Multiplex Cyclic Fluorescent Immunohistochemistry
Summary
Multiplex cyclic immunohistochemistry allows in situ detection of multiple markers simultaneously using repeated antigen-antibody incubation, image scanning, and image alignment and integration. Here, we present the operating protocol for identifying immune cell substrates with this technology in lung cancer and paired brain metastasis samples.
Abstract
The tumor microenvironment involves interactions between host cells, tumor cells, immune cells, stromal cells, and vasculature. Characterizing and spatially organizing immune cell subsets and target proteins are crucial for prognostic and therapeutic purposes. This has led to the development of multiplexed immunohistochemistry staining methods. Multiplex fluorescence immunohistochemistry allows the simultaneous detection of multiple markers, facilitating a comprehensive understanding of cell function and intercellular interactions. In this paper, we describe a workflow for the multiplex cyclic fluorescent immunohistochemistry assay and its application in the quantification analysis of lymphocyte subsets. The multiplex cyclic fluorescent immunohistochemistry staining follows similar steps and reagents as standard immunohistochemistry, involving antigen retrieval, cyclic antibody incubation, and staining on a formalin-fixed paraffin-embedded (FFPE) tissue slide. During the antigen-antibody reaction, a mixture of antibodies from different species is prepared. Conditions, such as antigen retrieval time and antibody concentration, are optimized and validated to increase the signal-to-noise ratio. This technique is reproducible and serves as a valuable tool for immunotherapy research and clinical applications.
Introduction
Brain metastases (BM) represent the most common central nervous system (CNS) tumors, occurring in nearly half of non-small cell lung cancer cases (NSCLC), with a poor prognosis1. An estimated 10%-20% of NSCLC patients already have BM at the time of initial diagnosis, and approximately 40% of NSCLC cases will develop BM during the course of treatment2. The tumor microenvironment (TME) is closely associated with NSCLC occurrence and BM, including various components, such as blood vessels, fibroblasts, macrophages, extracellular matrix (ECM), lymphoid, bone marrow-derived immune cells, and signaling molecules3,4. Microenvironmental immune cells play a crucial role in influencing cancer cell growth and development. Brain metastases present numerous potential treatment targets characterized by complex immunological microenvironments and signaling processes. For instance, PD-1 inhibitors have shown clinical efficacy for patients with lung cancer brain metastasis (LCBM) as an immune-checkpoint inhibitor (ICI). However, the frequency of responses to PD-1 therapy varies between primary NSCLC and LCBM5, suggesting that the tumor immune microenvironment acts as a critical ICI regulator.
Immunohistochemistry (IHC) is an invaluable tool in the fields of biology, foundation medicine, and pathology6. This detection method visualizes antigen expression through the interaction of antigen-antibody on a tissue slide7. IHC is used for diagnosing predictive markers, evaluating prognostic markers, guiding targeted therapies, and exploring the biological functions of tumor cells8. However, the traditional IHC method can only detect one biomarker at a time. To address this limitation, the innovation of immunohistochemical technology has led to the development of multiplex fluorescence immunohistochemistry (mfIHC), which allows for the simultaneous identification of multiple protein markers on the same tissue slide, both in bright field and fluorescent field9. This advancement provides accurate analysis of cell composition and molecular interactions among stromal cells, immune cells, and cancer cells within the TME.
In this study, we present a protocol for multiplex cyclic immunohistochemistry to analyze the spatial distribution of immune cells. Two primary antibodies of different species, such as rabbit and rat, are chosen for incubation simultaneously, followed by fluorescence-labeled secondary antibodies. Antigen retrieval is performed after each round of antigen-antibody reaction. Autofluorescence is blocked, and 4′, 6-diamidino-2-phenylindole (DAPI) is used for staining the nuclei. The panel includes sequential detection of CD3, CD8, CD20, and CK, cells are categorized according to the markers: tumor cells (CK+), mature T cells (CD3+), cytotoxic T cells (CD3+CD8+), B cells (CD20+)10,11.
Protocol
The research was approved by the medical ethics committee of Yunnan Cancer Hospital/the Third Affiliated Hospital of Kunming Medical University. All the subjects/legal guardians signed informed consent.
1. Slide preparation
- Cut sections of paired paraffin blocks containing primary lung tumor or lung cancer brain metastases cells at a thickness of 4 µm using a microtome. Remove sections to water and separate with tweezers, choose the best one and adhere it onto the polylysine-coated slide.
