Extracellular Protein Microarray Technology for High Throughput Detection of Low Affinity Receptor-Ligand Interactions
Summary
Here, we present a protocol to screen extracellular protein microarrays for identification of novel receptor-ligand interactions in high throughput. We also describe a method to enhance detection of transient protein-protein interactions by using protein-microbead complexes.
Abstract
Secreted factors, membrane-tethered receptors, and their interacting partners are main regulators of cellular communication and initiation of signaling cascades during homeostasis and disease, and as such represent prime therapeutic targets. Despite their relevance, these interaction networks remain significantly underrepresented in current databases; therefore, most extracellular proteins have no documented binding partner. This discrepancy is primarily due to the challenges associated with the study of the extracellular proteins, including expression of functional proteins, and the weak, low affinity, protein interactions often established between cell surface receptors. The purpose of this method is to describe the printing of a library of extracellular proteins in a microarray format for screening of protein-protein interactions. To enable detection of weak interactions, a method based on multimerization of the query protein under study is described. Coupled to this microbead-based multimerization approach for increased multivalency, the protein microarray allows robust detection of transient protein-protein interactions in high throughput. This method offers a rapid and low sample consuming-approach for identification of new interactions applicable to any extracellular protein. Protein microarray printing and screening protocol are described. This technology will be useful for investigators seeking a robust method for discovery of protein interactions in the extracellular space.
Introduction
The method reviewed here describes the printing of a collection of extracellular proteins in a microarray format, followed by a method for screening of a target of interest against this library. We have identified protein multimerization as a crucial step for detection of interactions characterized by low binding affinities. To enhance detection of these interactions, we describe a protocol based on multimerization of the query protein of interest using microbeads.
Secreted and cell surface-expressed proteins (collectively termed extracellular proteins) along with their interacting partners are key regulators of cellular communication, signaling and interaction with the microenvironment. They are, therefore, essential in regulating many physiological and pathological processes. Approximately a quarter of the human genome (≈5,000 proteins) encodes for extracellular proteins, which, given their significance and accessibility to systematically delivered drugs, represent key targets for drug development1. Consequently, extracellular proteins represent more than 70% of the protein targets with known pharmacological action for approved drugs on the market, known as the "druggable proteome". Despite their importance and abundance, the extracellular protein-protein interaction (ePPI) networks remain remarkably underrepresented in the available databases. This is fundamentally due to the complex biochemical nature of the extracellular proteins, which precludes their characterization using most available technologies2. Firstly, membrane proteins are difficult to solubilize, a process that often involves harsh washing conditions and detergents; secondly, extracellular proteins often lack relevant post-translational modifications such as glycosylation that are absent when these proteins are expressed in commonly used heterologous systems. Finally, interactions between receptors, such as co-receptors expressed on immune cells, are often transient and characterized by very low affinities (KD in the ~1 μM to >100 μM range). Altogether, the nature of these proteins and their binding partners render most widely utilized technologies, such as affinity purification/mass spectrometry (AP/MS) or yeast-two-hybrid, unsuitable for detection of interactions in the extracellular space2,3.
In an effort to overcome these technical challenges and accelerate the discovery of novel interactions for extracellular proteins, we have developed a high coverage extracellular protein microarray4,5. Microarrays offer the advantage of generating high-density surfaces with small amounts of sample, and are generally amenable to high throughput studies. Protein microarray-based studies have previously provided relevant insights into protein interactions for several model organisms, albeit mainly focusing on cytosolic interactions or on specific protein families6,7,8. In contrast, limited work has been done to investigate extracellular protein interactions using this technology. We have developed a protein microarray method to enable studies of ePPIs by building a comprehensive and highly diverse library of purified secreted proteins and single transmembrane (STM) receptors expressed as recombinant extracellular domains (ECD) fused to common tags for affinity purification4. The success of the protein microarray screens relies heavily on the establishment of a high quality protein library. For expression of both the library and query protein, mammalian cells or insect cells were preferentially chosen as heterologous expression systems, to ensure proper addition of post-translational modifications such as glycosylation or disulphide bonds. SDS-PAGE, size exclusion chromatography and multi-angle laser light scattering are techniques commonly utilized to assess recombinant protein quality. The protein library is then spotted onto epoxysilane slides and stored at -20 °C for long-term use. Protein concentrations above of 0.4 mg/mL are recommended for the protocol described below. Therefore, low-expressing proteins may require a concentration step prior to sample printing and storage. Nevertheless, a main advantage of this technique is the small volume of protein required (50 μg of protein is sufficient to perform >2,000 screens), alongside minimal query protein consumption (20-25 μg per duplicates screen). Using the protocol and equipment described here, and provided libraries are available, results for individual query proteins can be generated within one working day.
