Near Infrared Optical Projection Tomography for Assessments of β-cell Mass Distribution in Diabetes Research
Summary
We describe the adaptation of optical projection tomography (OPT)1 to imaging in the near infrared spectrum, and the implementation of a number of computational tools. These protocols enable assessments of pancreatic β-cell mass (BCM) in larger specimens, increase the multichannel capacity of the technique and increase the quality of OPT data.
Abstract
By adapting OPT to include the capability of imaging in the near infrared (NIR) spectrum, we here illustrate the possibility to image larger bodies of pancreatic tissue, such as the rat pancreas, and to increase the number of channels (cell types) that may be studied in a single specimen. We further describe the implementation of a number of computational tools that provide: 1/ accurate positioning of a specimen’s (in our case the pancreas) centre of mass (COM) at the axis of rotation (AR)2; 2/ improved algorithms for post-alignment tuning which prevents geometric distortions during the tomographic reconstruction2 and 3/ a protocol for intensity equalization to increase signal to noise ratios in OPT-based BCM determinations3. In addition, we describe a sample holder that minimizes the risk for unintentional movements of the specimen during image acquisition. Together, these protocols enable assessments of BCM distribution and other features, to be performed throughout the volume of intact pancreata or other organs (e.g. in studies of islet transplantation), with a resolution down to the level of individual islets of Langerhans.
Introduction
The insulin producing β-cells are key for the body’s ability to control blood glucose homeostasis. Therefore, assessments of pancreatic BCM distribution are imperative to many areas of pre-clinical diabetes research. In evaluations of therapeutic regimes for example, the impact of targeted gene ablation on endocrine cell differentiation or studies of diabetes aetiology in rodent models for the disease often depend on such analyses. Traditionally, these types of assessments have relied on time consuming stereological approaches that are difficult to perform due to the size and complex anatomical constitution of the pancreas. Most high resolution imaging approaches at present (typically optical), do not provide sufficient penetration depth to allow whole pancreas imaging in rodents. Conversely, imaging approaches that are not limited by their penetration depth (typically nuclear) provide to poor resolution to resolve the full BCM distribution and are hampered by the lack of adequate contrast agents4,5.
Optical projection tomography is a 3D imaging modality that enables high-resolution assessments of biomedical specimens on the mm to cm scale6. Hereby, information on the spatial position and volume of the individual insulin expressing islets of Langerhans may be extracted throughout the volume of the pancreas in normal and diabetic mice3,7-10. The aim of the current study is to further enhance the capacity of this technique for the assessment of pancreatic β-cells; their endogenous distribution, when grafted into other tissues, their relation to other pancreatic constituents (such as infiltrating cell types) and in larger pancreatic preparations than previously possible.
The near infrared optical projection tomography (NIR-OPT) setup
In the below protocols, an OPT scanner based on the original set up described by Sharpe et al1, adapted to imaging in the near infrared range is described and used. For single channel assessments of the mouse pancreas (e.g. of BCM), the SkyScan 3001 (Bioptonics) scanner may be used.
A metal halide lamp that provides higher excitation energy than a mercury arc lamp at wavelengths above 650nm, supplies the excitation light. The light is transferred through a liquid light guide. A useful combination of fluorochromes and band pass filters for NIR fluorescence imaging and channel separation are shown in Figure 3. The emitted light is detected with a back illuminated CCD camera, with high quantum efficiency in the NIR spectrum. The OPT scanning is automated using a LabView platform that controls the camera and stepper motor. To support samples in the size of intact rat pancreata, a protected silver coated mirror and a large cuvette is used. Finally, a sample holder that eliminates unwanted vertical movements of the sample during the scan was designed.
Protocol
1. Sample Preparation and Scanning
1.1 Sample preparation
The following procedure is performed essentially as described previously7.
- Harvest the pancreas. Use ice-cold PBS to avoid proteolytic degradation.
- Fix the tissue in 4% PFA in PBS on ice for 2-3 hr. Make sure the lobes are "spread out" during fixation. This will facilitate identification of anatomical landmarks after reconstruction.
- Wash in excess PBS for 30 min.
