The Zebrafish Tol2 System: A Modular and Flexible Gateway-Based Transgenesis Approach
Summary
This work describes a protocol for the modular Tol2 transgenesis system, a gateway-based cloning method to create and inject transgenic constructs into zebrafish embryos.
Abstract
Fetal alcohol spectrum disorders (FASD) are characterized by a highly variable set of structural defects and cognitive impairments that arise due to prenatal ethanol exposure. Due to the complex pathology of FASD, animal models have proven critical to our current understanding of ethanol-induced developmental defects. Zebrafish have proven to be a powerful model to examine ethanol-induced developmental defects due to the high degree of conservation of both genetics and development between zebrafish and humans. As a model system, zebrafish possess many attributes that make them ideal for developmental studies, including large numbers of externally fertilized embryos that are genetically tractable and translucent. This allows researchers to precisely control the timing and dosage of ethanol exposure in multiple genetic contexts. One important genetic tool available in zebrafish is transgenesis. However, generating transgenic constructs and establishing transgenic lines can be complex and difficult. To address this issue, zebrafish researchers have established the transposon-based Tol2 transgenesis system. This modular system uses a multisite Gateway cloning approach for the quick assembly of complete Tol2 transposon-based transgenic constructs. Here, we describe the flexible Tol2 system toolbox and a protocol for generating transgenic constructs ready for zebrafish transgenesis and their use in ethanol studies.
Introduction
Prenatal ethanol exposure gives rise to a continuum of structural deficits and cognitive impairments termed fetal alcohol spectrum disorders (FASD)1,2,3,4. The complex relationships between multiple factors make studying and understanding the etiology of FASD in humans challenging. To resolve this challenge, a wide variety of animal models have been used. The biological and experimental tools available in these models have proven crucial in developing our understanding of the mechanistic basis of ethanol teratogenicity, and the results from these model systems have been remarkably consistent with what is found in human ethanol studies5,6. Among these, zebrafish have emerged as a powerful model to study ethanol teratogenesis7,8, in part due to their external fertilization, high fecundity, genetic tractability, and translucent embryos. These strengths combine to make zebrafish ideal for real-time live imaging studies of FASD using transgenic zebrafish lines.
Transgenic zebrafish have been extensively used to study multiple aspects of embryonic development9. However, creating transgenic constructs and subsequent transgenic lines can be exceedingly difficult. A standard transgene requires an active promoter element to drive the transgene and a poly A signal or "tail", all in a stable bacterial vector for general vector maintenance. The traditional generation of a multi-component transgenic construct requires multiple time-consuming sub-cloning steps10. PCR-based approaches, such as Gibson assembly, can circumvent some of the issues associated with sub-cloning. However, unique primers must be designed and tested for the generation of every unique transgenic construct10. Beyond transgene construction, genomic integration, germline transmission, and screening for proper transgene integration have been difficult as well. Here, we describe a protocol for using the transposon-based Tol2 transgenesis system (Tol2Kit)10,11. This modular system uses multisite Gateway cloning to quickly generate multiple transgenic constructs from an ever-expanding library of "entry" and "destination" vectors. Integrated Tol2 transposable elements greatly increase the rate of transgenesis, allowing for the rapid construction and genomic integration of multiple transgenes. Using this system, we show how the generation of an endoderm transgenic zebrafish line can be used to study the tissue-specific structural defects underlying FASD. Ultimately, in this protocol, we show that the modular setup and the construction of transgenic constructs will greatly aid zebrafish-based FASD research.
Protocol
All zebrafish embryos used in this procedure were raised and bred following established IACUC protocols12. These protocols were approved by the University of Louisville.
NOTE: The wild-type zebrafish strain, AB, and the bmp4st72;smad5b1100 double mutant line were used in this study. All the water used in this procedure was sterile reverse osmosis water. Confocal images were taken under a laser-scanning confocal microscope. The endoderm measurements were made using the measure tool in ImageJ. All the statistical analyses were performed using statistical software.
1. Making the solutions and media
- Bacterial media: Dissolve LB broth or agar as per the manufacturer's instructions, and autoclave. Before pouring the media into 100 mm Petri dishes, add either ampicillin (50 µg/mL), kanamycin (50 µg/mL), or an ampicillin/chloramphenicol combination (50 µg/mL and 30 µg/mL, respectively).
- To make 20x embryo media stock, dissolve the following in 1 L of water: 17.5 g of NaCl, 0.75 g of KCl, 2.9 g of CaCl2, 0.41 g of K2HPO4, 0.142 g of Na2HPO4, and 4.9 g of MgSO4·7H2O. Filter-sterilize the stock solution using a 0.22 µm vacuum filtration system, and store at 4 °C.
NOTE: Ignore the white precipitate that forms; this will not impact the media. - In 19 L of water, dissolve 1.2 g of NaHCO3 and add 1 L of the 20x embryo media stock to generate the working embryo media solution, and maintain this solution at 28 °C.
- For 2% phenol red solution, dissolve 20 mg of phenol red sodium salt in 1 mL of RNase-free water, and store at room temperature.
2. Embryo injection molds
- To make the agarose injection plate, dissolve agarose in the embryo media to a final concentration of 3% by heating to a boil.
- Pour 50 mL of agarose into 100 mm Petri dishes, and while the agarose is still liquid, gently place the injection mold in the agarose to prevent bubble formation under the mold.
- Let the agarose plates cool. Once the agarose is solid, remove the injection mold, and store at 4 °C until ready to use.
3. Injection pipettes
- Pull 1.0 mm (OD) capillaries containing a filament into needles using a vertical pipette puller with the solenoid set to 2.5 and the heating element set to 14.6.