A major challenge in detecting protein interactions in the extracellular environment arises from their characteristically weak or transient nature, which precludes identification by most commonly used methodologies. Increasing the binding avidity greatly improves sensitivity for detection of weak protein interactions9,10,11. Based on this principle we developed a method to multimerize the query proteins (expressed as Fc fusion) using protein A-coated beads4,5. To avoid any potential inactivation of the query protein by random labeling, we instead label an irrelevant human immunoglobulin G with Cy5 and add it along with the query protein to the protein A beads, thus eliminating any artifacts due to the direct conjugation of a dye to the protein of interest. Given the micromolar affinities of several co-receptor pairs, the multivalent complexes greatly enhance signal to noise ratio, compared to Fc-fusion query proteins screened as soluble proteins4.
In summary, the goal of this protocol is to describe the preparation of microarray slides containing a pre-existing extracellular protein library for identification of receptor-ligand interactions. We review the steps for slide printing, followed by a protocol for screening of a protein of interest against the extracellular protein library. Moreover, we describe a method for enhanced detection of ePPIs based on microbeads to achieve increased avidity of the protein under study. The extracellular protein microarray technology described here represents a fast, robust and effective approach for screening and detecting novel ePPI with low false-positive ratios, and by utilizing only microgram quantities of the query protein under investigation. This technology has fueled multiple studies that have provided relevant insights into previously unknown cellular functions and signaling pathways for a variety of receptors12,13, including viral immunoregulators14, and can be utilized to de-orphanize any extracellular protein of interest.
Protocol
1. Generation of a Library of Extracellular Human Proteins
- Compile a list of cell surface receptors or secreted proteins of interest to build the protein microarray library. Specific protein families (for example, the immunoglobulin superfamily) or proteins selectively expressed in particular cell types can be selected for the study.
- For cell surface receptors, determine the extracellular domain (ECD) boundaries by identifying the signal peptide and transmembrane regions using software tools. Some of the relevant tools, freely available online, are referenced in the Table of Materials15,16,17,18.
- Synthesize the ECD for the genes of interest and clone into the relevant vector. Secreted proteins or the ECD of STM receptors fused to a number of common affinity tags can be purified from the conditioned media of cells transfected with the appropriate vector, or from baculovirus-insect cell expression systems.
Note: Mammalian cell-based systems (such as HEK/293 or CHO cells) are recommended to maximize the likelihood of proper protein folding and addition of relevant posttranslational modifications such as glycosylation. - Purify the proteins by standard affinity purification methods. Previous efforts have described automated or semi-automated procedures suitable for purification of hundreds of proteins, which can be scaled to generate the set of proteins of interest9,19,20,21.
Note: SDS-PAGE, size exclusion chromatography (S), or multi-angle laser light scattering (MALLS) are recommended to assess any non-covalent aggregation and to control for the overall quality of the protein preparations. - Adjust proteins to 200 or 400 μg/mL whenever possible, and dilute stocks with 80% glycerol for long-term storage in cryogenic vials at -20 °C. These represent master stocks and should be accessed only when necessary.
- Transfer aliquots of each protein (100 μL) to 96-well plates (stock plates), seal with adhesive foil, and store at -20 °C until microarray slide preparation. Stock plates are generated to minimize freeze-thaw cycles and ensure protein stability.