- Dehydrate the pancreas stepwise in methanol (33, 66%, 100%), 15 min/step. This minimizes the formation of bubbles during bleaching (see step 5) and prevents cell destruction during freeze-thawing (see step 7).
- Incubate the tissue in freshly prepared MeOH:DMSO:H2O2 bleaching buffer in a 2:1:3 ratio at RT for 24 hr to quench endogenous tissue fluorescence. For larger samples, exchange to new bleaching buffer and incubate for another 24 hr.
- Wash in excess MeOH, ON.
- Freeze-thaw for at least 5 cycles in -80 °C – RT to facilitate antibody penetration.
- Rehydrate stepwise back to TBST (33, 66%, 100%), 15 min/step.
- Block in TBST with 10% serum (preferably from the same species in which the secondary antibody was generated), 5% DMSO and 0.01% NaAz for 12-24 hr at RT
- Incubate with primary antibodies in blocking buffer for 48 hr, at RT, extend to 72 hr for larger samples (antibodies used here are listed in the table of reagents).
- Wash in excess TBST, ON.
- Incubate with fluorescently conjugated secondary antibodies for 48 hr, at RT, extend to 72 hr for larger samples.
- Wash in excess TBST, ON.
1.2
The following procedure describes how to mount the sample in agarose and attach it to the custom made sample holder (see Figure 7) prior to OPT scanning.
- Separate the splenic, duodenal and gastric lobes according to Figure 3A in Hörnblad et al3. The relationship between the lobes is further clarified in Hörnblad et al11.
- Prepare 1.5% (w/v) low melting temperature agarose in dH2O, filter, allow to cool to 37 °C and rinse the tissue in dH2O to wash away the detergent and remove bubbles before agarose-embedding on ice.
- Cut out an agarose block enclosing your sample and leave a ~1 cm spacer between the sample and the base of the agarose. Trim sharp edges (≤ 90 °) of the agarose block to reduce light scattering.
- Dehydrate the sample stepwise in MeOH (33, 66%, 100%), allowing time to equilibrate between each step. When the specimen sinks it is considered to be equilibrated.
- Clear the sample in a 1:2 solution of Benzyl Alcohol: Benzyl Benzoate (BABB) until it becomes transparent. Exchange BABB solution and incubate for an additional 12 hr.
- Position the cleared sample in the sample holder and secure it by inserting 2 needles through the agarose spacer via the drilled holes in the flanges of the holder.
- Place the sample in the scanner and submerge it into a cuvette filled with BABB clearing solution. When comparing a series of pancreata the same magnification should be used for all scans. The magnification should be optimized for the largest sample in the series.
1.3 Positioning of specimen at the AR
The following protocol describes the procedure to precisely position a sample using the COM-AR algorithm. This procedure is only applicable when the ROI includes the entire specimen. For detailed descriptions of the algorithms, please see Cheddad et al2.
- Acquire images of the specimen in two positions for both the anatomy and signal channels. Position1 at 0 ° (associated with the X-axis), displaying the largest projection area, and position2 at 90 ° (associated with the Z-axis). We are using the GFP channel to visualize the anatomy.
- Apply the expectation-maximization (EM) algorithm on the anatomy images to threshold the ROI.
- Calculate the COM points (x-coordinates) of the binary images obtained in step 2 for both the 0 ° and 90 ° projections.
- Superimpose a vertical line passing through the identified COM point calculated in step 3 on the 0 ° and 90 ° images of the signal channel.
- Use the images acquired in step 4 as references to move the sample so that the centre line of the Field of View passes through the found COM points of the sample.
1.4 Scanning
- Adjust exposure times to achieve the highest signal to noise ratio possible without saturating any areas of the projection images. Repeat for all channels to be scanned.
- Select the filter set for the first fluorescence channel to be scanned. Emission from shorter λ fluorophores may excite fluorophores with longer λ and thereby cause photo bleaching. To minimize this possibility, scan the fluorophore with longest excitation λ first.
