Using SCOPE to Identify Potential Regulatory Motifs in Coregulated Genes
Summary
A straight-forward and robust method to identify potential regulatory motifs in co-regulated genes is presented. SCOPE does not require any user parameters and returns motifs that represent excellent candidates for regulatory signals. The identification of such regulatory signals helps to understand the underlying biology.
Abstract
SCOPE is an ensemble motif finder that uses three component algorithms in parallel to identify potential regulatory motifs by over-representation and motif position preference1. Each component algorithm is optimized to find a different kind of motif. By taking the best of these three approaches, SCOPE performs better than any single algorithm, even in the presence of noisy data1. In this article, we utilize a web version of SCOPE2 to examine genes that are involved in telomere maintenance. SCOPE has been incorporated into at least two other motif finding programs3,4 and has been used in other studies5-8.
The three algorithms that comprise SCOPE are BEAM9, which finds non-degenerate motifs (ACCGGT), PRISM10, which finds degenerate motifs (ASCGWT), and SPACER11, which finds longer bipartite motifs (ACCnnnnnnnnGGT). These three algorithms have been optimized to find their corresponding type of motif. Together, they allow SCOPE to perform extremely well.
Once a gene set has been analyzed and candidate motifs identified, SCOPE can look for other genes that contain the motif which, when added to the original set, will improve the motif score. This can occur through over-representation or motif position preference. Working with partial gene sets that have biologically verified transcription factor binding sites, SCOPE was able to identify most of the rest of the genes also regulated by the given transcription factor.
Output from SCOPE shows candidate motifs, their significance, and other information both as a table and as a graphical motif map. FAQs and video tutorials are available at the SCOPE web site which also includes a “Sample Search” button that allows the user to perform a trial run.
Scope has a very friendly user interface that enables novice users to access the algorithm’s full power without having to become an expert in the bioinformatics of motif finding. As input, SCOPE can take a list of genes, or FASTA sequences. These can be entered in browser text fields, or read from a file. The output from SCOPE contains a list of all identified motifs with their scores, number of occurrences, fraction of genes containing the motif, and the algorithm used to identify the motif. For each motif, result details include a consensus representation of the motif, a sequence logo, a position weight matrix, and a list of instances for every motif occurrence (with exact positions and “strand” indicated). Results are returned in a browser window and also optionally by email. Previous papers describe the SCOPE algorithms in detail1,2,9-11.
Protocol
1. Prepare a list of names for genes that you believe are co-regulated for analysis by SCOPE.
Save the list as a text file or copy it to the clipboard to paste into SCOPE in step 3. The file should contain one gene name per line with no additional information. Alternatively, you can prepare the list as a FASTA file containing the actual sequences to be analyzed.
2. Start your web browser and connect to the URL: http://genie.dartmouth.edu/SCOPE/
3. Enter the information that SCOPE needs to perform the analysis.
The initial SCOPE page is shown in Figure 1. Different sections are addressed in this step.
- Use the ‘Species’ popup menu to choose the species you will be examining. It is important to choose the correct species because SCOPE refers to the genome to calculate background frequencies of occurrence for any candidate motif it is examining.
- Use the ‘upstream sequence” radio buttons to choose either intergenic or fixed length. Intergenic will analyze all the sequence between of the gene you are looking at and the previous (upstream) gene. This will mean that different upstream lengths will be used for each gene. Choosing fixed length will look at exactly that number of nucleotides upstream from the start of the current gene. In this case, SCOPE will examine the same length of upstream sequence for each gene, even if that extends into the previous gene (or not). Typically, 800 nts is the best length to choose, but this can vary with species.
- Next tell SCOPE what gene set to analyze either by pasting in your gene list into the gene list text box, or by pressing the ‘choose file’ button to select the file containing the list of genes that you created earlier. You may, alternatively, paste in a FASTA sequence file into the same text box.
- The next section of the page contains a checkbox for ‘Examine genome for other genes containing found motif(s)?’ This option can add considerable analysis time since SCOPE has to evaluate every other gene in the genome. However, this can be very useful in identifying other genes that are good candidates for being co-regulated with the genes in the starting gene set. Since SCOPE analyses are relatively quick, it is suggested that you leave this off in your initial analysis. It can always be turned on from the results page to rerun the analysis, as explained in the results section.
- The ‘Results must include’ section can be used to enter a motif that you want SCOPE to include in its analysis. You might want to do this if you are looking for a specific motif.