2. Extracellular Protein Microarray Printing
- Use a standard microarrayer for slide printing. The instrument utilized for the protocol described here has a capacity of 57 slides/run and uses a printhead holding 48 spotting pins. These pins generate spots of ~100 μm diameter separated by a spot-to-spot distance of ~350 μm and draws 0.25 μL per load. Under this configuration, up to 8,000 spots per slide can be accommodated.
- Generate working plates (384 well plates, 10 μL sample/well) from the stock plates. Perform this step manually prior to slide printing using the microarrayer. Then, spot proteins with quill-type spotting pins onto epoxysilane slides at 60% humidity (to prevent dehydration of the protein spots).
Note: Cy3-labeled Bovine serum albumin (BSA) can be spotted in duplicates between each protein sample (5 μg/mL in PBS/40% glycerol) to visualize the array for mask fitting (see section 4). Although recommended, this step is optional. - Subsequent to printing, remove the protein microarray slides from the humidified environment and block them overnight with 5% milk in PBST to inactivate the surface.
Note: The preferred approach is to use an ultrasonic fogger to generate a fine mist of blocking solution that settles onto the slide surface. - Store slides at -20 °C in 50% glycerol to prevent freezing.
3. Preparation of Multivalent Bait Complexes
Note: Interactions between extracellular proteins are often characterized by low affinities. To enable detection of these interactions by increasing binding avidity, a multivalent approach based on capturing the query protein, expressed as Fc-tagged ECD, on protein A-coated microbeads was developed4.
- Label the carrier IgG used for detection with Cy5 monoreactive dye and separate the free dye using desalting columns. Determine the dye to protein ratios by measuring ultraviolet absorbance at 280 and 650 nm.
Note: Protein-dye ratios between 2.0 and 4.0 are normally used. It is recommended to spin the Cy5 conjugates at 100,000 x g for 15 min to remove potential soluble aggregates due to the labeling process. - Determine the optimal microbead-to-protein ratio by titration of protein A against a constant amount of the query protein. The minimal saturating volume of beads where no free Fc-tagged protein remains, as measured using a competitive assay determined by biolayer interferometry, is used for the screening.
- Form the microbead-protein complexes by incubating the Fc-tagged query and the Cy5-IgG with protein A microbeads and mix on a tube rotator in PBS for 30 min at room temperature protected from light.
- To form these complexes, use the query protein at a final concentration of 20 μg/mL. To calculate the molar ratio of the query protein and Cy5-IgG, divide the molecular weight of the query protein by the molecular weight of the IgG (150,000 Da) and multiply by 40 μg/mL to determine the concentration of Cy5-IgG needed.
- Supplement samples with soluble protein A (1 mg/mL) immediately prior to incubation with the microarray slides (see 4.2 section below) to prevent binding of any free protein A beads to the Fc fusion proteins that may be present on the array.
Note: The final reaction volume may vary depending on the hybridization station or incubation chamber utilized. The instrumentation described in the Table of Materials allows sample incubation using relatively small volumes (~200 μL per slide).
4. Extracellular Protein Microarray Screening and Processing.
Note: There are a number of manufacturers that provide automatic processing platforms. If a hybridization station is not available, the following steps can be performed manually, ensuring that there is sufficient volume of buffer to keep the slides submerged at all times.
- Warm the slides at room temperature and rinse with PBS/0.1% Tween 20 (PBST) to remove residual glycerol before loading onto the hybridization station.
- Use the following screening protocol:
- Wash with PBST for 1 min.
- Load 200 μL of 1 mg/mL protein A in PBS/5% milk and incubate for 30 min to prevent uncomplexed protein A microbeads from binding to Fc-tagged proteins that may be present in the microarray.
- Wash five times with PBST for 1 min.
- Load 200 μL of the query:microbeads complex in the presence of 1 mg/mL protein A and incubate for 30 min.
- Wash with PBST for 1 min.
- After hybridization, rinse slides with water, place in individual 50-mL conical tubes, and dry by spinning at 900 x g for 5 min.
- Finally, scan the slides with a microarray scanner appropriate to detect Cy3 (if BSA-Cy3 has been printed) and Cy5 fluorescence by exciting at 532 and 635 nm, respectively.