- Open the shutter to illuminate the sample and collect the fluorescence signal over 360 °, rotating the sample along the vertical axis for each channel. The step angle used for the NIR-OPT setup is 0.9 ° and for the Bioptonics 3001 scanner 0.45 °.
- Select the appropriate filter for the next channel and proceed as above.
2. Computational Processing and Reconstruction
2.1 Post-acquisition misalignment detection and correction (A-value tuning)
In projection tomography, it is in general necessary to assign a post-alignment value to the projections to fine-tune the images position along the axis of rotation prior to reconstruction. However, a small aberration in the angle of the camera towards the optical axis can cause non-uniform A-values along the length of the sample and thus induce geometric distortions. To avoid such distortions, a computational method to find the accurate and unified post-alignment value (A-value) throughout the entire specimen may be applied2.
- Use a discrete Fourier transform to divide a projection of the specific signal at 0 ° and 180 ° into 8 pixels height blocks and calculate the shift along the x-axis (the A-value) between each block.
- Apply a linear least squares regression to help calculate the angle θ', which describes the slope of the x-axis-shift along the length of the specimen, and to find a rotational centre point.
- Correct for misalignments during scanning by rotating all projections θ'/2 around the rotational centre point.
2.2 Contrast limited adaptive histogram equalization (CLAHE)
To facilitate detection and segmentation of objects (islets) exhibiting very weak signals, which are at risk to be "thresholded out" during reconstruction and/or segmentation for quantitative assessments, a CLAHE algorithm may be applied to the projection images. The CLAHE operation is performed with two major intensity transformations:
- The local contrast is estimated and equalized within non-overlapping blocks in the projection image.
- The intensities are then normalized at the border regions between blocks through bilinear interpolation.
The name contrast-limited refers to the clip limit, which is set to avoid saturating pixels in the image. In this protocol, the MATLAB built-in function "adapthisteq" was used and applied with the default clip limit of 0.01 and a tile size of 256. Note, the optimal tile size needs to be tested empirically and can vary depending on the specimen analyzed. More details on the algorithm and examples can be found in Hörnblad et al3.
Note! The above listed computational processing steps (including COM-AR, A-Value tuning and CLAHE, see 1.3-2.2) are built upon standard algorithms and are executed in MATLAB (Mathworks).
2.3 Tomographic reconstruction and iso-surface rendering
- Using a filtered back-projection algorithm, the corrected and normalized images may now be reconstructed with unified misalignment compensation and minimal requirement for optimization of dynamic range. In this protocol, all reconstructions are carried out using the filtered back projection method available in the NRecon software (Skyscan), version 1.6.8 (http://www.skyscan.be/products/downloads.htm). Note, the magnification of an imaged object depends on its distance from the focal point of the lens unless parallel beam geometry is implemented in the imaging setup. Therefore, when importing a projection dataset to the NRecon software it is important to include the correct object to source distance (mm) and rotational direction of the scanner (for counter clockwise input "cc" and for clockwise input "cw") in the accompanying log file, to avoid cone beam induced artifacts during reconstruction.
- To visualize and quantify the stack of virtual sections obtained, generate 3D iso-surfaces using suitable image processing software such as Imaris or Volocity.
Murine islet isolation and transplantation procedures were performed at the Diabetes Research Institute's Preclinical Cell Processing and Translational Model Core under protocols reviewed and approved by the University of Miami Institutional Animal Care and Use Committee. The ethical committee for animal research, northern Sweden, approved all other experiments involving animals.
Representative Results
In the current report we describe a protocol for the extraction and computational processing of BCM data in rodent pancreata (and other tissues) using NIR-OPT (Figure 1). As illustrated in Figure 2, tissue autofluorescense from pancreatic specimen is as expected markedly decreased in the NIR spectrum. This leads to a significant increase in mean signal to noise (S:N) ratio for the assessment of insulin labeled islets of Langerhans. By the adaptations of OPT to imaging in the NIR part of the spectrum, as described herein, at least three specific channels may be visualized with sufficient S:N ratio to enable assessments of antibody labeled cell types throughout the volume of the murine pancreas with distinct channel separation (see Figure 3 and 4). Applied to imaging of diabetogenic processes and/or BCM assessments in general, the technique thus allows for the visualization and quantification of insulin positive areas in relation to surrounding and/or interacting cell types (see Figure 4). Such assessments are thanks to the increased tissue penetration depth obtained in the NIR range possible to perform in much larger specimens than previously, including the rat pancreas, which is 3-5 times larger than its mouse counterpart (see Figure 5). Regardless of whether visible or NIR wavelengths are utilized, the implementation of CLAHE may significantly facilitate OPT based assessments of BCM during different genetical and physiological conditions by increasing the detection sensitivity of the technique (see Figure 6). A blueprint for the developed sample holder is shown in Figure 7.