- Place the slides of tissues in an oven at 65 °C for 30 min to enhance tissue adhesion.
- Immerse the slides in xylene through three changes, each lasting for 10 min.
- Gradually reduce the alcohol concentration and incubate slides in 100% ethanol for 5 min, 90% ethanol for 5 min, 75% ethanol for 5 min, and in deionized water for 3 min.
2. Heat-induced epitope retrieval (HIER)
- Dilute 100x sodium citrate buffer solution (pH 6.0) to 1x (10 mM) in deionized water, preparing enough buffer solution to completely submerge the slides.
- Place the slides in a pressure cooker, subjecting them to high heat (100 °C) and pressure (~30 psi) for 2 min. After heating, allow the slides to cool to room temperature in distilled water for 3 min.
- Dissolve one packet of 52 g of phosphate-buffered saline (PBS; powdered) in 5 L of deionized water for preparing PBS buffer solution (pH 7.0). Place the slides in PBS buffer solution for 5 min, with 3 changes.
3. Peroxidase blocking
- Cover the sections with 3% hydrogen peroxide and incubate for 10 min at room temperature. Rinse the slides in PBS, 3x for 5 min each.
- Use filter paper to absorb excess water away from the perimeter of the tissue. Meanwhile, make sure that the tissue is moist.
4. Primary antibody incubation for first round
- Prepare a working mixture of the primary antibodies for CD8 (Rabbit monoclonal antibody, clone SP16) and CD20 (mouse monoclonal antibody, clone L26), diluted 1:50 in Bond primary antibody diluent.
- Add the antibody complex onto the sections and incubate for 1 h at room temperature.
- Prepare a solution of 0.1% Tween/phosphate-buffered saline: 1 mL of Tween in 1 L of PBS buffer solution.
- Wash the sections with 0.1% Tween/phosphate-buffered saline, 3x for 5 min each. Use filter paper to draw excess water away from the perimeter of the tissue, meanwhile, make sure the tissue is moist.
5. Secondary antibody incubation for first round
- Prepare a mixture of fluorescence-labeled goat anti-rabbit antibody (Excitation (Ex): 495 nm) and goat anti-mouse antibody (Ex: 578 nm), diluted 1:50 in phosphate buffer saline. The goat anti-rabbit antibody attaches to the primary antibody of CD8, and the goat anti-mouse antibody attaches to the primary antibody of CD20.
- Add the secondary antibody mixture dropwise and incubate at room temperature for 1 h.
6. Heat-induced epitope retrieval and peroxidase blocking
- Dilute 100x sodium citrate (pH 6.0) to 1x (10 mM) in deionized water, preparing enough buffer solution to completely submerge the slides.
- Place the slides in a pressure cooker and subject them to high heat (100 °C) and pressure (~30 psi) for 1 min. After heating, place the slides in distilled water to cool to room temperature for 3 min.
- Perform peroxidase blocking as described in step 3.
7. Primary antibody incubation for second round
- Prepare a working mixture of the primary antibodies for CD3 (Rabbit monoclonal antibody, clone SP7) and CK (mouse monoclonal antibody, MX005), diluted 1:50 in primary antibody diluent.
- Add the antibody complex onto the sections and incubate for 1 h at room temperature.
- Wash with 0.1% Tween/phosphate-buffered saline, 3x for 5 min each. Use filter paper to draw excess water away from the perimeter of the tissue, meanwhile, make sure the tissue is moist.
8. Secondary antibody incubation for second round
- Prepare goat anti-rabbit (Ex: 652 nm) and goat anti-mouse antibody (Ex: 590 nm) mixture, dilution 1:50 in phosphate buffer saline. The goat anti-rabbit antibody attaches to the primary antibody of CD3, and the goat anti-mouse antibody attaches to the primary antibody of CK.
- Add secondary antibody mixture dropwise, incubate at room temperature for 1 h. Wash with 0.1% Tween/phosphate-buffered saline, 3x for 5 min each.
9. Autofluorescence quenching and DAPI staining
- Add reagent (0.15 M/L KMnO4) to the tissue section for 1 min. Rinse with running water for 5 min.