NOTE: Any heated puller will work if it pulls the capillaries into two needles cleanly and evenly, but the injection needles need to be pulled within 1 week of injecting the zebrafish embryos for consistent injection results.- Place the capillary in the puller, tighten the clamp at each end, and then activate the heating element.
- Pull the capillaries to create two injection needles.
- Remove the capillaries from the puller, and store them in an empty Petri dish with a strip of modeling clay that acts as the needle holder.
4. Transposase mRNA preparation
- Linearize the Tol2Kit plasmid containing transposase with a NotI restriction endonuclease by combining the following in a 0.5 mL microcentrifuge tube: 10 µL of transposase plasmid (150 ng/µL), 1.5 µL of restriction endonuclease reaction buffer, 0.3 µL of NotI endonuclease.
- Fill the reaction up to 20 µL with RNase-free water, and then mix and digest at 37 °C overnight.
- Stop the transposase digestion by adding 1 µL of 0.5 M EDTA (pH 8.0), 2 µL of 5 M NH4OAc, and 80 µL of 100% EtOH to the digestion reaction, and then mix and chill at −20 °C overnight.
- Centrifuge the solution at 14,000x g for 20 min at 4 °C, and then aspirate the supernatant and resuspend the DNA pellet in RNase-free water.
- Determine the linear plasmid concentration in nanograms per microliter (ng/µL) using a fluorometer by first running 2 µL of an RNase-free water blank and then running 2 µL of the linearized plasmid.
NOTE: The plasmid concentrations typically range between 100-200 ng/µL - Set up the SP6 mRNA production by combining the following components in order in a 0.5 mL microcentrifuge tube: 10 µL of 2x NTP/CAP, 2 µL of 10x reaction buffer, 0.1-1 µg of linear DNA template, and 2 µL of SP6 enzyme mix.
- Fill the reaction up to 20 µL with RNase-free water, and then mix and incubate at 37 °C for 2 h.
- After incubating for 2 h, add 1 µL of DNase, mix well, and incubate for an additional 15 min at 37 °C.
- To stop the SP6 reaction, add 30 µL of LiCl precipitation solution to the reaction tube, and then mix and chill at −20 °C overnight.
- Centrifuge the solution at 14,000 x g for 20 min at 4 °C to pellet the mRNA, and then aspirate the supernatant and wash the mRNA pellet with 1 mL of 80% EtOH (diluted with RNase-free water).
- Centrifuge the solution at 14,000 x g for 20 min at 4 °C to pellet the mRNA, and then aspirate the supernatant. Allow the pellet to air dry.
- Resuspend the mRNA pellet in 20 µL of RNase-free water, determine the mRNA concentration in nanograms per microliter (ng/µL) using a fluorometer as described in step 4.5 (should be at least 100 ng/µL), aliquot 100 ng of mRNA per tube for single use, and store at −80 °C.
NOTE: To make sure the mRNA is not degraded, 2 µL of mRNA can be quickly run on a DNA electrophoresis gel to confirm a single band at 1,950 bps.
5. Multisite Gateway cloning to create entry vectors for transgenesis
NOTE: This protocol is modified from Kwan et al.10, with the LR reaction written as a half-LR reaction and with a total volume of 5 µL. To generate new entry elements, BP reactions using 5', middle, and 3' donor vectors need to be used10,13.
- To perform the reaction, gather 10 fmol each of p5E, pME, and p3E and 20 fmol of the destination vector (pDest).
NOTE: Multisite LR recombination uses four different plasmid vectors: a 5' entry vector (p5E), a middle entry vector (pME), a 3' entry vector (p3E), and a destination vector (pDest), to generate a unique transgenic construct flanked by Tol2 repeats ready to be injected into zebrafish embryos.- Identify the size of all four vectors in base pairs from the plasmid source (Table 1, Tol2Kit Wiki page14 or Addgene11; see the Table of Materials).
NOTE: For novel vectors, full plasmid sequencing via commercial sequencing companies can provide the exact plasmid size in bps. - Determine the concentration of all four vectors in nanograms per microliter (ng/µL) using a fluorometer, as described in step 4.5.
- Using the weight of DNA as 660 g/mol and the plasmid size (in bps), calculate the total nanograms (ng) of plasmid needed to reach either 10 fmol (entry vectors) or 20 fmol (destination vector).
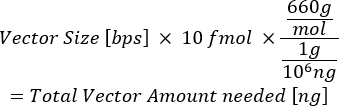
NOTE: The Mosimann lab at the University of Colorado, Anschutz Medical Campus, has generated a freely available spreadsheet to calculate the amount of plasmid needed (in ng) and the volume (in µL) of the vectors needed in the LR reaction15.
- Identify the size of all four vectors in base pairs from the plasmid source (Table 1, Tol2Kit Wiki page14 or Addgene11; see the Table of Materials).
- To generate the construct described below, sox17:EGFPCAAX, use the following vectors: a p5E vector containing the zebrafish sox17 promoter, which is 6,918 bp in size16; a pME vector containing the prenylated EGFP, which is 3,345 bp in size10; a p3E vector containing the SV40 late poly A signal sequence, which is 2,838 bp in size10; and the pDest, which is 5,883 bp in size10.
- Using the plasmid concentration determined in step 5.1.2 and the amount in nanograms (ng) for each vector (step 5.1.3), calculate the total microliters (µL) of each plasmid needed to reach either 10 fmol (entry vectors) or 20 fmol (destination vector), and then divide this by two for use in the 5 µL half-LR reactions (all the values listed below are listed in Table 2).