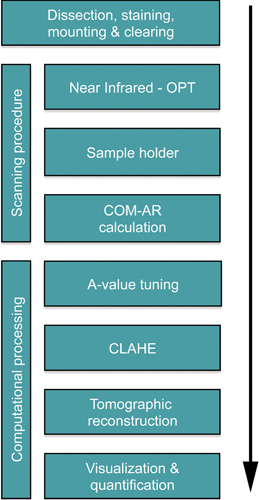
Figure 1. Flowchart depicting the critical steps for OPT based analyses of BCM in the murine pancreas. The time required to assess a typical mouse pancreas is 13-14 days. The majority of the time is consumed during tissue processing and immunohistochemical staining (10 days), tissue clearing requires approximately 2 days whereas the length of the scanning is dependent on the exposure time required (normally around 1 hr). The subsequent computational processing typically is performed within a day. Note, the relatively lengthy staining protocol is ideally suited for batch processing of larger amounts of specimens.
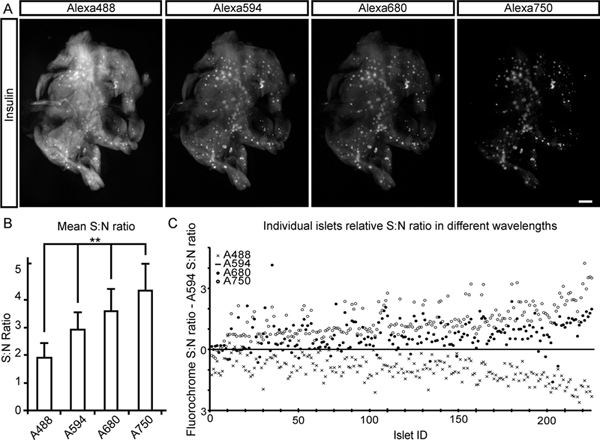
Figure 2. Signal to noise ratios for BCM assessments at different wavelengths. A mouse duodenal pancreatic lobe, stained for insulin and with a cocktail of fluorochrome-conjugated secondary antibodies (Alexa 488, 594, 680 and 750), was used to determine S:N ratios at different wavelengths. A, Images show the first projection frame for each signal channel. B, Graph illustrating the mean S:N for each signal channel. The ratios were determined as the mean islet intensity (based on 215 islets) divided by the background intensity (the endogenous tissue fluorescence from the exocrine tissue). C, Graph showing S:N ratios for the individual islets in each channel normalized to the S:N obtained for the Alexa 594 channel. One way ANOVA was used for statistical analyses. Significance levels indicated correspond to **p<0.01. Scale bar in (A) corresponds to 1 mm. Click here to view larger figure.
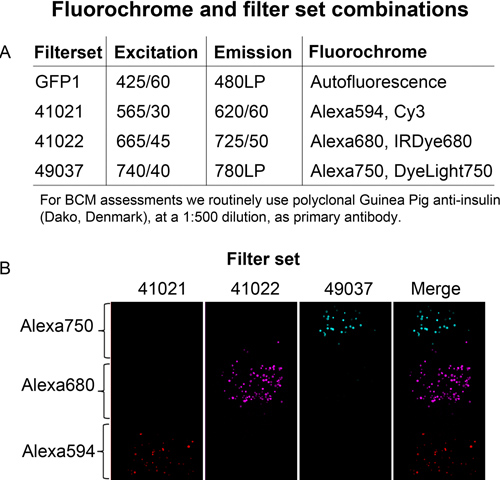
Figure 3. Channel separation. A, Secondary antibodies conjugated with Alexafluor dyes listed in the table were immobilized separately on proteinG-sepharose beads. B, The fluorescent beads were then embedded at different levels in an agarose phantom and imaged using indicated filters.