CAUTION: KMnO4 is toxic and damages the skin. Make sure to wear gloves when handling the slides. If the liquid drips onto the skin, wipe it off quickly with clean napkins and flush with flowing water. - Dry the slide with increasing concentrations of alcohol (70%, 90%, 100%), for 3 min in each concentration.
- Add DAPI and coverslip for multispectral imaging. The amount of DAPI depends on the tissue size. Make sure the tissue is completely covered by DAPI after the coverslip is added.
10. Slide scanning
- Place the slides on a tray and push until it cannot be pushed further.
- To start the program, double-click on the Program Icon on the desktop. During start-up, the mode selection window is displayed. The selection window displays two brightfield and fluorescent modes: automatic mode and manual mode. In fluorescent scan mode, click Automatic Mode.
- Move the mouse cursor over ? button to display the information about the settings. Click Filter Setting > Filter Channel to choose the Channel Number > Pseudo Color. Define the color and save.
- Click Routine Work > Scanning Mode > Full Automatic. Click Routine Work > Slide Name to define slide name for output. Click Routine Work > Channel Settings to choose filters.
- Click Routine Work > Scan Options to determine the resulting quality and storage location of the virtual slide.
- Click Vorschau for threshold setting and selection of the range to be scanned.
- Click Hardware > Filters > Live > Auto Focus > Auto Exposure > Digital Gain > Tick use manual exposure time > Tick limiting the range > Set Current.
- Click Routine Work > Start Scan. Select magnification level as 20x or 40x. Choose 20x for appropriate file sizes. MRXS extension is defined by 3D Pannoramic MIDI scanner. Image extension can also be changed to TIFF image.
- Click Slide Viewer > Multiview Toolbox to choose the image for confocal, then align and integrate the image.
11. Quantitative evaluation of cell densities
- Quantify percentages of positive immune cells (CD3+, CD3+CD8+, CD20+) in tumor and stroma areas with Halo 10 scanner software. Validate CK staining to define tumor tissue.
Representative Results
We present a protocol for cyclic antigen detection using 5-color multiplex fluorescence on a single slide. Through our optimization of the assay, we enable the incubation of two antibodies from different species (Figure 1). The necessary devices for the experiment procedure include a pressure cooker and immunostaining box (Figure 2A).
After completing the assay, we define pseudo color of the four markers before scanning the slides. The pseudo colors of CD3, CD8, CD20 and CK are yellow, red, green, and cyan, respectively. The nuclei are labeled with DAPI. Representative regions of tumor and stroma are selected for analysis. Fluorescent spectrum at 495 nm, 578 nm, 652 nm, and 590 nm are captured using 3D automatic digital slide scanner. A representative stack image and single-labeled images in lung cancer brain metastases tissue are shown in Figure 2B. Based on images containing single marker fluorescence signal, the scanner software extracted cell phenotype characteristics based on pseudo color. Corresponding numbers of positive immune cells and total immune cells were counted as well. Each stained protein levels are quantified in the detected tissues by calculating H-score12. After these procedures, the number of each tumor-infiltrating lymphocytes type and the proportion of each cell type in the total number of target cells are counted and analyzed. The proportion of each immune cell type presents the effective percentage of tumor-infiltrating lymphocytes. The percentage of CD3+, CD3+CD8+ T cells and CD20+ B cells was analyzed in lung cancer brain metastases compared to primary lung cancer sections. Representative results are shown in Figure 3. The density of immune cells (CD3+ T cells, CD3+ CD8+ T cells) is lower in lung cancer brain metastases than primary lung cancer. CD20+ B cells are higher in the brain metastasis group than the primary lung cancer group. However, the results are not different significantly between these two groups for the limited samples.
