Mass Histology to Quantify Neurodegeneration in Drosophila
Summary
Drosophila is widely used as a model system to study neurodegeneration. This protocol describes a method by which degeneration, as determined by vacuole formation in the brain, can be quantified. It also minimizes effects due to the experimental procedure by processing and sectioning control and experimental flies as one sample.
Abstract
Progressive neurodegenerative diseases like Alzheimer's disease (AD) or Parkinson's disease (PD) are an increasing threat to human health worldwide. Although mammalian models have provided important insights into the underlying mechanisms of pathogenicity, the complexity of mammalian systems together with their high costs are limiting their use. Therefore, the simple but well-established Drosophila model-system provides an alternative for investigating the molecular pathways that are affected in these diseases. Besides behavioral deficits, neurodegenerative diseases are characterized by histological phenotypes such as neuronal death and axonopathy. To quantify neuronal degeneration and to determine how it is affected by genetic and environmental factors, we use a histological approach that is based on measuring the vacuoles in adult fly brains. To minimize the effects of systematic error and to directly compare sections from control and experimental flies in one preparation, we use the 'collar' method for paraffin sections. Neurodegeneration is then assessed by measuring the size and/or number of vacuoles that have developed in the fly brain. This can either be done by focusing on a specific region of interest or by analyzing the entire brain by obtaining serial sections that span the complete head. Therefore, this method allows one to measure not only severe degeneration but also relatively mild phenotypes that are only detectable in a few sections, as occurs during normal aging.
Introduction
With the increase in life expectancy, neurodegenerative diseases like Alzheimer's or Parkinson's have become an increasing health threat for the general population. According to the National Institutes of Health, 115 million people worldwide are predicted to be affected by dementia in 2050. Although significant progress has been made in identifying genes and risk factors involved in at least some of these diseases, for many of them, the underlying molecular mechanisms are still unknown or not well understood.
Simple invertebrate model organisms like Caenorhabditis elegans and Drosophila melanogaster offer a variety of experimental advantages to study the mechanisms of neurodegenerative diseases, including a short life cycle, large number of progeny, and the availability of well-established and sometimes unique genetic and molecular methods1-12. Furthermore, these organisms are amenable to unbiased interaction screens that can identify factors contributing to these diseases by their aggravating or ameliorating effects on neurodegenerative phenotypes.
Analyzing such genetic interactions and assessing aging effects requires quantitative protocols to detect neurodegeneration and to measure its severity. This assessment can be done relatively easily when measuring behavioral aspects in Drosophila, such as olfactory learning, negative geotaxis, or fast phototaxis, which provide a numeric performance value13-21. It is also possible to determine the effects on neuronal survival by counting neurons. However, this is only possible when focusing on a specific population that is clearly identifiable, like the dopaminergic neurons that are affected in PD, and even then, the results have been controversial22-24.
The protocol described here uses the collar method to perform paraffin serial sections, a method that was originally developed by Heisenberg and Böhl, who used it to isolate anatomical brain mutants in Drosophila25. The use of the collar method has subsequently been adapted, including in cryosections, vibratome sections, and plastic sections26-28. Here, this method is employed to obtain serial sections of the entire fly head, which can then be used to measure the vacuoles that develop in flies with neurodegenerative phenotypes16,21,29-32. These measurements can be done in specific brain areas or can cover the entire brain; the latter approach allows one to identify even weak degenerative phenotypes, as observed during aging. Finally, when using the collars, up to 20 flies can be processed as one preparation, which is not only less time-consuming, but also allows for the analysis of control and experimental flies in the same preparation, minimizing artifacts due to slight changes in the preparation.
Protocol
1. Fixing the Head on Collars and Embedding in Paraffin
Note: All of the steps in the fixation process should be done in a fume hood. Methylbenzoate, while not posing a health risk, has a highly distinct odor, which can be overwhelming if not handled in a fume hood.