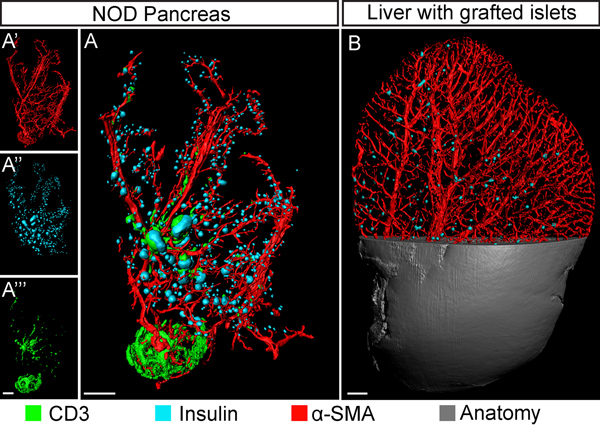
Figure 4. OPT based multichannel imaging in diabetes research. A, OPT based iso-surface reconstruction of a pancreas (12 weeks, duodenal lobe) from the Non Obese Diabetic (NOD) model for type 1 diabetes. The specimen is stained for insulin (islet β-cells, pseudo colored blue); smooth muscle α-actin (blood vessels, red) and CD3 (infiltrating T-lymphocytes, green). The corresponding secondary antibodies used were; Cy3, IRDye-680 and DyeLight-750 respectively. The insets (A’-A”’) show the individual signal channels. B, OPT image (blow up view) of a mouse liver lobe (lobus sinister lateralis) grafted with syngenic islets and imaged with NIR-OPT two weeks post transplantation. The insulin expressing islets are pseuodocolored in blue and the smooth muscle α-actin positive vessels are in red. The approach enables assessments of islet graft distribution within the vascular network. Scale bars correspond to 1 mm.
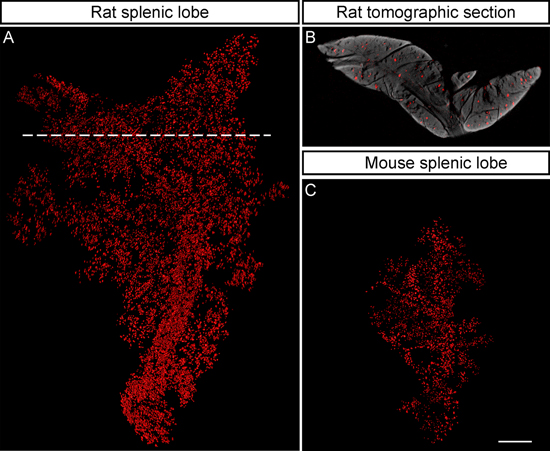
Figure 5. NIR-OPT facilitates the imaging of larger specimens. A, Iso-surface rendering of the BCM distribution in a rat pancreas from the Zucker Fatty model for type 2 diabetes (splenic lobe at 9 months), exemplifying the possibility to image specimen on the rat pancreas scale by NIR-OPT. As determined by this technique the displayed lobe is ~6 times larger (v/v) than its mouse counterpart and harbors 10139 insulin expressing islets of Langerhans whose β-cell volume makes up 1.32 % of the total lobular volume. B, Tomographic section corresponding to the broken line in (A) illustrating that islets from all depths of the tissue are detected. C, Iso-surface rendering of the BCM distribution in a mouse pancreas (splenic lobe at 8 weeks) displayed as a size reference. The displayed lobe harbors 2490 insulin expressing islets whose β-cell volume makes up 0.89% of the total lobular volume. The pancreata are stained with GP anti-insulin followed by Alexa594 conjugated goat anti-GP (mouse) and IRDye 680 Conjugated Donkey anti-GP (rat) antibodies respectively. The specimens in (A-C) are depicted to scale and the scale bar in (C) corresponds to 2 mm.