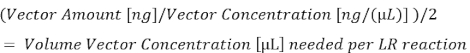
- To generate 10 fmol of p5E-sox17, use 45.66 ng (at a concentration of 140.3 ng/µL) and dilute with sterile water by a factor of 4 to get 0.65 µL.
- To generate 10 fmol of pME-EGFPCAAX, use 22.08 ng (at a concentration of 213.5 ng/µL) and dilute with sterile water by a factor of 10 to get 0.52 µL.
- To generate 10 fmol of p3E-poly A, use 15.73 ng (at a concentration of 276.1 ng/µL) and dilute with sterile water by a factor of 20 to get 0.57 µL.
- For 20 fmol of the destination vector, use 77.66 ng of pDest (at a concentration of 307.3 ng/µL) and dilute with sterile water by a factor of 5 to get 1.63 µL.
- To set up a 5 µL half-LR reaction, combine the volumes of the p5E, pME, p3E, and pDest determined in step 5.3 in a 0.5 mL microcentrifuge tube, and then add sterile water to reach a total reaction volume of 4 µL.
- Vigorously vortex the LR enzyme mix 2x for 1 min each to ensure proper mixing, and then add 1 µL to the reaction and mix thoroughly.
- Incubate at 25 °C overnight (16+ h).
- Stop the LR reaction by adding 0.5 µL of proteinase K (2 µg/µL), incubate at 37 °C for 10 min, and then cool to room temperature.
- Transform 3-4 µL of plasmid into thawed, chemically competent cells by adding the plasmid and letting the cells sit on ice for 25-30 min.
- While the cells sit on ice, warm two LB-ampicillin agar plates to room temperature.
- Heat-shock the cells in a 42°C water bath for precisely 30 s.
- After the heat shock, add 250 µL of antibiotic-free, rich liquid media to the cells, and incubate with shaking at 37 °C for 1.5 h.
- Spread 300 µL of bacteria on one ampicillin plate, and incubate at 37°C overnight.
- The next morning, screen the colonies for the presence of two phenotypes, clear and opaque, pick the clear colonies (as described in Kwan et al.10) one at a time, inoculate the liquid culture, and then shake at 37 °C overnight.
NOTE: Clear colonies almost always contain the correct LR colony (>85% of the time). - Using a commercial kit as per the manufacturer's instructions, perform a plasmid prep on each liquid culture, and diagnostically digest each plasmid to confirm the appropriate size of the transgenic construct, which indicates that the LR gateway reaction has worked properly.
- Re-transform the correct transgenic constructs using the standard bacterial transformation protocol as described in steps 5.4-5.12 using 0.5-1 µL of the plasmid, and only incubate at 37 °C for 1 h.
- Re-streak the colonies, and confirm that each has the LR transgenic construct as in step 5.14.
- Determine the plasmid concentration as described in step 4.5 (at least 150 ng/µL is needed for the embryo injection).
6. Injection of the transgene into the embryos
- Thaw a single-use transposase mRNA sample and Tol2 transgenic construct on ice and prewarn an agarose injection plate to room temperature.
- Combine the following on ice: 150 ng of plasmid, 100 ng of transposase mRNA, and 2% phenol red (0.3 µL). Then, add RNA-free water to make up the volume to 3 µL.
- Transfer one cell-stage embryos using transfer pipettes to the injection plate (described in step 2) filled with EM completely covering the agar, and then gently press approximately 50-75 embryos per slot of the injection plate.
- Add the mRNA/plasmid/phenol red mixture to the open end of a capillary injection needle, place the capillary needle in the injection rig armature, and turn on the injection rig.
NOTE:The injection rig uses compressed gas, such as air, to pulse the desired volume (described in step 6.5) of the mRNA/plasmid/phenol red mixture into the embryo when activated. - Lower the capillary needle into the EM of the injection plate, and break the tip using forceps to allow only a small amount of mRNA/plasmid/phenol red mixture to be pumped out by the injection rig.
- Inject the embryos with a small 3 nL bolus of the mRNA/plasmid/phenol red mixture into the cell body of the embryo.
NOTE: A 3 nL bolus is a volume of mRNA/plasmid/phenol red mixture at which seven boluses can theoretically fit equally across the midline of the embryo. - When finished injecting the embryos, remove them from the injection plate by gently popping them out of the slot and transfer-pipetting them to a 100 mm Petri dish with fresh EM (approximately 40 mL), and then incubate at 28.5 °C to allow the embryos to develop.
7. Screening embryos for transgenic insertion
- For the expression of fluorescent proteins, pick the appropriate developmental stage when the fluorescent transgene should be expressed, and screen for the presence of fluorescence under a fluorescent dissecting microscope.
NOTE: These embryos will be mosaic in their expression and show a genomic insertion of the transgenic construct. - Screen for non-fluorescent transgenes by screening for a fluorescent transgenic marker (as described in step 7.1), such as GFP driven by the cardiac-specific promoter for the gene cmlc2 or mCherry driven by the lens-specific promoter of αcyrstallin, which are contained in distinct destination vectors.
- Allow the embryos positive for transgenic insertion to fully develop to the adult zebrafish stages.
- Once potential transgenic fish reach breeding age, screen them individually for both germline transmission and transgene expressivity by breeding them to wild-type zebrafish.
- Those adults whose embryos develop normally and give the strongest and most appropriate transgene expression are kept as unique transgenic zebrafish lines for future work and analyses.
NOTE: As an alternative approach to screening for the genomic insertion of transgenic constructs, design PCR primers that only amplify the transgenic construct and not wild-type DNA.