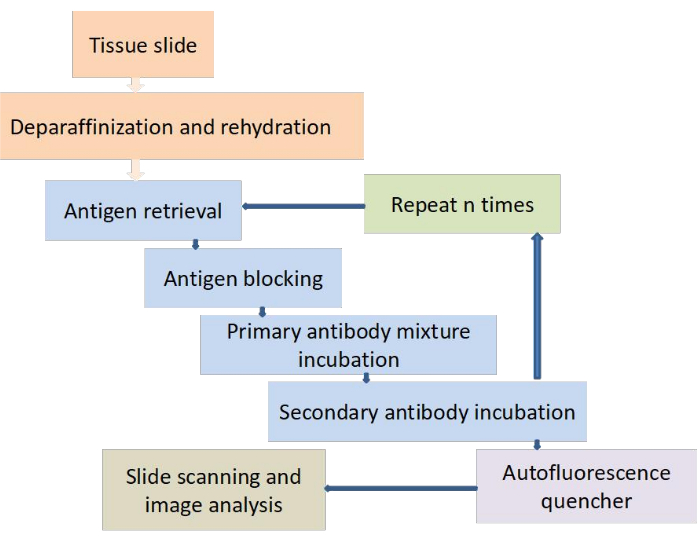
Figure 1: Workflow of multiplex cyclic fluorescence immunohistochemistry staining. The 4 µm sections are cut and adhered on adhesive slide. The following operating steps are performed: deparaffinization and rehydration, antigen retrieval, antigen retrieval, antigen blocking, primary antibody mixture incubation, secondary antibody incubation, then repeating the steps of antigen retrieval, antigen blocking, primary antibody mixture incubation and secondary antibody incubation, followed by reducing autofluorescence, selecting the appropriate filter for slide scanning and analyzing the results. Please click here to view a larger version of this figure.
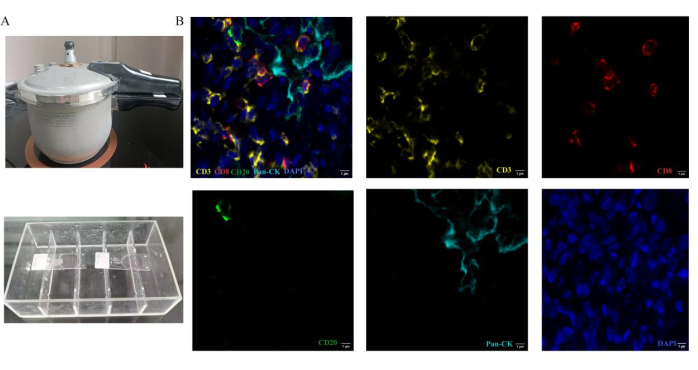
Figure 2: Operation methods of multiplex fluorescence immunohistochemistry staining. Antigen treatment is performed by heating sections in a pressure cooker. (A) The pressure cooker is used for Heat-induced epitope retrieval. In the process of antibody incubation, the slides are placed in the immunostaining box. (B) Representative composite and single-stained images for the panel used in lung cancer brain metastases tissue. Please click here to view a larger version of this figure.
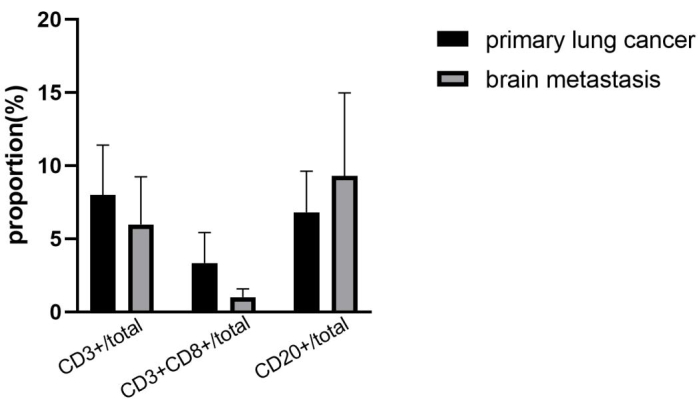
Figure 3: Immune cells proportion within primary lung cancer and lung cancer brain metastases. CD3+, CD3+CD8+, CD20+ immune cells proportion in four lung cancer brain metastases tissues are lower than paired primary lung cancer tissues (error bars: Standard deviation). Please click here to view a larger version of this figure.
Discussion
We have described the process for multiplex cyclic fluorescence immunohistochemistry staining. The primary antibody selection is a crucial aspect of the fluorescence immunohistochemistry assay, and monoclonal antibodies are recommended for better specificity and repeatability. To optimize the working concentration of the primary antibody, a series of dilutions have been tested through immunohistochemistry experiments. Both positive controls (to assess target antigen expression) and negative controls (no primary antibody incubation) are essential and should be set up.
