alexander Graham Bell CGS D awarded to E.M.A. Figures were created on BioRender.com.
Materials
| Equipment | |||
| C18 reverse phase column – AdvanceBio Peptide column (100 mm x 2.1 mm 2.7 µm) | Agilent | LC-MS data acquisition | |
| Heating mantle controller, Optichem | Fisher | 50-401-788 | for 4% SDS boil |
| Heating Mantle, 1000mL Hemispherical | Fisher | CG1000008 | for 4% SDS boil |
| Incubator, 37°C | for sacculi purification and MS sample prep | ||
| Leibig condenser, 300MM 24/40, | Fisher | CG121805 | for 4% SDS boil |
| Lyophilizer | Labconco | for lyophilization of sacculi | |
| Magentic stirrer | Fisher | 90-691-18 | for 4% SDS boil |
| mass spectrometer Q-Tof model UHD 6530 | Aglient | LC-MS data acquisition | |
| microcentrifuge filters, Nanosep MF 0.2 µm | Fisher | 50-197-9573 | cleanup of sample before MS injection |
| Retort stand | Fisher | 12-000-102 | for 4% SDS boil |
| Retort clamp | Fisher | S02629 | for 4% SDS boil |
| round bottom flask – 1 liter pyrex | Fisher | 07-250-084 | for 4% SDS boil |
| Sonicator model 120 | Fisher | FB120 | for sacculi purification |
| Sonicator – micro tip | Fisher | FB4422 | for sacculi purification |
| Ultracentrifuge | Beckman | sacculi wash steps | |
| Ultracentrifuge bottles, Ti45 | Fisher | NC9691797 | sacculi wash steps |
| Water supply | City | for water cooled condenser | |
| Software | |||
| Chemdraw | Cambridgesoft | molecular editor for muropeptide fragmentation prediction | |
| Excel | Microsoft | viewing lists of annotated muropeptides, abundance, isotopic patterns, etc. | |
| MassHunter Acquisition | Aglient | running QTOF instrument | |
| MassHunter Mass Profiler Professional | Aglient | bioinformatic differential analysis | |
| MassHunter Personal Compound Database and Library Manager | Aglient | muropeptide m/z MS database | |
| MassHunter Profinder | Aglient | recursive feature extraction | |
| MassHunter Qualitative analysis | Aglient | viewing MS and MS/MS chromatograms | |
| Prism | Graphpad | Graphing software | |
| Perseus | Max Plank Institute of Biochemistry | 1D annotation | |
| Material | |||
| Acetonitrile | Fisher | A998-4 | |
| Ammonium acetate | Fisher | A637 | |
| Amylase | Sigma-Aldrich | A6380 | |
| Boric acid | Fisher | BP168-1 | |
| DNase | Fisher | EN0521 | |
| Formic acid | Sigma-Aldrich | 27001-500ML-R | |
| LC-MS tuning mix – HP0321 | Agilent | G1969-85000 | |
| Magnesium chloride | Sigma-Aldrich | M8266 | |
| Magnesium sulfate | Sigma-Aldrich | M7506 | |
| Mutanolysin from Streptomyces globisporus ATCC 21553 | Sigma-Aldrich | M9901 | |
| Nitrogen gas (>99% purity) | Praxair | NI 5.0UH-T | |
| Phosphoric acid | Fisher | A242 | |
| Pronase E from Streptomyces griseus | Sigma-Aldrich | P5147 | |
| RNase | Fisher | EN0531 | |
| Sodium azide | Fisher | S0489 | |
| Sodium borohydride | Sigma-Aldrich | 452890 | |
| Sodium dodecyl sulfate (SDS) | Fisher | BP166 | |
| Sodium hydroxide | Fisher | S318 | |
| Sodium Phosphate (dibasic) | Fisher | S373 | |
| Sodium Phosphate (monobasic) | Fisher | S369 | |
| Stains-all | Sigma-Aldrich | E9379 |
Referenzen
- Vollmer, W., Blanot, D., de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiology Reviews. 32, 149-167 (2007).
- Pazos, M., Peters, K. Peptidoglycan. Sub-cellular Biochemistry. 92, 127-168 (2019).
- Typas, A., Banzhaf, M., Gross, C. A., Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature Reviews. Microbiology. 10 (2), 123-136 (2011).
- Yadav, A. K., Espaillat, A., Cava, F. Bacterial strategies to preserve cell wall integrity against environmental threats. Frontiers in Microbiology. 9, 2064 (2018).
- Bera, A., Herbert, S., Jakob, A., Vollmer, W., Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Molecular Microbiology. 55 (3), 778-787 (2005).
- Brott, A. S., Clarke, A. J. Peptidoglycan O-acetylation as a virulence factor: its effect on lysozyme in the innate immune system. Antibiotics. 8 (3), 94 (2019).
- Putty, S., Vemula, H., Bobba, S., Gutheil, W. G. A liquid chromatography-tandem mass spectrometry assay for D-Ala-D-Lac: A key intermediate for vancomycin resistance in vancomycin-resistant Enterococci. Analytical Biochemistry. 442 (2), 166-171 (2013).