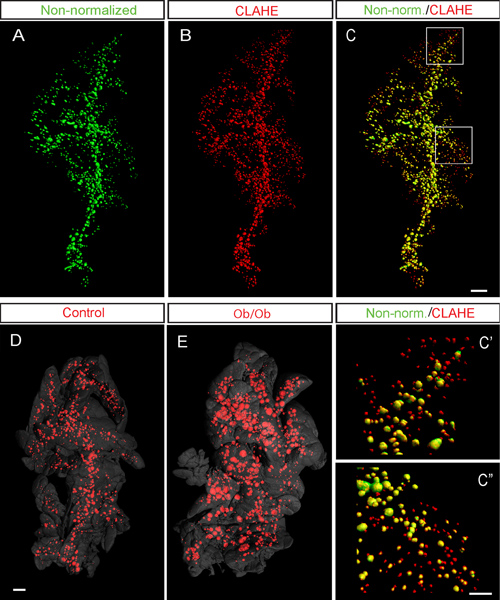
Figure 6. CLAHE facilitates detection of islets in the murine pancreas by OPT imaging. A-C, Representative iso-surface rendered OPT images of a C57Bl/6 mouse pancreas (splenic lobe at 8 weeks) labeled for insulin. Iso-surface reconstructions of OPT images were performed before (A, pseudo colored green) and after the CLAHE protocol was applied (B, pseudo colored red). C, Overlay of the non-normalized data in (A) and the CLAHE processed data in (B). C’-C”, Representative high magnification overlay of the non-normalized (A) and CLAHE processed (B) images. As shown by the presence of “red-only” islets, the CLAHE script facilitates the detection of small and low signal intensity islets. In the current example the depicted specimen (after CLAHE processing) harbored 2419 islets with a volume of 1.74 mm3 (Numbers based on the corresponding unprocessed projection data was 1057 islets with a volume of 1.77 mm3). D and E, Example data from control (D) and the ob/ob mouse model for type 2 diabetes12 (E) at 6 months implementing the CLAHE protocol. Note the massive general increase in islet size in the ob/ob pancreas (E). In (D) and (E) the pancreas outline (gray) is based on the signal from tissue autofluorescence. Scale bar in C is 500 μm in A-C. Scale bar in C” corresponds 200 μm in C’ and C”. Scale bar in E corresponds to 1 mm in (D) and (E). Images in (A-C) are adapted from Hörnblad et al3 and were generated using the Bioptonics 3001 scanner.
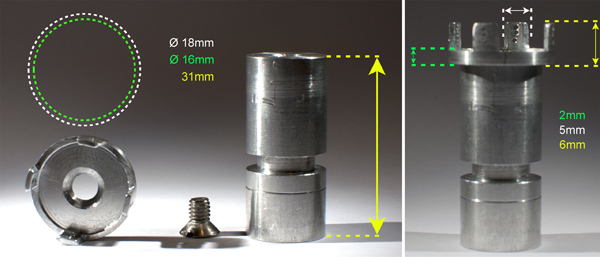
Figure 7. Sample holder for attachment of OPT specimens. The specimen is secured by inserting needles through the agarose spacer via the pre-drilled holes in the flanges. The holder is hinged to the stepper motor via a strong magnet located in its base. This setup omits the use of unstable glues and prevents unwanted movements of the specimen during scanning.
Discussion
The described techniques for OPT imaging enables extraction of spatial and quantitative parameters throughout the volume of the murine pancreas. Due to limitations in achievable resolution for this type of mesoscopic imaging it should be noted that, as for most imaging modalities, the larger the specimen the lower the resolution (Although the use of a higher resolution CCD should increase the resolution of the OPT scan). Hence, for the assessment of intact pancreatic mouse lobes, the technique at present does not provide single cell resolution although close (approx. 15-20 μm)7. Still, for the extraction of BCM distribution in the mouse pancreas the protocols have provided data that more than well match those obtained by e.g. point counting morphometry3,13 It should be noted that although implementation of the CLAHE protocol allows for detection of significantly more islets, these islets are generally smaller and do not contribute substantially to the overall β-cell volumes.