- Perform data analysis using the accompanying microarray data analysis. The relevant array list (as a .gal file) is loaded into the data extraction software. At this stage, utilize the Cy3-BSA spots to find blocks and use the auto-fitting options in the software, followed by manual alignment of the features when necessary. Save files as .gpr files for further analysis.
5. Data Analysis
- Save data as GPR files and process in R using the limma package, commonly utilized for analysis of microarray data4,5.
- For preprocessing, do background correction and within-slide normalization. Since each protein is printed in duplicated spots, combine both replicate measurements to create a single score for that protein in the library.
- Calculate scores for each slide and analyze results for the intersection of high-scoring candidates between slides.
- Use duplicate microarrays from separate print-runs to control for slide variability and use intersection plots representing data from both screens to call final hits.
- Finally, perform an additional level of filtering to exclude promiscuous proteins within the array. Such non-specific binders are identified as proteins exceeding a specific threshold hit frequency as defined by the user across independent screens.
Note: This protocol is described in detail in Tom et al. 20135. In our case the non-specific binder calling is based on the determination of the cumulative prey hit rate, and a data-driven elimination threshold of 5% is used.
Representative Results
A schematic of the workflow for the extracellular protein microarray technology is shown in Figure 1. Once the microarray slides containing the extracellular protein library are available, the screening of the protein of interest and data analysis can be completed within one day. Many physiologically relevant interactions between membrane-embedded receptors are characterized by very weak binding strengths (KD in the micromolar range). To improve detection of such interactions, a method based on query protein (expressed as Fc fusion) multimerization on protein A-microbeads was developed. This protocol is described in the text, and a representative example of the performance of this approach for enhanced detection of PPI is shown in Figure 2.
A representative example of a protein microarray screen is presented in Figure 3. In the example shown, an orphan human adenovirus immunomodulatory protein is screened for interactions against a library consisting of more than 1,500 receptor ECD or secreted proteins14. As described, the ECD of the viral protein is recombinantly expressed as an Fc fusion protein, and then incubated with protein A-coated microbeads in order to form multivalent complexes. The protein complexes were screened using a hybridization station to minimize manual operations, however, similar screens can be performed using a hybridization chamber or similar devices. It is advised to run duplicate screens using slides printed in separate spotting runs in order to control for any variability during the print run. The figure shows the resultant intersection plots from duplicate, independent screens. Positive interactions and overall slide background are readily visualized upon scanning of the plates (Cy5 fluorescence), facilitating hit calling. Note that it is recommended to spot each receptor in the library in duplicates so as to increase hit confidence. For most query proteins, none, one, or few hits are observed, demonstrating the specificity of the method for detection of PPIs.
Certain query proteins show high background as detected by binding to an unusually high number of proteins within the array and/or to the slide. In our experience, such non-specific background is observed for only a small number of proteins. Different factors relating to the nature of the protein could influence non-specific interactions, such as recognition of glycosylation motifs. Figure 3 shows an example of a viral protein screened against the microarray library that showed high background, precluding identification of specific hits. While minor modifications in the protocol can be considered to decrease background (such as increased salt or detergent concentration), in these instances it is recommended to explore alternative methods for deorphanization of the protein under study.
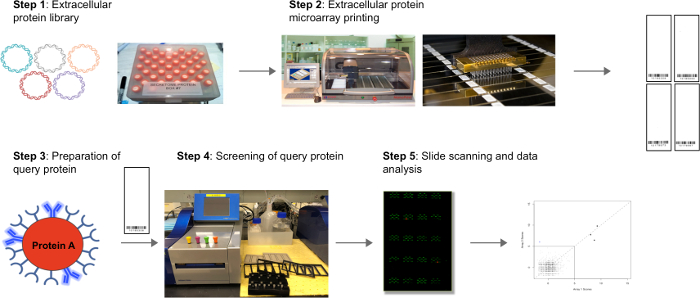
Figure 1: Extracellular protein microarrays for rapid and robust identification of receptor-ligand interactions. (Step 1) A library of extracellular proteins of interest is compiled. (Step 2) The purified proteins are printed on epoxysilane slides using a microarray printer and stored at -20 °C. (Step 3) The query protein of interest is multimerized on microbeads for increased avidity and detection of transient protein-protein interactions. A fluorescent IgG is complexed with the query:microbeads as inert labeled carrier. (Step 4) Screening for binding partners of a query protein. The fluorescent query protein complex is screened against the extracellular protein microarray using a hybridization station for automated slide processing. (Step 5) Slides are then visualized using a microarray scanner. The BSA-Cy3 spots can be observed in the 535 nm channel and the hits yielded from the screen can be observed as duplicate spots at 635 nm. Data analysis for hit calling is performed by creating intersection plots of two independent experiments. Hits are called based on mutually high scores above background, as described in the protocol section. Please click here to view a larger version of this figure.