- Arthur, M., et al. Evidence for in vivo incorporation of D-lactate into peptidoglycan precursors of vancomycin-resistant Enterococci. Antimicrobial Agents and Chemotherapy. 36 (4), 867-869 (1992).
- Mardarowicz, C. Isolierung und Charakterisierung des Murein-Sacculus von Brucella. Z. Naturforsdig. 21, 1006-1007 (1966).
- Glauner, B., Höltje, J. V., Schwarz, U. The composition of the murein of Escherichia coli. The Journal of Biological Chemistry. 263 (21), 10088-10095 (1988).
- Glauner, B. Separation and quantification of muropeptides with high-performance liquid chromatography. Analytical Biochemistry. 172, 451-464 (1988).
- Anderson, E. M., et al. Peptidoglycomics reveals compositional changes in peptidoglycan between biofilm- and planktonic-derived Pseudomonas aeruginosa. The Journal of Biological Chemistry. 295 (2), 504-516 (2020).
- van der Aart, L. T., et al. High-resolution analysis of the peptidoglycan composition in Streptomyces coelicolor. Journal of Bacteriology. 200 (20), 00290 (2018).
- Bern, M., Beniston, R., Mesnage, S. Towards an automated analysis of bacterial peptidoglycan structure. Analytical and Bioanalytical Chemistry. 409, 551-560 (2017).
- Brott, A. S., Sychantha, D., Clarke, A. J. Assays for the enzymes catalyzing the O-acetylation of bacterial cell wall polysaccharides. Methods in Molecular Biology. 1954, 115-136 (2019).
- Clarke, A. J. Compositional analysis of peptidoglycan by high-performance anion-exchange chromatography. Analytical Biochemistry. 212 (2), 344-350 (1993).
- Rusconi, F., Douard Valton, &. #. 2. 0. 1. ;., Nguyen, R., Dufourc, E. Quantification of sodium dodecyl sulfate in microliter-volume biochemical samples by visible light spectroscopy. Analytical Biochemistry. 295, 31-37 (2001).
- Verwer, R. W. H., Beachey, E. H., Keck, W., Stoub, A. M., Poldermans, J. E. Oriented fragmentation of Escherichia coli sacculi by sonication. Journal of Bacteriology. 141 (1), 327-332 (1980).
- Benjamini, Y., Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological). 57 (1), 289-300 (1995).
- Article, R., Alonso, A., Marsal, S., Julià, A., James Carroll, A. Analytical methods in untargeted metabolomics: state of the art in 2015. Frontiers in Bioengineering and Biotechnology. 3, 23 (2015).
- Xi, B., Gu, H., Baniasadi, H., Raftery, D. Statistical analysis and modeling of mass spectrometry-based metabolomics data. Methods in Molecular Biology. 1198, 333-353 (2014).
- Tyanova, S., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. 13 (9), 731-740 (2016).
- Cox, J., Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 13, 12 (2012).
- Chang, J. D., Foster, E. E., Thadani, A. N., Ramirez, A. J., Kim, S. J. Inhibition of Staphylococcus aureus cell wall biosynthesis by desleucyl-oritavancin: A quantitative peptidoglycan composition analysis by mass spectrometry. Journal of Bacteriology. 199 (15), 00278 (2017).
- Glauner, B., Höltje, J. V. Growth pattern of the murein sacculus of Escherichia coli. The Journal of Biological Chemistry. 265 (31), 18988-18996 (1990).
- Espaillat, A., et al. Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. Journal of the American Chemical Society. 138 (29), 9193-9204 (2016).
- Quintela, J. C., Caparros, M., de Pedro, M. A. Variability of peptidoglycan structural parameters in Gram-negative bacteria. FEMS Microbiology Letters. 125, 95-100 (1995).
- Quintela, J. C., Zollner, P., Portillo, F. G., Allmaier, G., de Pedro, M. A. Cell wall structural divergence among Thermus spp. FEMS Microbiology Letters. 172, 223-229 (1999).
- Torrens, G., et al. Comparative analysis of peptidoglycans from Pseudomonas aeruginosa isolates recovered from chronic and acute infections. Frontiers in Microbiology. 10, 0868 (2019).
- Antignac, A., et al. Detailed structural analysis of the peptidoglycan of the human pathogen Neisseria meningitidis. The Journal of Biological Chemistry. 278 (34), 31521-31528 (2003).
- De Jonges, B. L. M., Changi, Y. S., Gage, D., Tomaszs, A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. The Journal of Biological Chemistry. 267 (16), 11248-11264 (1992).
- Lee, M., et al. Muropeptides in Pseudomonas aeruginosa and their role as elicitors of β-lactam-antibiotic resistance. Angewandte Chemie – International Edition. 55, 6882-6886 (2016).
- Schumann, P. Peptidoglycan Structure. Methods in Microbiology. 38, 101-129 (2011).
- Wientjes, F. B., Woldringh, C. L., Nanninga, N. Amount of peptidoglycan in cell walls of Gram-negative bacteria. Journal of Bacteriology. 173 (23), 7684-7691 (1991).