The immunohistochemical protocols involved are relatively lengthy (up to two weeks), but the actual hands on time for specimen preparation is short and therefore the technique is well suited for the study of large cohorts of animals9. If the potential of heterogenic distribution patterns is a focus for the investigation, it should be stressed that care should be taken in the steps concerning fixation and mounting to avoid that the pancreatic tissue becomes fixed in an unfavorable way and a flat (“spread out”) mount of the tissue should be strived for to facilitate such assessments.
An important issue when performing OPT is that the sample’s COM is fixed at the axis of rotation and that it does not move, either vertically or horizontally, during the scanning procedure. Therefore it is essential to have a stable mechanical setup and a well-functioning system for attaching the sample. We solved this issue by constructing a new mount (Figure 7).
Parallel geometry was not true for our NIR-OPT or Bioptonics 3001 scanner, which was detected as a vertical shift between the back and front positions of peripheral objects in the projection images recorded. By adjusting the object to source distance in the log file of the respective scanner (see 2.3.1) we could significantly improve the quality of our data and correct for geometric distortions at the far edges of the projection images, which is of particular importance when assessing larger specimens.
In the current protocol, we provide a suggestion of filter sets that allow visualization of three different specific channels and an “anatomy” channel in assessments of intact pancreatic preparations. Obviously these settings could be modulated to better fit the fluorochromes utilized for a given study although, as with all forms of fluorescent microscopy, the potential hazard of signal bleed-through should be carefully evaluated. The study of insulin labeled islets with fluorochromes that are excited above 750 nm has not yet been possible by us using the metal halide lamp that our set up utilizes. It is possible that a camera with even higher quantum efficiency in the pertinent wavelengths in combination with alternative light sources (e.g. diode lasers) could increase the potential of NIR-OPT further and allow for imaging at even higher wavelengths.
OPT imaging is a highly versatile technique for spatial and quantitative assessments of biomedical specimen on the mm-cm scale. Although the protocols presented here have been developed for the main purpose of pancreas/diabetes research they should be possible to translate to research on other species, specimen types and markers. By the potential to visualize several distinct channels in intact pancreatic preparations, NIR-OPT imaging further holds the potential as a tool to evaluate the uptake specificity of contrast agents intended for non-invasive assessments by other imaging modalities, as long as these contrast agents could be designed to carry also a fluorophore detectable by OPT.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
Dr. P. Lindström is acknowledged for providing ob/ob mice. J. Lehtonen is acknowledged for assistance with video production and J. Gilbert for help with editing. This study was supported by grants from the Diabetes Research Institute Foundation (A.P.), the Juvenile Diabetes Research Foundation (A.P. and U.A.), the European Commission (FP-7, Grant agreement no.: CP-IP 228933-2) (J.S. and U.A.), the Kempe Foundations, Umeå University and the Swedish research council to U.A.