- Before anesthetizing the flies, make up 50 mL of Carnoy solution by adding 15 mL of chloroform and 5 mL of glacial acetic acid to 30 mL of 99% ethanol (do not mix the chloroform and acetic acid). Pour it in a glass container with a flat bottom, such as a crystalizing dish, to ensure that the collars can lay flat and are completely covered by the solution.
- Anesthetize the flies with CO2 or ether.
- Thread flies (up to 20 with most collars) by their necks into the collars using forceps. Remember to align all of the heads in the same orientation, as seen in Figure 1A, and be gentle to ensure that no damage occurs to the head or eyes.
- Include sine oculis flies (arrows, Figure 1A) at random positions so that the order of the flies can be easily identified in the sections. In addition, if the experimental flies have a light or white eye color, thread some red-eyed flies, such as wild type, between them to ensure that sufficient pigment is present to stain the slide. Record the order of the flies on a protocol sheet together with the collar number if using more than one collar.
- Once a collar has been finished, place it in the prepared Carnoy solution for 3.5 – 4 h.
- Dump out the Carnoy solution into the appropriate disposal canister and begin the ethanol washes. Make sure to pour slowly so as not to disturb the placement of the collars in the container.
- Wash the collars for 30 min in 99% ethanol twice.
- Wash the collars in 100% ethanol for 1 h. Be sure to change the washes on time to prevent overdehydration.
- Put the collars in methylbenzoate O/N at RT. Seal the container with parafilm to prevent the evaporation of the methylbenzoate.
- Pour the methylbenozate into the proper disposable container in the fume hood. Add a previously-prepared mixture of 1:1 low melting point (56 – 57 °C) paraffin wax and methylbenzoate for 1h. From this point on, the collars need to be kept in an incubator at 65 °C to make sure that the paraffin does not harden.
- Pour out the methylbenozate and paraffin mixture into the proper disposable container, and pour molten pure paraffin wax, kept at 65 °C, onto the collars.
- Change the paraffin after 30 min and repeat this at least 5 times. At least 6 – 8 washes should be performed.
- Once the washes are complete, place the collars into a rubber ice cube tray with slots approximately the size of the collars. Pour molten paraffin over them until completely covered and allow it to harden O/N (try to avoid air bubbles).
- Remove the paraffin blocks containing the collars from the ice cube tray. Separate the paraffin block from the collar using a razor blade, gently breaking off the collar. The heads will be in the paraffin block while the bodies will stay in the collar. The blocks can be kept at room temperature.
- To clean the collars, soak them in a deparafinization agent at 65 °C to remove the paraffin, clean with light scrubbing, and wash in ethanol before reusing.
2. Sectioning and Mounting
- Warm a heating plate to 50 °C. Place the object holders (or metal mounting blocks) and razor blades on the plate and let them warm up.
- Depending on the desired orientation for sectioning, attach the paraffin block either with the heads towards the side (for horizontal sections) or facing upwards (for frontal sections) to a heated mounting block (briefly melting the block at the contact side). Remove the block from the heating plate and allow it to cool for at least 10 min to ensure that the paraffin is hardened enough for a proper seal onto the mounting block. Keep the row of heads aligned in parallel with the surface of the block as much as possible to prevent uneven sections.
- Take a razor blade and trim the excess paraffin away from the fly heads so that only a small row with the embedded heads remains (the razor blade can be warmed up for easier trimming). Make sure not to trim too much so that the paraffin does not break during sectioning (more trimming can be done during sectioning).
- Place the mounting block into the object holder of the microtome and ensure that the alignment of the row of heads is as parallel as possible to the edge of the blade.
- Prepare microscope slides by covering them with a thin layer of poly-L-Lysine (PLL) solution and let them dry for 5 min. Cover them with water shortly before use.
- Cut 7 µm sections and transfer the ribbon of sections to the slide floating on the water.