In this protocol, the primary antibodies are diluted and prepared for a mixture from different species. The fluorescence-labeled secondary antibodies are also pooled and incubated in the same way, derived from different species. So, the critical step is choosing the species for different primary antibodies based on the antigens, and the monoclonal antibody is preferred compared with polyclonal antibody. These can ensure that the combination between primary antibody and secondary antibody is specific. The mixed liquid should achieve the working concentration for both primary antibodies and cross-reactivity should not exist between the two antibodies simultaneously. If the primary antibodies are of the same species, one secondary antibody is incubated first, then the second one. When determining the sequence of primary antibody incubation in a panel, priority should be given to antibodies that are sensitive to the antigen-antibody reaction, for the low expression antigens, the intensity would strengthen during second epitope retrieval. Before the second round of antibody incubation, repeating antigen retrieval takes less time (1 min) compared to the first operation (2 min). The fluorescence intensity from previous cyclic staining will not decrease even though the sections undergo heat-induced retrieval twice. For avoiding non-specific immunoreactivity, the incubation conditions need to be optimized including antibody working concentration, incubation time, and environmental temperature.
Various methods of epitope retrieval have been developed in recent decades, primarily divided into heat-induced epitope retrieval (HIER) and protease-induced epitope retrieval (PIER). Heating is an efficient antigen retrieval method that exposes antigen epitopes, making them more effectively detected by antibodies13. The two main options for antigen retrieval are based on citrate buffer and high pH EDTA buffer14. The optimized retrieval condition is identified according to the target antigen.
Autofluorescence can interfere with fluorescent imaging on sections caused by endogenous fluorophores and reagents used in tissue processing15. The use of an associated reagent is required for elution. Fluorophores are selected with emission peaks avoiding autofluorescence peaks (around 490 nm)16,17. Sudan Black B and NaBH4 have been reported for quenching tissue autofluorescence18,19. The combination of Sudan Black B and NaBH4 reduced fluorescence background in targeted renal formalin-fixed paraffin-embedded tissue20. In this protocol, the tissue treatment of KMnO4 in a concentration of 0.15 M/L is time saving for 1 min. KMnO4 covers a layer on the tissue for shielding spontaneous fluorescence and reduces background fluorescence, the specific staining of detected protein is more visualized.
The whole slides are scanned in four different filters channels, image alignment analysis is necessary, it is a great challenge to align the localization of single-cell and subcellular structures. For multispectral images from this technique, professional light imaging equipment and quantitative analysis software are needed for avoiding spectral crosstalk. The expensive cost of the instrument limits its application. Co-incubation of two antibodies saves time, especially when dealing with a 6-marker panel, which requires 3 cycles of incubation. This technique is used to visualize more detailed characterization of immune cells in tumor immune microenvironments. In future, the method will be applied for quantitative analysis of tumor-associated tertiary lymphoid structures.
In summary, multiplex cyclic fluorescence immunohistochemistry enables multiple targets to be stained by individually labeled fluorophores on a single slide. This assay provides an improved understanding of the spatial distribution of cells in the tumor immune microenvironment, and the spatial proximity of tumor-immune cells contributes to screening patients who will benefit from immunotherapy.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NO.81860413, 81960455), Yunnan Science and Technology Department Fund (202001AY070001-080), Scientific Research Foundation of Education Department of Yunnan Province(2019J1274).
Materials
| 0.15 mol/L KmnO4 | Maixin Biotechnology Co. Ltd. | MST-8005 | |
| 100x sodium citrate | Maixin Biotechnology Co., Ltd | MVS-0100 | |
| 3% hydrogen peroxide | Maixin Biotechnology Co., Ltd | SP KIT-A1 | |
| 3D Pannoramic MIDI | 3D histech Ltd | Pannoramic MIDI 1.18 | |
| Alexa Fluor 488 | Abcam | ab150113 | |
| Alexa Fluor 568 | Abcam | ab175701 | |
| Alexa Fluor 594 | Abcam | ab150116 | |
| Alexa Fluor 647 | Abcam | ab150079 | |
| Bond primary antibody diluent | Lecia | AR9352 | |
| CD20 | Maixin Biotechnology Co., Ltd | kit-0001 | |
| CD3 | Maixin Biotechnology Co., Ltd. | kit-0003 | |
| CD8 | Maixin Biotechnology Co., Ltd | RMA-0514 | |
| CK | Maixin Biotechnology Co. Ltd. | MAB-0671, | |
| DAPI | sig-ma | D8417 | |
| ethanol | Sinopharm Group Chemical reagent Co., LTD | 10009218 | |
| Histocore Multicut | lecia | 2245 | |
| PBS(powder) | Maixin Biotechnology Co., Ltd | PBS-0061 | |
| slide viwer | 3D histech Ltd | ||
| xylene | Sinopharm Group Chemical reagent Co., LTD | 10023418 |
Referenzen
- Wanleenuwat, P., Iwanowski, P. Metastases to the central nervous system: Molecular basis and clinical considerations. J Neurol Sci. 412, 116755 (2020).