Materials
| Name of Reagent/Material | Company | Catalog Number | Comments |
| Methanol | Scharlau | ME03162500 | |
| 30% H2O2 | Scharlau | HI01362500 | |
| Benzyl Alcohol | Scharlau | AL01611000 | |
| Benzyl Benzoate | Scharlau | BE01851000 | |
| Low-meltingpoint agarose | LONZA | 50100 | |
| Paraformaldehyde (PFA) | Sigma-Aldrich | 158127 | |
| DMSO | Sigma-Aldrich | D5879 | |
| Triton-X100 | Sigma-Aldrich | T8787 | |
| Mouse anti-aSMA-Cy3 | Sigma-Aldrich | C6198 | Primary antibody |
| Rabbit anti-CD3 | Sigma-Aldrich | C7930 | Primary antibody |
| Guinea Pig anti-Ins | DAKO | A0564 | Primary antibody |
| Donkey anti GP-IRDye680 | LI-COR Biosciences | 926-32421 | Secondary antibody |
| Goat anti Rb-DyeLight750 | Thermo Scientific | 35570 | Secondary antibody |
| Goat anti GP-Alexa594 | Molecular Probes | A-11076 | Secondary antibody |
| Goat anti GP-Alexa488 | Molecular Probes | A-11008 | Secondary antibody |
| Goat anti GP-Alexa594 | Molecular Probes | A-11012 | Secondary antibody |
| Goat anti GP-Alexa680 | Molecular Probes | A-21076 | Secondary antibody |
| Goat anti GP-Alexa750 | Molecular Probes | A-21039 | Secondary antibody |
| OPT Skyscan 3001 | Bioptonics | OPT-Scanner | |
| Leica MZ FLIII | Leica Microsystems | Stereomicroscope | |
| Leica Objective 0.5x | Leica Microsystems | 10446157 | |
| Leica Camera adapter 1.0x | Leica Microsystems | 10445930 | |
| EL6000 Metal Halide | 11504115 | Lightsource | |
| Liquid Light Guide | 11504116 | ||
| Cuvette | Hellma Analytics | 6030-OG | 55 x 55 x 52.5 mm |
| Mirror | Edmund Optics | F68-334 | 50 x 50 mm |
| Andor Ikon-M | Andor Technology | DU934N-BV | Back-illuminated CCD |
| Filterset | Chroma Technology | 41021-MZFLIII | TXR, Alexa-594, Cy3 |
| Filterset | Chroma Technology | 41022-MZFLIII | IRDye680, Alexa-680 |
| Filterset | Chroma Technology | 49037-MZFLIII | Dylight750, Alexa-750 |
| ProteinG-Sepharose beads | GE Healthcare | 17-0618-01 | Protein G Sepharose 4 Fast Flow |
| Sodium Azide | Sigma-Aldrich | 08591 | Sodium azide 0.1 M solution |
Referenzen
- Sharpe, J., et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 296, 541-545 (2002).
- Cheddad, A., Svensson, C., Sharpe, J., Georgsson, F., Ahlgren, U. Image Processing Assisted Algorithms for Optical Projection Tomography. IEEE Trans. Med. Imaging. , (2012).
- Hornblad, A., Cheddad, A., Ahlgren, U. An improved protocol for optical projection tomography imaging reveals lobular heterogeneities in pancreatic islet and beta-cell mass distribution. Islets. 3, 204-208 (2011).
- Holmberg, D., Ahlgren, U. Imaging the pancreas: from ex vivo to non-invasive technology. Diabetologia. 51, 2148-2154 (2008).
- Ahlgren, U., Gotthardt, M. Approaches for imaging islets: recent advances and future prospects. Adv. Exp. Med. Biol. 654, 39-57 (2010).
- Sharpe, J. Optical projection tomography. Annu. Rev. Biomed. Eng. 6, 209-228 (2004).
- Alanentalo, T., et al. Tomographic molecular imaging and 3D quantification within adult mouse organs. Nat. Methods. 4, 31-33 (2007).
- Alanentalo, T., et al. High-resolution three-dimensional imaging of islet-infiltrate interactions based on optical projection tomography assessments of the intact adult mouse pancreas. J. Biomed. Opt. 13, 054070 (2008).
- Alanentalo, T., et al. Quantification and Three-Dimensional Imaging of the Insulitis-Induced Destruction of beta-Cells in Murine Type 1 Diabetes. Diabetes. 59, 1756-1764 (2010).
- Sun, G., et al. Ablation of AMP-activated protein kinase alpha1 and alpha2 from mouse pancreatic beta cells and RIP2.Cre neurons suppresses insulin release in vivo. Diabetologia. 53, 924-936 (2010).
- Hornblad, A., Eriksson, A. U., Sock, E., Hill, R. E., Ahlgren, U. Impaired spleen formation perturbs morphogenesis of the gastric lobe of the pancreas. PLoS One. 6, e21753 (2011).
- Lindström, P. The physiology of the Obese-Hyperglycemic Mice (ob/ob Mice). The Scientific World JOURNAL. 7, 665-685 (2007).
- Bock, T., Pakkenberg, B., Buschard, K. Genetic background determines the size and structure of the endocrine pancreas. Diabetes. 54, 133-137 (2005).