NOTE: To obtain the entire brain for horizontal sections, we collect the ribbon from when starting to cut into the eye until the head has been completely cut (cutting from the proboscis into the brain). More than one slide may be needed for the entire head. - Place the slide on a heat plate at 37 °C and allow the ribbon to expand for about 1 min.
- Remove excess water (by pouring it off or using a tissue) and dry the slides O/N.
- Remove the paraffin wax from the slide by placing the slides in a tall, vertical slide-staining jar filled with a deparafinization agent (completely covering the sections). Perform 3 washes of 30 min – 60 min each.
- Remove the slide from the final wash. Place 2 drops of embedding media onto the slide and cover it with a large coverslip.
3. Photographing and Analyzing the Sections
- Allow the prepared slides to dry for 1 – 2 d. Then, examine them on a fluorescence microscope under blue light.
- Use a lower magnification to determine the orientation of the flies and to find the region of interest if focusing on a specific region.
NOTE: For the sws flies (see Figure 2), we find the section that contains the great commissure and take an image (usually at 40X magnification). When analyzing the entire brain (as in Figure 3), we scroll through all the sections from a head and either photograph the section with the most severe phenotype or all sections that show vacuoles. - For a double-blind analysis, take and number images without knowing the genotype and record the collar number and position of the head in the row to identify them later.
- Once the images have been taken, analyze them using an imaging software.
- Count the number of vacuoles per section or per head. To measure the vacuole size, open the images in a software program and select the vacuoles with a selection tool. Determine the amount of pixels in the selected vacuoles.
- For conversion to µm2, take a photo of a stage micrometer calibration slide at the magnification used for acquiring photos. Determine the amount of pixels in 100 µm2 to calculate a conversion factor.
- Convert the total pixel number into µm2 by dividing the number of pixels by the conversion factor calculated in the above step.
Representative Results
Using the described method results in serial sections stained by the eye pigment33 that encompass the entire fly head. A part of this is shown in Figure 1B, where the sections from an individual head are shown from top to bottom. The sections from different flies are seen left to right in this example. To facilitate orientation and identification of the flies, an eyeless fly (sine oculis) is inserted as a marker at position 3 (arrow, Figure 1B).
To quantify neurodegeneration, we measure the formation of vacuoles that can be detected in these sections. Vacuoles are defined as round, dark spots that are within the green fluorescent neuropil (arrowheads in Figure 2 and 3) or cortex and that are visible in at least 2 consecutive sections of the fly brain. Quantifying neurodegeneration by measuring vacuoles can either be done by focusing on a specific brain region or by analyzing the entire brain. Limiting the analysis to a specific region of the brain is useful in cases where a mutation only affects a specific region, like the olfactory lobes in the futsch'olk mutant34, but it can also be used when there is severe degeneration in all or many regions of the brain. An example for the latter is the swiss cheese (sws) mutant (Figure 2), where measuring all vacuoles would be too time consuming. We therefore took only one image and, to ensure that the measurements were always done at the same level, we took all images at the level of the great commissure (gc, Figure 2A), which is only contained in one or two sections. Whereas we did not detect vacuole formation in 1-day-old sws'1 flies (data not shown), a loss-of-function allele35, some vacuoles were detectable in 7-day-old sws'1 flies (arrowheads, Figure 2A). Aging the flies to 14 d (Figure 2B) and 21 d (Figure 2C) further increased this phenotype, showing its progressive nature. Counting the number of vacuoles in the deutocerebral neuropil (dn) using the described method confirmed a significant increase in the number of vacuoles with age. Also, the combined area encompassed by vacuoles was significantly increased with aging (Figure 2D).
However, not all mutants show such a severe phenotype as sws, and in those cases, differences in degeneration are difficult to determine when focusing on a small area. Similarly, the degeneration that occurs during aging is quite mild (Figure 3A – C) and therefore, we analyzed the entire brain when quantifying this phenotype. Determining the sum of all vacuoles in the brain revealed a significant increase with age, and this was also the case when measuring the combined area of these vacuoles (Figure 3D).