- The last section on the page can be used to enter your email address and a comment to be saved with the analysis. If this is filled in, SCOPE will send an email with a link back to the web page containing results, and it will also include two attachments. One is a plain text file that has all the analysis results in human readable format. The second attachment contains an XML file that has every result that SCOPE has found in a computer readable format. If you want to do some additional analysis on the results, the XML file is very useful. Both files are “zipped” before being sent with the email.
- For this demo, we will start with the same information. This can be easily achieved by pressing the ‘Sample Search’ button which will fill in the necessary information. Press this button now. Three genes will be entered for you and appropriate choices made for the other fields. Leave these as they are set. The three genes are involved in telomere maintenance in Saccharomyces cerevisiae. The filled in form is shown in Figure 2. Press the ‘Run SCOPE’ button at the bottom of the page to start the analysis.
4. Representative Results:
The main results of the analysis are shown in Figure 3. The top of the page contains a table of information about the motifs that were found by SCOPE. The first column contains a list of motifs that were found and small colored squares serve as a legend for the graphical motif map shown below. The display of any given motif may be toggled on or off by clicking in the colored box (or where the colored box would be). This can be very useful to hide the display of highly repeated motifs that might make it difficult to see the less prevalent motif patterns.
Other columns of data are Count (the number of occurrences of that motif in the entire gene set), Sig value (an indication of the significance of that motif), Coverage (the percentage of the submitted genes that contain at least one instance of that motif), and Algorithm (which of the three component algorithms was used to detect the motif).
Clicking on any of the listed motifs will take the user to a page containing detailed information for that motif. The results details are shown for the cyan motif (atgnnnnttg) in Figure 4. On this page, the motif is represented in three ways: a sequence logo, a position weight matrix, and a list of all motif instances with their positions, strands and genes.
A little further down the page are some additional details about the results of looking for other genes containing this motif. As can be seen, in this case there were 1344 other genes containing the motif, all of which actually improved the Sig value when added to the original gene set. Pressing ‘Add checked genes to search’ will return to the SCOPE setup page with these genes added to the original gene set and the parameters set as they were previously. In this case, 10 extra genes are added to the original three.
Figure 5 shows the results of the analysis containing the extra genes for this motif. The original three genes are on the bottom of the results (in lower case). Looking at the pattern of motifs in the upstream region of these extra genes clearly shows that they are similar. In fact, many of these genes are involved in telomere maintenance as were the original three genes. Note also that the original motif is now the highest scoring motif in this set.
Another set of SCOPE results is shown in Figure 6. In this case, the set of genes are those that are involved in ribosome biogenesis in Saccharomyces cerevisiae. These genes are not actually part of the ribosome but are responsible for assembling ribosomes and include a number of modification enzymes. What is clear in the figure is that the red and green motifs form a reliable pattern that is likely to be involved in regulation of the genes in this set. We are investigating this pattern of “modules” in more detail and will report on it in a later publication.
Figure 1. Main SCOPE input page. This page is used to enter the genes to be analyzed and to define the species and the length of upstream region to be examined. Optionally, the user can request the results by email or restrict the search to any specified motif. Video help is also available.
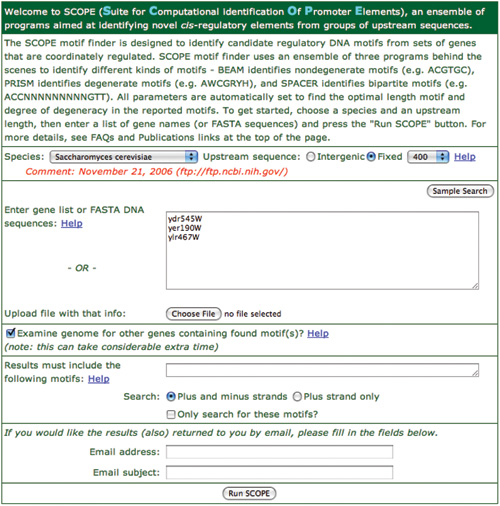
Figure 2. Main SCOPE input page with values filled in for performing a search. These parameters are the result of pressing the ‘Sample Search’ button. In this case, the check box to find other genes containing the motifs found by SCOPE is checked. This option takes longer to compute (every gene in the genome has to be examined) but can provide interesting insights.