Representative Results
To generate the transgenic constructs, we used the Tol2 transgenesis system. Three entry vectors, including p5E, which holds the gene promoter/enhancer elements, pME, which holds the gene to be expressed by the promoter/enhancer elements, and p3E which, at minimum, holds the polyA tail, were used to generate the transgenic construct via multisite gateway LR cloning. The destination vector, pDest, provides the Tol2 repeats for the genomic insertion of the transgenic construct in zebrafish embryos and contains all the essential genetic information for bacterial growth (Figure 1A). For the purposes of this manuscript, we created and injected a sox17:EGFPCAAX transgenic construct. The promoter of the endoderm marker, sox17, was used to drive membrane-tagged EGFP in the developing endoderm of the zebrafish. For transgenic constructs that express non-fluorophore proteins, a pDest vector that contains a fluorescent transgenic marker such as cmlc2:EGFP can be used to aid in genomic insertion (Figure 1A).
Using the values described in the protocol above (Table 2), the sox17:EGFPCAAX transgenic construct was generated, and 4 µL of this construct was transformed into chemically competent Escherichia coli cells. After incubating at 37 °C overnight, the agar plate contained approximately 250 colonies (LR reactions typically average 150-300 colonies per plate in our lab). The screening of these colonies was performed based on opacity. In our hands, colonies that were clear had the correct sox17:EGFPCAAX product >85% of the time, whereas the opaque colonies never contained the correct recombination product (Figure 1B, arrowhead vs. arrow). After isolating the plasmid from several colonies, the restriction digestion of the recombinant products was performed with the restriction enzyme, NcoI. Two different clear colonies and one opaque colony were digested. Both clear colonies had a single band at the correct size of 9,544 bp (undigested plasmids loaded as a digest control), while the opaque colony did not have any bands at all (Figure 2A). These positive sox17:EGFPCAAX constructs were retransformed, the subsequent colonies were re-streaked, those colonies were confirmed to have the plasmid, and −80 °C bacterial freezer stocks were generated. The concentrations of both the sox17:EGFPCAAX plasmid and the capped-transposase mRNA were determined so they could both be used for injection (Figure 2B).
The embryos were prepared for injection by placing one cell-stage embryos into a pre-warmed embryo injection mold (Figure 3A–C). Once placed in the injection mold, the injection needle containing the sox17:EGFPCAAX mRNA was placed in a micropipette holder on the micromanipulator (Figure 3D,E). The embryos were injected with 100 ng of transposase mRNA, 150 ng of sox17:EGFPCAAX plasmid, and phenol red (injection tracking dye). A 3 nL bolus (determined by size as described in the protocol, step 6.6) was injected into the cell body and not into the yolk of the embryo for the best chance of integration (Figure 3F)10.
After injection, the embryos were removed from the injection mold and allowed to develop for 24 h. At 24 h post fertilization (hpf), the embryos were screened for the endoderm expression of EGFPCAAX. Mosaic endoderm EGFPCAAX expression was observed in ~75% of the injected embryos (Figure 4A,A'). To screen the embryos injected with non-fluorescent transgenes such as Gal4 or CreERT2, a destination vector that also contains a transgenic marker (i.e., cmlc2:EGFP) (Figure 1A) can be used (Figure 4B,B'). Adult zebrafish that had germline transgenesis of sox17:EGFPCAAX generated fluorescent embryos with the endoderm fully labeled with EGFPCAAX (Figure 4C).
Using the newly created sox17:EGFPCAAX transgenic line, we were able to directly assess the impact of ethanol on the formation of the endodermal pouches. The pouches are protrusions that form on the lateral edge of the endoderm and are important for the formation of the facial skeleton and multiple organ systems17. We have previously shown that blocking bone morphogenetic protein (Bmp) signaling results in hypoplasia of the pouches18. We now show that the ethanol treatment of Bmp mutants from 10-18 hpf has a subtle yet significant impact on pouch size. The dorsal-ventral length of each pouch from pouches 1-5 (zebrafish have six pouches, but the sixth had yet to fully form at the developmental stage imaged) was measured in control and ethanol-treated wild-type Bmp mutant embryos (Figure 5A). As a control for general growth impacts due to ethanol exposure, the anterior-posterior length of the entire endoderm was measured (Figure 5A). The overall length of the endoderm was not impacted by genotype or treatment (Figure 5B). However, pouches 1 and 3 showed significant increases in pouch length between the untreated and ethanol-treated wild-type embryos but significant decreases in length in between the untreated and ethanol-treated Bmp mutants (Figure 5C; two-way ANOVA; pouch 1: F(1,54) = 10.39, p = 0.0021; pouch 3: F(1,54) = 12.70, p = 0.0008). Pouch 2 showed significant increases in pouch size between the untreated and ethanol-treated wild-type and Bmp mutant groups, respectively (Figure 5C; two-way ANOVA; F(1,54) = 18.94, p < 0.0001).
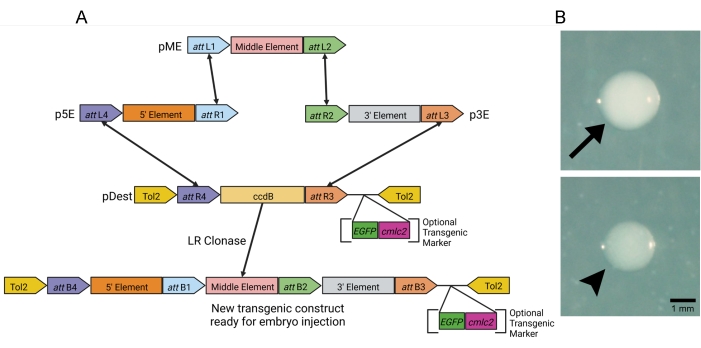
Figure 1: LR gateway reaction for transgene construction. (A) Schematic of the modular four-part LR gateway cloning reaction. LR Clonase recombines the three entry vectors, p5E, pME, and p3E, and the destination vector, pDest, in a highly specific reaction to generate a novel transgenic product ready for embryo injection. For screening non-fluorescent transgenes, pDest vectors can contain optional transgenic markers (cmlc2:EGFP, as an example). (B) Example bacterial colonies obtained from the transformation of the LR recombination products. Clear colonies contained a correct LR recombination product >85% of the time (arrowhead), whereas opaque colonies never contained a correct recombination product (arrow). Please click here to view a larger version of this figure.