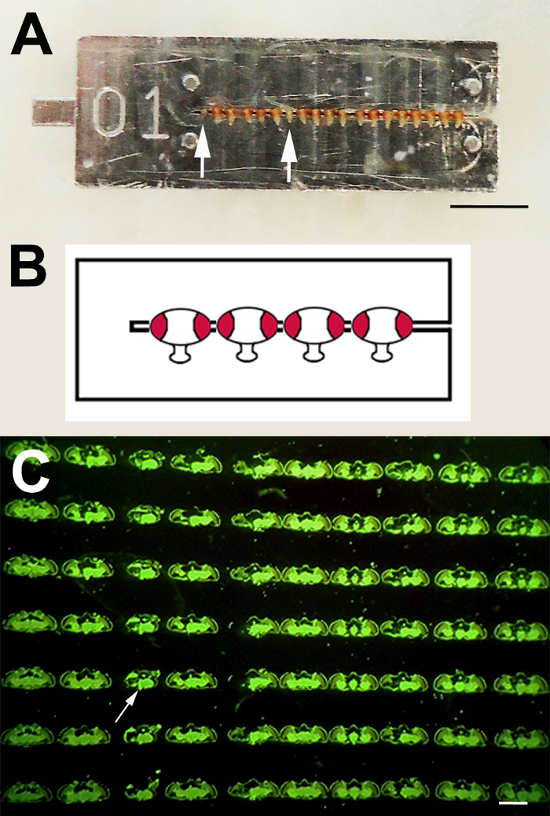
Figure 1. Paraffin Serial Sections. A) Using the collar method, experimental and control flies can be processed as one sample by threading them onto one collar. Eyeless sine oculis flies are inserted for orientation (arrows). B) Schematic showing the orientation of the fly heads in the collar. C) In this image, sections from different fly heads are oriented left to right on the slide. From top to bottom on the slide, serial sections from the same fly head can be seen. In this case, a sine oculis fly was inserted at position three (arrow). The sections are stained by the fluorescent eye pigment that washes over the sections after cutting. Scale bar in A = 5 mm and in C = 0.5 mm. Please click here to view a larger version of this figure.
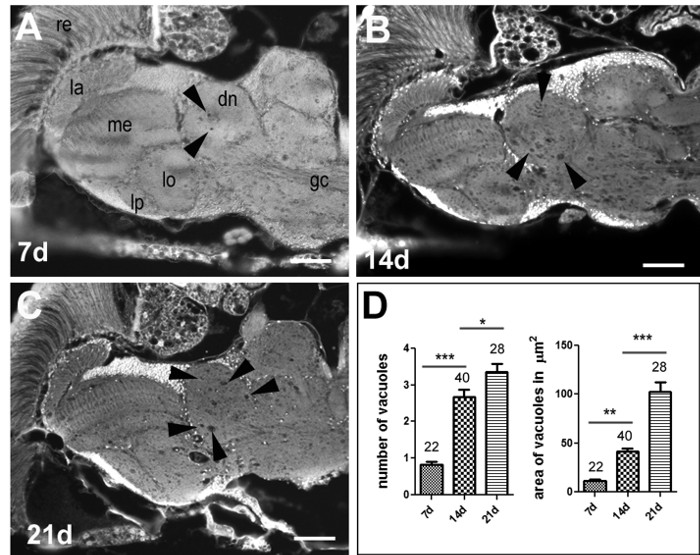
Figure 2. Progressive Neurodegeneration in the Swiss-cheese Mutant. Paraffin head section from 7-day- (A), 14-day- (B) and 21-day- (C) old sws'1 flies. The arrowheads point to vacuoles that have developed with aging. The age-related degeneration is quantified by counting the number of vacuoles and measuring their combined area (D). The SEM and number of analyzed flies is indicated. The scale bar = 25 µm. ***p <0.001. Please click here to view a larger version of this figure.