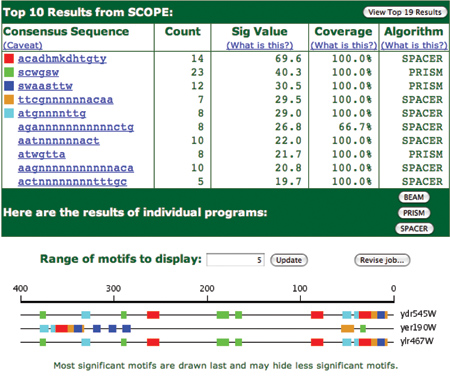
Figure 3. Main SCOPE results page. This page summarizes the results of the SCOPE search. A list of all high scoring motifs is provided and a color coded motif map shows the positioning of the identified motifs in the set of genes analyzed. Clicking on a colored box next to a motif will toggle the display of that motif on or off in the motif map. In addition to a significance score (Sig value), the fraction of genes containing the motif (coverage), and the algorithm used to find that motif are also provided.
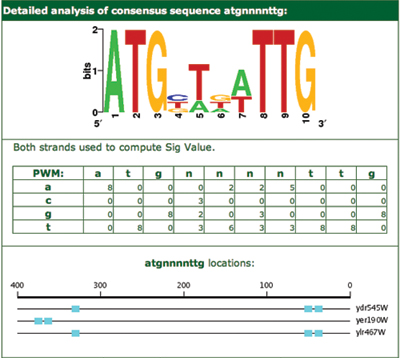
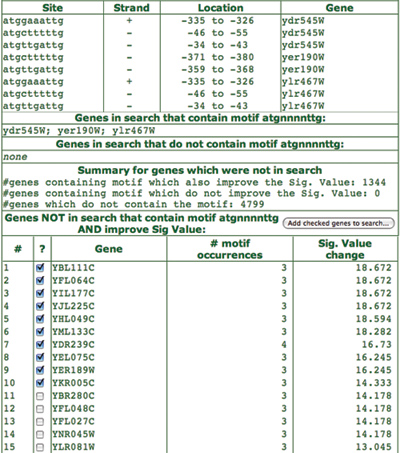
Figure 4. This results detail page is brought up when a specific motif is clicked in the main results page. It shows details of the individual motif. The sequence logo, the position weight matrix, and the consensus sequence each represent a different kind of summary of the list of motif instances also on the page. Since ‘find extra genes’ was checked in the original search setup, there is also information on this page about any other genes in the genome that contain this motif. From this page it is also possible to start another SCOPE run including the extra genes identified on this page.
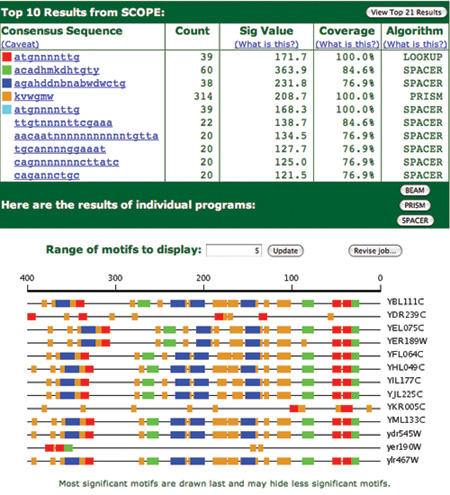
Figure 5. This figure shows the results of looking for extra genes for the motif ‘atgnnnnttg’ shown in Figure 4. The original three genes are in lower case at the bottom of the motif map. The additional genes are shown in upper case. There is a clear pattern to the motifs in the upstream regions of these genes. Notice also that the specified motif shows an algorithm as ‘LOOKUP’ because that is how it was identified. It actually matches the 5th motif found by SPACER in this analysis.