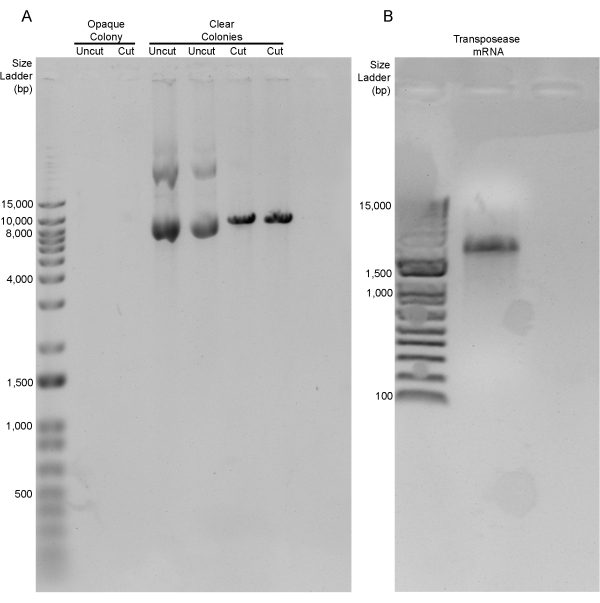
Figure 2: Analysis of the plasmid DNA and transposase mRNA. (A) Diagnostic digestion of the three colonies from the transformation of the LR recombination products. The single opaque colony did not contain any plasmid to screen, while the two clear colonies contained a single band at 9,544 bp (uncut vs. cut). (B) Transposase mRNA at 1,950 bp. Please click here to view a larger version of this figure.
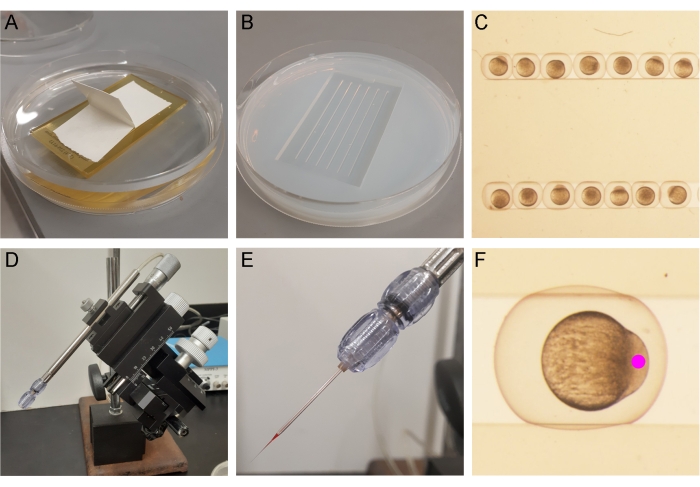
Figure 3: Zebrafish embryo injection setup. (A) An injection mold is placed in liquid agarose diluted in EM (3% v/v). (B) Once solidified, the mold is removed from the agarose plate. (C) The embryos are arranged in the injection molds. (D,E) The mRNA/plasmid/phenol red mixture is pipetted into the injection needle, which is placed in the micropipette holder in the micromanipulator. (F) A 3 nL bolus is injected into the cell body of the embryo at the one-cell stage. Please click here to view a larger version of this figure.
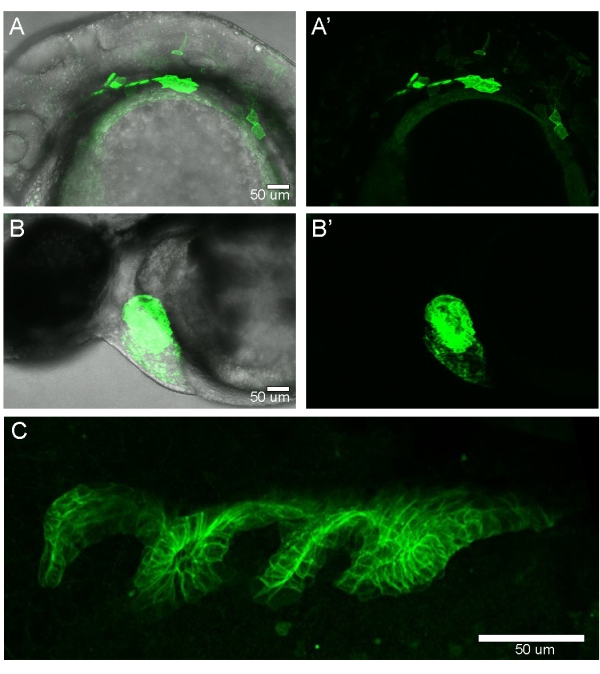
Figure 4: Screening zebrafish injected with the transgenic construct. (A,A') Confocal image of an embryo imaged at 24 hpf shows the mosaic expression of the sox17:EGFPCAAX transgene. (B,B') Transgenic marker expression of cmlc2:EGFP in the developing heart at 24 hpf. Lateral views of the embryos, with anterior to the left and dorsal at the top. (C) Confocal image of an embryo generated from a transgene-carrying adult zebrafish. The entire endoderm is expressing EGFPCAAX. Lateral view of the embryo, with anterior to the left and ventral at the top. Please click here to view a larger version of this figure.