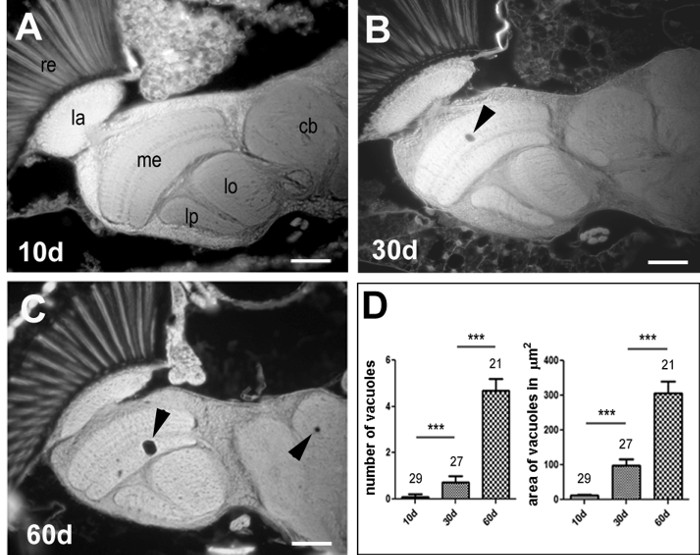
Figure 3. Neurodegeneration Occurs with age. Paraffin head section from 10-day- (A), 30-day- (B) and 60-day- (C) old wild-type flies. The arrowheads point to vacuoles that have developed in aged flies. The age-related degeneration is quantified by counting the number of vacuoles and measuring their combined area (D). The SEM and number of analyzed flies is indicated. The scale bar = 25 µm. ***p <0.001. Please click here to view a larger version of this figure.
Discussion
The described method provides a means to quantify neurodegeneration in the brain of Drosophila. While other methods, like counting a specific cell type, can be used to identify neurodegeneration, the advantage of this method is that it can be applied more generally. Counting cells requires that these cells can be reliably identified using either a specific antibody or the expression of a cell-specific marker, which is not always available. Furthermore, it has been shown that dramatically different results can be obtained with that method24, presumably due to the conditions used for labelling and for detection. Another method to detect degenerating cells is the use of cell death markers, like anti-activated caspase 3. However, this only identifies cells actively undergoing cell death and once the cells have died, they are no longer detectable. Another advantage of the method proposed here is that no staining is required due to the autofluorescence caused by the eye pigment, which saves time and minimizes artifacts caused by changes in the staining conditions. It is important to note that, when using this method, the flies' eyes have enough pigment to stain the slide. If the eye color is too light, adding some wild-type flies to the collar would be advisable to ensure sufficient and even staining across the slide. An additional advantage of this method is that multiple areas can be examined, even in the same head. Although we do not show the data, we have used these sections to examine neurodegeneration in the retina and glial cell loss in the lamina cortex30,36. As described here, this method provides horizontal sections, but by turning the paraffin block by 90° when melting it onto the object holder, one can also obtain frontal sections. Thus, this method is a versatile procedure that allows for the timely and efficient measurement of neurodegeneration in the fly brain. Because the formation of vacuoles has been observed in many fly models of human neurodegenerative diseases, including models for AD, PD, Amyotrophic Lateral Sclerosis (ALS), and diseases caused by polyglutamine-repeats16,37-40, this method can be used to quantify neurodegenerative phenotypes in a variety of disease models.