- Schoenmaekers, J., Dingemans, A. C., Hendriks, L. E. L. Brain imaging in early stage non-small cell lung cancer: still a controversial topic. J Thorac Dis. 10, S2168-S2171 (2018).
- Vilariño, N., Bruna, J., Bosch-Barrera, J., Valiente, M., Nadal, E. Immunotherapy in NSCLC patients with brain metastases. Understanding brain tumor microenvironment and dissecting outcomes from immune checkpoint blockade in the clinic. Cancer Treat Rev. 89, 102067 (2020).
- Babar, Q., Saeed, A., Tabish, T. A., Sarwar, M., Thorat, N. D. Targeting the tumor microenvironment: Potential strategy for cancer therapeutics. Biochim Biophys Acta Mol Basis Dis. 1869 (6), 166746 (2023).
- Goldberg, S. B., et al. Pembrolizumab for management of patients with NSCLC and brain metastases: long-term results and biomarker analysis from a non-randomised, open-label, phase 2 trial. Lancet Oncol. 21 (5), 655-663 (2020).
- Sukswai, N., Khoury, J. D. Immunohistochemistry Innovations for Diagnosis and Tissue-Based Biomarker Detection. Curr Hematol Malig Rep. 14 (5), 368-375 (2019).
- Janardhan, K. S., Jensen, H., Clayton, N. P., Herbert, R. A. Immunohistochemistry in Investigative and Toxicologic Pathology. Toxicol Pathol. 46 (5), 488-510 (2018).
- Torlakovic, E. E., Nielsen, S., Vyberg, M., Taylor, C. R. Getting controls under control: the time is now for immunohistochemistry. J Clin Pathol. 68 (11), 879-882 (2015).
- Tan, W. C. C., et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond). 40 (4), 135-153 (2020).
- Wong, P. F., et al. Multiplex quantitative analysis of tumor-infiltrating lymphocytes and immunotherapy outcome in metastatic melanoma. Clin Cancer Res. 25 (8), 2442-2449 (2019).
- Sanchez, K., et al. Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and PD-L1 in early-stage breast cancer. Breast Cancer Res. 23 (1), 2 (2021).
- Zhang, W., et al. Multiplex immunohistochemistry indicates biomarkers in colorectal cancer. Neoplasma. 68 (6), 1272-1282 (2021).
- Salameh, S., Nouel, D., Flores, C., Hoops, D. An optimized immunohistochemistry protocol for detecting the guidance cue Netrin-1 in neural tissue. MethodsX. 5, 1-7 (2018).
- McClellan, P., Jacquet, R., Yu, Q., Landis, W. J. A Method for the immunohistochemical identification and localization of Osterix in periosteum-wrapped constructs for tissue engineering of bone. J Histochem Cytochem. 65 (7), 407-420 (2017).
- Sun, Y., et al. Sudan black B reduces autofluorescence in murine renal tissue. Arch Pathol Lab Med. 135 (10), 1335-1342 (2011).
- Taube, J. M., et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 8 (1), 000155 (2020).
- Clarke, G. M., et al. A novel, automated technology for multiplex biomarker imaging and application to breast cancer. Histopathology. 64 (2), 242-255 (2014).
- Oliveira, V. C., et al. Sudan Black B treatment reduces autofluorescence and improves resolution of in situ hybridization specific fluorescent signals of brain sections. Histol Histopathol. 25 (8), 1017-1024 (2010).
- Ahrens, M. J., Dudley, A. T. Chemical pretreatment of growth plate cartilage increases immunofluorescence sensitivity. J Histochem Cytochem. 59 (4), 408-418 (2011).
- Zhang, Y., et al. Spectral characteristics of autofluorescence in renal tissue and methods for reducing fluorescence background in confocal laser scanning microscopy. J Fluoresc. 28 (2), 561-572 (2018).