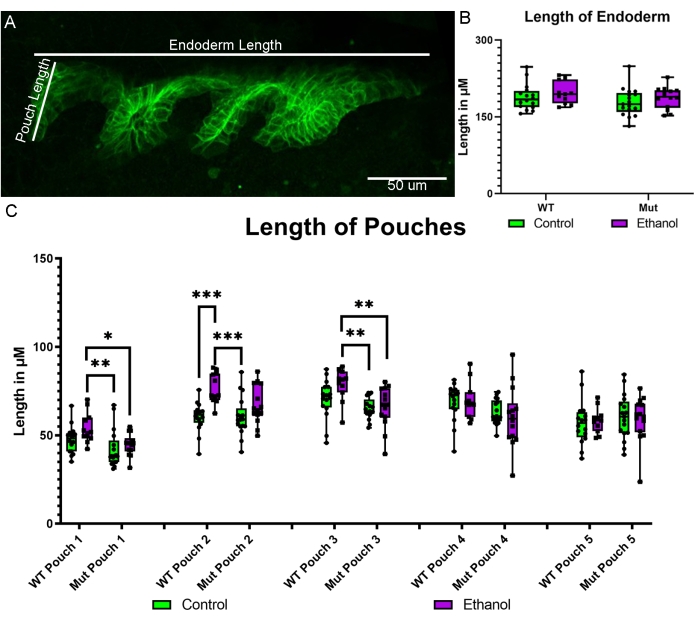
Figure 5: Pouch measures in ethanol-treated wild-type and Bmp mutant embryos. (A) Schematic showing the measurement of the overall length of the endoderm and the length of the pouches. (B) The measurements of the overall anterior-posterior endoderm length show no difference between untreated and ethanol-treated wild-type and Bmp mutant embryos. (C) The measurements of dorsal-ventral pouch length indicate that pouches 1 and 3 show increases in pouch length between the untreated and ethanol-treated wild-type embryos but decreases in length between the untreated and ethanol-treated Bmp mutants (two-way ANOVA; pouch 1: F(1,54) = 10.39, p = 0.0021; pouch 3: F(1,54) = 12.70, p = 0.0008). Pouch 2 shows an increase in length between the untreated and ethanol-treated wild-type and Bmp mutant groups (two-way ANOVA; F(1,54) = 18.94, p < 0.0001). No differences were observed in pouch length between pouches 4 and 5. Please click here to view a larger version of this figure.
Table 1: Tol2Kit components housed in the Lovely Lab. Every entry and destination vector available in the Lovely Lab, their descriptions, and their lab of origin. Please click here to download this Table.
Table 2: Calculation of the amount and concertation of each vector in the LR recombination reaction. The amount of each vector was calculated from the size (in bp) and the fmol needed for each component: 10 fmol of each entry vector and 20 fmol of the destination vector. From the plasmid concentration, each vector is diluted in sterile water to add 1 µL or less to the LR reaction. Sterile water is added to the vector pool to equal 4 µL, 1 µL of LR reaction mix is added to the reaction, and it is incubated at 25°C overnight. Please click here to download this Table.
Discussion
Zebrafish are ideally suited for studying the impact of ethanol exposure on development and disease states7,8. Zebrafish produce large numbers of translucent, externally fertilized, genetically tractable embryos, which allows for the live imaging of several transgene-labeled tissues and cell types simultaneously in multiple environmental contexts19,20. These strengths, combined with the strong developmental genetic conservation with humans, make zebrafish a powerful model system for imaging the impact of ethanol7,8. Here, we described the protocol for a modular approach to generating and injecting transgenic constructs and establishing transgenic zebrafish lines for future ethanol studies.
Many different approaches have been established to generate transgenic zebrafish. However, generating both transgenic constructs and transgenic lines can be daunting. While traditional sub-cloning techniques, BAC cloning, or PCR-based approaches such as Gibson assembly allow for generating transgenic constructs, they have low throughput and lack versatility in their construction10. In addition, these strategies need to be redesigned for every transgenic construct created. Sub-cloning requires multiple digestions and ligations, assuming all the restriction sites necessary are present and unique. BAC cloning requires the sequencing and testing of multiple BAC clones. For Gibson assembly, new PCR primers have to be designed for each transgenic construct. Furthermore, the injection of either uncut or linearized DNA leads to low germline transmission21,22,23.
The Tol2Kit is a modular system that uses gateway cloning to generate complete transgenic constructs flanked by Tol2 repeats that, when combined with transposase mRNA, greatly increase transgenesis and germline transmission10,11,24,25. Dozens of entry and destination vectors are available from the Kwan lab and the Cole Lab10,11. In addition, zebrafish labs that use the Tol2Kit have generated and curated many more entry and destination vectors. As an example of the breadth of vectors available, we have pooled and used over 70 vectors (Table 1). With all these community resources, one can quickly generate thousands of different transgenic constructs by simply combining the vectors of choice in an LR reaction.