Overall, this protocol is simple and easily completed once the initial setup of the equipment is carried out. Some notes to keep in mind are to carefully time the ethanol washes to avoid over-dehydration of the heads, to carefully trim the paraffin blocks so as not to lose sections or heads, and to not overexpand the ribbon on the water when the slide is on the heat block. If the ribbon is allowed to expand too far, tearing of the fly heads can result and the order of the heads in the ribbon can be lost. In addition, being blind to the genotype while doing the analyses is essential to avoid bias. This is best achieved by having one person preparing the slides and keeping the records while another person is taking the pictures and doing the measurements. One of the limitations of this method is that degeneration of only a few cells will be very difficult to detect. In that case, a specific stain of the affected cell population would be more informative. In addition, this method does not allow one to distinguish between different kinds of cell death, which requires more specific methods, such as a TUNEL staining to determine apoptotic cell death. Lastly, this method cannot differentiate between cell death and axonal degeneration, which would also be detectable as vacuoles in the neuropil.
As shown in our results, this method can be used to address degeneration in specific brain areas or in the entire brain. In our experience, it is useful to analyze only a specific area when the phenotype is quite strong, even when all brain regions are affected. This significantly reduces the work load and does not affect the outcome. We originally analyzed several areas in the sws mutant and observed very similar results in the progression of the phenotype when comparing only one area and when analyzing all areas (data not shown). However, it should be noted that a clearly-identifiable region should be chosen to avoid artifacts due to the analysis of different areas or areas at different levels.
In contrast, in cases where the phenotype is relatively mild, it is better to analyze the entire brain, because the likelihood of finding vacuoles in a specific region is low. For example, this is the case when determining age-related neurodegeneration, as shown in Figure 3, which results in only 4 – 5 vacuoles in the entire brain in 60-day-old flies. When counting vacuoles in the entire brain, one has to take into account that small vacuoles will only show up in one section, whereas larger ones will extend over several sections. Concerning the latter, the close proximity of adjacent sections when using this method (Figure 1) provides another advantage, because it is relatively easily to determine whether the same vacuole is present in several sections.
In conclusion, this protocol can prove useful for studying many Drosophila models of different neurodegenerative diseases. Identifying interacting proteins that ameliorate or aggravate the degenerative phenotype can provide crucial insights about the underlying mechanisms that cause or modify diseases such as Alzheimer's and Parkinson's. In this publication, we use this method to detect degeneration that is progressive, where the animals develop normally but show increasing degeneration during aging. Additionally, this method can also be adapted to determine degeneration that is caused by developmental defects, which should already be present in newly-eclosed flies.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants to D.K. from the Medical Research Foundation of Oregon and from NIH/NINDS (NS047663). E.S. was supported by a training grant from the NIH (T32AG023477).
Materials
| Name of the Reagent/Equipment | Company | Catalog Number | |
| Collar | Genesee Scientific | TS 48-100 | We are using custom made collars that are made from one piece of metal instead of layers as the ones by Genesee. A discription to make collars can be found at http://flybrain.neurobio.arizona.edu/Flybrain/html/atlas/fluorescent/index.html |
| Rubber ice cube tray for embedding | Household store | The size can be made to fit by glueing in additional walls | |
| Crystallizing dish | Fisher Scientific company | 08-762-3 | |
| Ether | Fisher Scientific Company | E138-1 | |
| Ethanol | Decon Laboratories Inc. | 2701 | |
| Choloroform | Fisher Scientific Company | C298-500 | |
| Glacial Acetic Acid | Fisher Scientific Company | A38-212 | |
| Methylbenzoate | Fisher Scientific Company | M205-500 | Distinct Odor |
| Use in fume hood | |||
| Low Melting Point Paraffin Wax | Fisher Scientific Company | T565 | Make sure to keep extra melted in a 65°C waterbath |
| Microtome | Leica Biosystems | Reichert Jung 2040 Autocut | |
| Microscope Slide | Fisher Scientific Company | 12-550D | |
| Microscope Cover Glass | Fisher Scientific Company | 12-545-M | |
| SafeClear | Fisher Scientific Company | 314-629 | Three different containers for washes |
| Vertical Staining Jar with Cover | Ted Pella Inc. | 432-1 | |
| Permount | Fisher Scientific Company | SP15-500 | |
| Poly-L-lysine Solution | Sigma Life Science | P8290-500 |
Referenzen
- Alexander, A. G., Marfil, V., Li, C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front Genet. 5, 279 (2014).