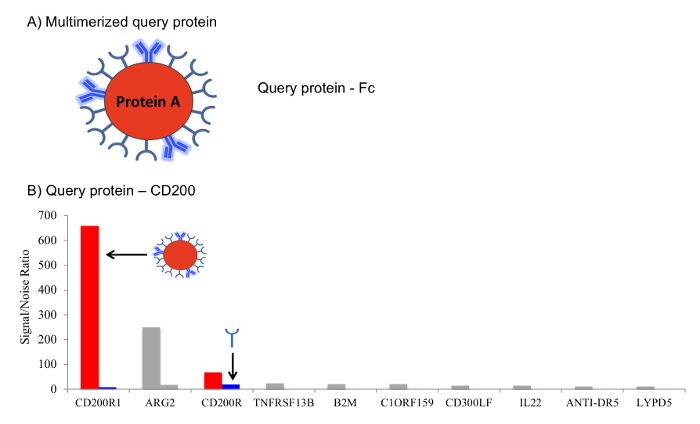
Figure 2: Enhanced signal through formation of multivalent complexes for the query protein. (A) Multimerization of query proteins. Query proteins are expressed as recombinant Fc fusions and coupled to protein A microbeads to form multimerized complexes. (B) High-avidity leads to detection of low affinity interactions. The avidity afforded by the multimerized query protein results in the stabilization of weak interactions on the protein microarray, leading to higher signal to noise ratios and enhanced hit calling. Plots shows screening results for CD200, screened as a microbead complex, or a soluble protein directly labeled with Cy5 dye to enable visualization of the hits. Multimerization (red bars) allows robust detection of the low affinity binding partner CD200R1, present in the protein microarray library, with significant signal/noise ratio in comparison with the same protein screened as a soluble product (blue bars). Grey bars indicate non-specific binders. Only the top 10 interactions have been represented in the plot. Please click here to view a larger version of this figure.
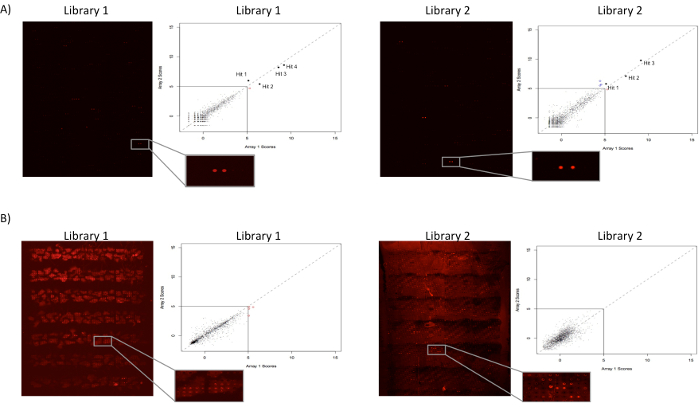
Figure 3: Protein microarray Technology for identification of novel receptor-receptor virus-host interactions. (A) Exemplary receptor discovery results for an orphan adenoviral immunomodulatory protein. Images represent actual microarray scans, showing hits from proteins spotted in duplicates (red). Assays are performed in duplicates to control for any slide variability, and results represented as intersection plots. In the plots, the red and blue dots represent hits called only on one array replicate, whereas black dots represent intersecting hits from both independent runs. The entire microarray library is printed on two slides for ease of use given the number of proteins present in the collection, and to ensure enough separation for each individual protein spotted on the array. (B) Viral protein screen exhibiting high background that precludes hit calling. Certain query proteins may display high background, as detected by binding to multiple proteins and/or the slides, therefore posing challenges for identification of high scoring hits. Please click here to view a larger version of this figure.