Optimal LR reactions do require exact calculations of plasmid size and concentration. The calculations for each vector are done in femtomole (fmol) values, so each reaction does not require a high plasmid volume to generate a transgenic construct. However, this small amount of plasmid makes dilutions and proper pipetting exceedingly critical to the success of the reaction10. Additional factors that can impact the LR reaction are the size of the inserts in the different entry vectors. For example, the p5E-sox17 element used in this study is over 4 kb, which, if combined with equally large pME and p3E elements, will result in a very large transgenic vector. A large plasmid in bacteria can be difficult to culture, thus decreasing the number of colonies generated from the bacterial transformation. This will also result in slower growth and decreased plasmid levels when isolated10. Importantly, having highly competent bacterial cells, as well as increasing the incubation of the bacterial cells during the transformation of the recombination plasmid, are key to generating enough colonies to capture proper transgene formation. In addition, using the correct LR reaction enzyme (listed in the Table of Materials) also plays a major role in the success of the LR reaction.
Beyond the generation of a transgenic construct and bacterial transformations, the embryo injection and genome integration can be difficult as well. The generation of high-quality transposase mRNA greatly increases the frequency of genomic integration10. The pulled capillaries need to be made just shortly before injecting the embryos as older needles can lose capillary action (i.e., the injection fluid fails to move to the tip of the needle) (Figure 3E). Both breaking the tip of the needle to only inject ~3 nL of fluid and injecting the cell body require practice. Our training protocol involves injecting the cell body with only phenol red injection dye until breaking the needle and injecting the embryos are mastered. Injecting the cell body drastically increases the rate of genome integration10. Combining all this, we average 75% or greater genome insertion and mosaic expression, but this can vary from construct to construct.
Since genomic insertion can be random, every embryo that shows mosaic expression is a unique transgenic insertion and requires continued screening. This continued screening is necessary to show that the integration site is not detrimental to the development of the embryo, that germline transmission occurs, and that the expression of the transgene is not attenuated or potentially silenced. Once a transgenic line has been established, it can be readily used for multiple analyses in ethanol studies. For those non-fluorescent transgenic constructs, commonly used transgenic markers include cmlc2:GFP and αcrystallin:RFP, which label the heart and retina, respectively, and allow for continued transgenic screening. In addition to transgenic markers for screening, these two transgenes can be used to directly study the impact of ethanol on heart and retina development.
Using the protocol described above, we generated a sox17:EGFPCAAX transgenic zebrafish line that had stable germline transmission of an endoderm-specific membrane-tagged EGFP construct. Using this line, we were able to measure the impact of ethanol on endodermal pouch formation in ethanol-sensitive Bmp mutant embryos. We showed that the sizes of pouches 1-3 but not of pouches 4 and 5 nor the overall endoderm length were impacted in ethanol-treated Bmp mutants (Fig 5). This pilot work suggests that the Bmp-ethanol interaction disrupts endoderm formation, in particular the cell behaviors underlying pouch formation. This work exemplifies the utility of generating transgenic zebrafish lines for ethanol studies. As a result, creating novel transgenic lines will greatly increase our understanding of ethanol-sensitive cellular processes and tissue events in FASD.
Ultimately, transgenic zebrafish, and zebrafish in general, have proven to be incredibly powerful in the study of FASD7,8. The Tol2Kit is an extremely versatile toolkit that enables researchers to quickly generate multiple transgenic constructs ready to inject into zebrafish. The modular design and the ease of generating new entry vectors result in extreme flexibility in generating transgenic constructs without having to redesign any components. Overall, this toolkit will greatly improve both zebrafish research and research in general aimed at improving the understanding of FASD.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The research presented in this article was supported by a grant from the National Institutes of Health/National Institute on Alcohol Abuse (NIH/NIAAA) R00AA023560 to C.B.L.
Materials
| Addgene Tol2 toolbox | https://www.addgene.org/kits/cole-tol2-neuro-toolbox/ | ||
| Air | Provided directly by the university | ||
| Ampicillin | Fisher Scientific | BP1760 | |
| Analytical Balance | VWR | 10204-962 | |
| Borosil 1.0 mm OD x 0.75 mm ID Capillary | FHC | 30-30-0 | |
| Calcium Chloride | VWR | 97062-590 | |
| Chloramphenicol | BioVision | 2486 | |
| EDTA | Fisher Scientific | BP118-500 | |
| Fluorescent Dissecting Microscope | Olympus | SZX16 | |
| Kanamycin | Fisher Scientific | BP906 | |
| Laser Scanning Confocal Microscope | Olympus | Fluoview FV1000 | |
| Lawsone Lab Donor Plasmid Prep | https://www.umassmed.edu/lawson-lab/reagents/lawson-lab-protocols/ | ||
| LB Agar | Fisher Scientific | BP9724 | |
| LB Broth | Fisher Scientific | BP1426 | |
| Low-EEO/Multi-Purpose/Molecular Biology Grade Agarose | Fisher Scientific | BP160-500 | |
| LR Clonase II Plus Enzyme | Fisher Scientific | 12538200 | |
| Magnesium Sulfate (Heptahydrate) | Fisher Scientific | M63-500 | |
| Micro Pipette holder | Applied Scientific Instrumentation | MIMPH-M-PIP | |
| Microcentrifuge tube 0.5 mL | VWR | 10025-724 | |
| Microcentrifuge tube 1.5 mL | VWR | 10025-716 | |
| Micromanipulator | Applied Scientific Instrumentation | MM33 | |
| Micropipette tips 10 μL | Fisher Scientific | 13611106 | |
| Micropipette tips 1000 μL | Fisher Scientific | 13611127 | |
| Micropipette tips 200 μL | Fisher Scientific | 13611112 | |
| mMESSAGE mMACHINE SP6 Transcription Kit | Fisher Scientific | AM1340 | |
| Mosimann Lab Tol2 Calculation Worksheet | https://www.protocols.io/view/multisite-gateway-calculations-excel-spreadsheet-8epv599p4g1b/v1 | ||
| NanoDrop Spectrophotometer | NanoDrop | ND-1000 | |
| NcoI | NEB | R0189S | |
| NotI | NEB | R0189S | |
| Petri dishes 100 mm | Fisher Scientific | FB012924 | |
| Phenol Red sodium salt | Sigma Aldrich | P4758-5G | |
| Pipetman L p1000L Micropipette | Gilson | FA10006M | |
| Pipetman L p200L Micropipette | Gilson | FA10005M | |
| Pipetman L p2L Micropipette | Gilson | FA10001M | |
| Potassium Chloride | Fisher Scientific | P217-500 | |
| Potassium Phosphate (Dibasic) | VWR | BDH9266-500G | |
| Pressure Injector | Applied Scientific Instrumentation | MPPI-3 | |
| QIAprep Spin Miniprep Kit | Qiagen | 27106 | |
| Sodium Bicarbonate | VWR | BDH9280-500G | |
| Sodium Chloride | Fisher Scientific | S271-500 | |
| Sodium Phosphate (Dibasic) | Fisher Scientific | S374-500 | |
| Stericup .22 µm vacuum filtration system | Millipore | SCGPU11RE | |
| Tol2 Wiki Page | http://tol2kit.genetics.utah.edu/index.php/Main_Page | ||
| Top10 Chemically Competent E. coli | Fisher Scientific | C404010 | |
| Vertical Pipetter Puller | David Kopf Instruments | 720 | |
| Zebrafish microinjection mold | Adaptive Science Tools | i34 |
Referenzen
- Denny, L., Coles, S., Blitz, R. Fetal alcohol syndrome and fetal alcohol spectrum disorders. American Family Physician. 96 (8), 515-522 (2017).