- Bonini, N. M., Fortini, M. E. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 26, 627-656 (2003).
- Calahorro, F., Ruiz-Rubio, M. Caenorhabditis elegans as an experimental tool for the study of complex neurological diseases: Parkinson’s disease, Alzheimer’s disease and autism spectrum disorder. Invert Neurosci. 11, 73-83 (2011).
- Chen, X., Barclay, J. W., Burgoyne, R. D., Morgan, A. Using C. elegans to discover therapeutic compounds for ageing-associated neurodegenerative diseases. Chemistry Central Journal. 9, 65 (2015).
- Gama Sosa, M. A., De Gasperi, R., Elder, G. A. Modeling human neurodegenerative diseases in transgenic systems. Hum Genet. 131, 535-563 (2012).
- Jaiswal, M., Sandoval, H., Zhang, K., Bayat, V., Bellen, H. J. Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu Rev Genet. 46, 371-396 (2012).
- Konsolaki, M. Fruitful research: drug target discovery for neurodegenerative diseases in Drosophila. Expert Opin Drug Discov. 8, 1503-1513 (2013).
- Kretzschmar, D. Neurodegenerative mutants in Drosophila: a means to identify genes and mechanisms involved in human diseases. Invert Neurosci. 5, 97-109 (2005).
- Kretzschmar, D., et al. Swiss cheese et allii, some of the first neurodegenerative mutants isolated in Drosophila. J Neurogenet. 23, 34-41 (2009).
- Li, J., Le, W. Modeling neurodegenerative diseases in Caenorhabditis elegans. Exp Neurol. 250, 94-103 (2013).
- Prussing, K., Voigt, A., Schulz, J. B. Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol Neurodegener. 8, (2013).
- Wentzell, J., Kretzschmar, D. Alzheimer’s disease and tauopathy studies in flies and worms. Neurobiol Dis. 40, 21-28 (2010).
- Ali, Y. O., Escala, W., Ruan, K., Zhai, R. G. Assaying locomotor, learning, and memory deficits in Drosophila models of neurodegeneration. J Vis Exp. , (2011).
- Barone, M. C., Bohmann, D. Assessing neurodegenerative phenotypes in Drosophila dopaminergic neurons by climbing assays and whole brain immunostaining. J Vis Exp. , e50339 (2013).
- Dutta, S., Rieche, F., Eckl, N., Duch, C., Kretzschmar, D. Glial expression of Swiss-cheese (SWS), the Drosophila orthologue of Neuropathy Target Esterase, is required for neuronal ensheathment and function. Dis Model Mech. , (2015).
- Iijima, K., et al. Abeta42 mutants with different aggregation profiles induce distinct pathologies in Drosophila. PLoS One. 3, e1703 (2008).
- Krishnan, N., et al. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol Dis. 45, 1129-1135 (2012).
- Liu, H., et al. Automated rapid iterative negative geotaxis assay and its use in a genetic screen for modifiers of Abeta(42)-induced locomotor decline in Drosophila. Neurosci Bull. 31, 541-549 (2015).
- Papanikolopoulou, K., Skoulakis, E. M. Temporally distinct phosphorylations differentiate Tau-dependent learning deficits and premature mortality in Drosophila. Hum Mol Genet. 24, 2065-2077 (2015).
- Ping, Y., et al. Linking abeta42-induced hyperexcitability to neurodegeneration, learning and motor deficits, and a shorter lifespan in an Alzheimer’s model. PLoS Genet. 11, e1005025 (2015).