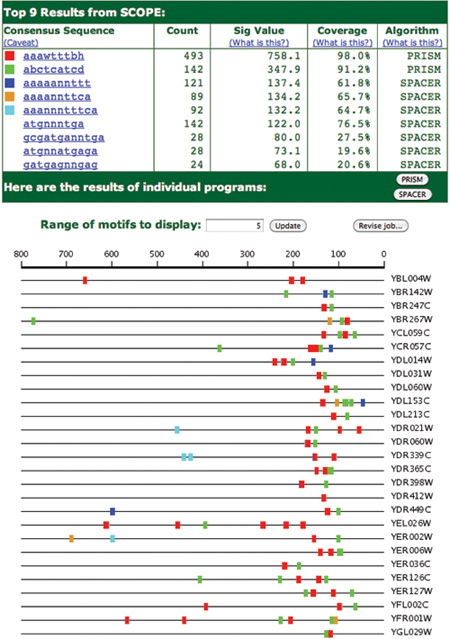
Figure 6. SCOPE output for genes involved in ribosome biogenesis in Saccharomyces cerevisiae. Note the conserved pattern of modules consisting of the motifs ‘aaawtttbh’ (red) and ‘abctcatcd’ (green) separated by about 10-30 nts and present at 100-200 nucleotides upstream of transcription start for the gene.
Discussion
SCOPE provides the researcher with a powerful tool to use for the identification of potential regulatory motifs in sets of coordinately regulated genes. The user is not required to guess at the size of the motif or the number of occurrences of the motif as many other motif finding sites require. These parameters are basically unknowable until the motif is identified. The interface is very simple both for entering sequences or gene names and for viewing the output.
SCOPE output provides detailed information about all of the motifs that are identified, using three different ways of motif representation. Each instance of the motif in all of the genes is listed with position and “strand” information. Graphical results in the form of motif maps provide a visual display that is easy to understand and provides an intuitive way to see patterns in the motifs that are present.
SCOPE is very robust to the presence of noise in the data. Typically, this takes the form of extra genes being present in the starting set that might not actually be co-regulated with the rest of the genes. This often happens when starting with genes that are co-expressed in microarray experiments. Sometimes the experiment is noisy, or there may be several transcription factors activated in the experimental conditions used for the microarray experiment. These different transcription factors will likely have different target sites on the DNA. Even in the presence of 4-fold extraneous genes (noise:signal ratio is 4:1), SCOPE is still maintains 50% of its accuracy in predicting sites1.
Although SCOPE contains over 2 million synonyms for gene names, it sometimes fails to identify some genes names. We are constantly updating our synonym lists, but sometimes find that different synonyms refer to the same gene. In those cases, we do not include the synonyms because of the ambiguity. if you have a gene name that is not found by SCOPE, it is recommended that you refer to the genome specific site to find an alternative gene name to use in SCOPE. Examples of appropriate gene names for each species are provided by SCOPE.
SCOPE currently contains 72 species with new species being added all the time. The web site contains video help as well as FAQs. Source code is freely available to academic users by writing to RHG.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This research was supported by a grant to RHG from the National Science Foundation, DBI-0445967.
Referenzen
- Chakravarty, A., Carlson, J. M., Khetani, R. S., Gross, R. H. A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics. 8, 249-249 (2007).
- Carlson, J. M., Chakravarty, A., DeZiel, C. E., Gross, R. H. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 35, 259-264 (2007).
- Blom, E. J., Roerdink, J. B., Kuipers, O. P., Hijum, S. A. v. a. n. MOTIFATOR: detection and characterization of regulatory motifs using prokaryote transcriptome data. Bioinformatics. 25, 550-551 (2009).
- Blom, E. J. DISCLOSE : DISsection of CLusters Obtained by SEries of transcriptome data using functional annotations and putative transcription factor binding sites. BMC Bioinformatics. 9, 535-535 (2008).
- Bushey, A. M., Ramos, E., Corces, V. G. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23, 1338-1350 (2009).
- Znaidi, S. Identification of the Candida albicans Cap1p regulon. Eukaryot Cell. 8, 806-820 (2009).
- Sharma, D., Mohanty, D., Surolia, A. RegAnalyst: a web interface for the analysis of regulatory motifs, networks and pathways. Nucleic Acids Res. 37, W193-W201 (2009).
- Znaidi, S. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell. 7, 836-847 (2008).
- Carlson, J., Chakravarty, A., Gross, R. B. E. A. M. A beam search algorithm for the identification of cis-regulatory elements in groups of genes. J Comput Biol. 13, 686-701 (2006).
- Carlson, J., Chakravarty, A., Khetani, R., Gross, R. Bounded search for de novo identification of degenerate cis-regulatory elements. BMC Bioinformatics. 7, 254-254 (2006).
- Chakravarty, A., Carlson, J. M., Khetani, R. S., DeZiel, C. E., Gross, R. H. SPACER: identification of cis-regulatory elements with non-contiguous critical residues. Bioinformatics. 23, 1029-1031 (2007).