- Popova, S., et al. Comorbidity of fetal alcohol spectrum disorder: A systematic review and meta-analysis. The Lancet. 387 (10022), 978-987 (2016).
- Wilhoit, L. F., Scott, D. A., Simecka, B. A. Fetal alcohol spectrum disorders: Characteristics, complications, and treatment. Community Mental Health Journal. 53, 711-718 (2017).
- Wozniak, J. R., Riley, E. P., Charness, M. E. Clinical presentation, diagnosis, and management of fetal alcohol spectrum disorder. The Lancet Neurology. 18 (8), 760-770 (2019).
- Patten, A. R., Fontaine, C. J., Christie, B. R. A Comparison of the different animal models of fetal alcohol spectrum disorders and their use in studying complex behaviors. Frontiers in Pediatrics. 2, 93 (2014).
- Lovely, C. B. Animal models of gene-alcohol interactions. Birth Defects Research. 112 (4), 367-379 (2020).
- Fernandes, Y., Lovely, C. B. Zebrafish models of fetal alcohol spectrum disorders. Genesis. 59 (11), 23460 (2021).
- Fernandes, Y., Buckley, D. M., Eberhart, J. K. Diving into the world of alcohol teratogenesis: a review of zebrafish models of fetal alcohol spectrum disorder. Biochemistry and Cell Biology. 96 (2), 88-97 (2018).
- Choe, C. P., et al. Transgenic fluorescent zebrafish lines that have revolutionized biomedical research. Lab Animal Research. 37 (1), 26 (2021).
- Kwan, K. M., et al. The Tol2kit: A multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Developmental Dynamics. 236 (11), 3088-3099 (2007).
- Don, E. K., et al. A Tol2 gateway-compatible toolbox for the study of the nervous system and neurodegenerative disease. Zebrafish. 14 (1), 69-72 (2017).
- Westerfield, M. . The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). , (2000).
- Protocols. UMass Chan Medical School Available from: https://www.umassmed.edu/lawson-lab/reagents/lawson-lab-protocols (2017)
- Mosimann, C. Multisite gateway calculations: Excel spreadsheet. protocols.io. , (2022).
- Chung, W. -. S., Stainier, D. Y. R. Intra-endodermal interactions are required for pancreatic β cell induction. Developmental Cell. 14 (4), 582-593 (2008).
- Grevellec, A., Tucker, A. S. The pharyngeal pouches and clefts: Development, evolution, structure and derivatives. Seminars in Cell & Developmental Biology. 21 (3), 325-332 (2010).
- Lovely, C. B., Swartz, M. E., McCarthy, N., Norrie, J. L., Eberhart, J. K. Bmp signaling mediates endoderm pouch morphogenesis by regulating Fgf signaling in zebrafish. Development. 143 (11), 2000-2011 (2016).
- Silva Brito, R., Canedo, A., Farias, D., Rocha, T. L. Transgenic zebrafish (Danio rerio) as an emerging model system in ecotoxicology and toxicology: Historical review, recent advances, and trends. Science of The Total Environment. 848, 157665 (2022).
- Lai, K. P., Gong, Z., Tse, W. K. F. Zebrafish as the toxicant screening model: Transgenic and omics approaches. Aquatic Toxicology. 234, 105813 (2021).
- Stuart, G. W., McMurray, J. V., Westerfield, M. Stable lines of transgenic zebrafish exhibit reproducible patterns of transgene expression. Development. 109 (3), 577-584 (1988).
- Stuart, G. W., McMurray, J. V., Westerfield, M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 103 (2), 403-412 (1990).
- Thermes, V., et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mechanisms of Development. 118 (1-2), 91-98 (2002).
- Kawakami, K., et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental Cell. 7 (1), 133-144 (2004).
- Kawakami, K., Asakawa, K., Muto, A., Wada, H. Tol2-mediated transgenesis, gene trapping, enhancer trapping, and Gal4-UAS system. Methods in Cell Biology. 135, 19-37 (2016).