- Sujkowski, A., Rainier, S., Fink, J. K., Wessells, R. J. Delayed Induction of Human NTE (PNPLA6) Rescues Neurodegeneration and Mobility Defects of Drosophila swiss cheese (sws) Mutants. PLoS One. 10, e0145356 (2015).
- Barone, M. C., Sykiotis, G. P., Bohmann, D. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson’s disease. Dis Model Mech. 4, 701-707 (2011).
- Coulom, H., Birman, S. Chronic exposure to rotenone models sporadic Parkinson’s disease in Drosophila melanogaster. J Neurosci. 24, 10993-10998 (2004).
- Navarro, J. A., et al. Analysis of dopaminergic neuronal dysfunction in genetic and toxin-induced models of Parkinson’s disease in Drosophila. J Neurochem. 131, 369-382 (2014).
- Heisenberg, M., Böhl, K. Isolation of anatomical brain mutants of Drosophila by histological means. Zeitschrift für Naturforschung C. 34, 143 (1979).
- Han, P. L., Meller, V., Davis, R. L. The Drosophila brain revisited by enhancer detection. J Neurobiol. 31, 88-102 (1996).
- Lin, C. W., et al. Automated in situ brain imaging for mapping the Drosophila connectome. J Neurogenet. 29, 157-168 (2015).
- Strausfeld, N. J., Sinakevitch, I., Vilinsky, I. The mushroom bodies of Drosophila melanogaster: an immunocytological and golgi study of Kenyon cell organization in the calyces and lobes. Microsc Res Tech. 62, 151-169 (2003).
- Bettencourt da Cruz, A., Wentzell, J., Kretzschmar, D. Swiss Cheese, a protein involved in progressive neurodegeneration, acts as a noncanonical regulatory subunit for PKA-C3. J Neurosci. 28, 10885-10892 (2008).
- Bolkan, B. J., Kretzschmar, D. Loss of Tau results in defects in photoreceptor development and progressive neuronal degeneration in Drosophila. Dev Neurobiol. 74, 1210-1225 (2014).
- Cook, M., Mani, P., Wentzell, J. S., Kretzschmar, D. Increased RhoA prenylation in the loechrig (loe) mutant leads to progressive neurodegeneration. PLoS One. 7, e44440 (2012).
- Wittmann, C. W., et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 293, 711-714 (2001).
- Rasmuson, B., Green, M. M., Ewertson, G. QUALITATIVE AND QUANTITATIVE ANALYSES OF EYE PIGMENTS AND PTERIDINES IN BACK-MUTATIONS OF THE MUTANT wa IN DROSOPHILA MELANOGASTER. Hereditas. 46, 635-650 (1960).
- Bettencourt da Cruz, A., et al. Disruption of the MAP1B-related protein FUTSCH leads to changes in the neuronal cytoskeleton, axonal transport defects, and progressive neurodegeneration in Drosophila. Mol Biol Cell. 16, 2433-2442 (2005).
- Kretzschmar, D., Hasan, G., Sharma, S., Heisenberg, M., Benzer, S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 17, 7425-7432 (1997).
- Dutta, S., Rieche, F., Eckl, N., Duch, C., Kretzschmar, D. Glial expression of Swiss cheese (SWS), the Drosophila orthologue of neuropathy target esterase (NTE), is required for neuronal ensheathment and function. Dis Model Mech. 9, 283-294 (2016).
- Davis, M. Y., et al. Glucocerebrosidase Deficiency in Drosophila Results in alpha-Synuclein-Independent Protein Aggregation and Neurodegeneration. PLoS Genet. 12, e1005944 (2016).
- Dias-Santagata, D., Fulga, T. A., Duttaroy, A., Feany, M. B. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J Clin Invest. 117, 236-245 (2007).
- Kretzschmar, D., et al. Glial and neuronal expression of polyglutamine proteins induce behavioral changes and aggregate formation in Drosophila. Glia. 49, 59-72 (2005).
- Li, Y., et al. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 107, 3169-3174 (2010).