Discussion
A significant number of orphan receptors remain in the human genome, and novel interacting partners continue to emerge for extracellular proteins with previously characterized ligands. Defining the receptor-ligand interactions in human and model organisms is essential to understand the mechanisms that dictate cellular communication during homeostasis, as well as dysregulation leading to disease, and therefore inform new or improved therapeutic options. Nevertheless, detection of extracellular protein interactions by widely used technologies has represented a significant barrier mainly due to the technical challenges associated to the biochemistry of the cellular receptors and their interacting partners. In this report we describe the use of an extracellular protein microarray for screening of query proteins of interest, with emphasis on utilization of microbead complexes for enhanced detection of PPIs.
Probing receptor-receptor interactions is particularly challenging, as many physiologically relevant pairs are characterized by very weak interaction with binding affinities in the micromolar range2,3. Avidity enhancement through multimerization has proven to be a useful strategy to increase sensitivity for detection of low affinity PPIs. We have developed a method based on Fc fusion proteins, for efficient formation of multivalent microbead-protein query complexes4. Multimerization of the query protein is a key step to detecting low affinity PPIs. As shown in the example in Figure 2, this method significantly enhances signal in comparison with the same query protein screened as a soluble (non-complexed) analyte, while retaining minimal background. Alternatively, the protein microarray screens can be performed with soluble Cy5-labeled query protein, an approach that is compatible with any fusion tag and that may suffice for identification of more stable, higher affinity interactions, such as those between some soluble ligands and their receptors. It should be noted that because the avidity is increased through multimerization of the query on microbeads, the microarray signal is not a quantitative measure of the interaction strength. Rather, binding affinities should be calculated with the monomeric versions of the proteins under evaluation. In addition, it is highly advisable that any hits identified through the microarray technology are validated through independent methods. We routinely use surface plasmon resonance as a gold standard biophysical method for PPI studies and measurement of the binding kinetics.
Although out of the scope of this report, it should be noted that the success of any screening efforts using the microarray technology heavily relies on selection and availability of a high quality protein library. Relevant criteria for selection of extracellular proteins have been previously published4,22. A number of useful tools for prediction of relevant protein features (such as transmembrane helices or signal peptides) are freely available online, including the following servers: SignalP15, TMHMM16, Phobious17, and TOPCONS18. Excellent methods papers describing affinity purification methods are available elsewhere. It should also be mentioned that the microarray protein library generation relies on the production of protein ECDs as soluble recombinant products, and therefore this approach allows studies of type I, type II and GPI-anchored cell surface receptors and secreted proteins, but is generally not suitable for multitransmembrane-containing proteins such as G protein-coupled receptors. The purification of hundreds of proteins as recombinant soluble proteins demands significant logistic efforts and can be very resource- and time-consuming. Nevertheless, with the decreased costs of gene synthesis and increased throughput of the protein purification systems, library generation is now relatively faster and more affordable. Alternatively, commercial sources offer purified extracellular proteins that can be purchased in microgram amounts, which suffice for generation of a durable protein microarray library given the small requirements for slide printing. These limitations notwithstanding, microarray technology offers great advantages such as minimal consumption of protein reagents, fast readouts (new PPIs can be identified within one day) and affordable instrumentation. In addition, once the constructs and purification conditions have been established, small-scale purifications can produce enough material to prepare thousands of arrays.
Finally, in some instances, relatively high background is observed, appearing as binding to many proteins on the microarray library. There are multiple factors that might influence non-specific binding. For example, some proteins may be highly charged, or interact with carbohydrates, accounting for non-specific background via recognition of common motifs. Similarly, non-specific binding may also be due to the nature of the query protein, such as for example recognition of sialic acid motifs, found in multiple other proteins. As the microarray libraries expand, it is more likely that that a common list of non-specific binders might be present. Such non-specific binders can be identified by tracking their behavior across various screens against unrelated query proteins. It is advisable to generate a list of non-specific binders across screens, to improve specific hit calling. In our case, we have found that applying a 5% cutoff (i.e., a protein is flagged as non-specific if detected as binder is 5% or more of unrelated query protein screens) is important to reduce false positives. It should be noted that if the main purpose of the assays is to run only a few selected screens, it may not be worthwhile to develop a statistical analysis. In this case, it may not be feasible to detect false positives due to a limited dataset, and therefore we recommend that special emphasis is placed on confirmation of the hits using alternative methods.
We anticipate the extracellular protein microarrays, especially in combination with multimerization methods for detection of transient receptor-ligand interactions, will continue to offer a unique platform for rapid and robust identification of novel PPI in the extracellular space.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank Philamer Calses and Kobe Yuen for critically reading the manuscript. We are thankful to Randy Yen for excellent technical advice.
Materials
| Ultra Pure MB Grade glycerol | USB Corporation | 56-81-5 | Protein storage |
| SeptoMark blocking buffer | Zeptosens | BB1, 90-40 | Blocking buffer microarray slides |
| Bovine serum albumin | Roche | 03-117-957-001 | Slide control for mask fitting (optional) |
| Polypropylene multiwell plates | Greiner Bio One | 82050-678 | Protein storage |
| Polypropylene multiwell plates | Arrayit | MMP384 | Slide printing |
| NanoPrint LM60, or similar contact microarrayer | Arrayit | NanoPrint LM60, or similar contact microarrayer | Slide printing |
| Micro spotting pins | Arrayit | Micro spotting pins | Slide printing |
| ZeptoFOG blocking station | Zeptosens | ZeptoFOG blocking station, 1210 | Block slides after printing |
| Skim milk powder | Thermo Fisher | LP0031 | Blocking solution |
| Epoxysilane-coated glass slide | Nextrion Slide E | 1064016 | Microarray slides |
| Glass holder and slide rack set | Wheaton | 900303 | Slide storage |
| Cy5 monoreactive dye | GE Healthcare | PA23031 | Albumin labeling |
| Cy5 monoreactive dye | GE Healthcare | PA25001 | Human IgG labeling |
| Pro-spin desalting column | Princeton Separations | CS-800 | Remove free dye |
| Adhesive aluminum foil seal | AlumaSeal | F-384-100 | Seal stock plates |
| Polypropylene cryogenic vials | Corning | 430658 | Master vials for protein library storage |
| Protein A microbeads | Miltenyi | 120-000-396 | Query protein multimerization |
| Human IgG | Jackson Immunoresearch | 009-000-003 | Irrelevant IgG for labeling |
| Protein A | Sigma | P7837 | Microarray slide blocking |
| Hybridization station, a-Hyb or similar | Miltenyi | Hybridization station, a-Hyb or similar | Automated microarray processing (optional) |
| GenePix 4000B scanner or similar | Molecular Devices | GenePix 4000B scanner or similar | Slide scanning |
| GenePix Pro or equivalent data extraction software | Molecular Devices | GenePix Pro or equivalent data extraction software | Data processing |
| Signal P4.1 | DTU Bioinformatics, Technical University of Denmark | online software | Prediction tool to determine presence and location of signal peptide cleavage sites |
| TMHMM 2.0 server | DTU Bioinformatics, Technical University of Denmark | online software | Prediction of transmembrane helices in proteins |
| Phobius | Stockholm Bioinformatics Center | online software | A combined transmembrane topology and signal peptide predictor |
| TOPCONS | Stockholm University | online software | Prediction of membrane topology and signal peptides |
Referenzen
- Overington, J. P., Al-Lazikani, B., Hopkins, A. L. How many drug targets are there?. Nature Reviews Drug Discovery. 5 (12), 993-996 (2006).
- Wright, G. J., Martin, S., Bushell, K. M., Sollner, C. High-throughput identification of transient extracellular protein interactions. Biochemical Society Transactions. 38 (4), 919-922 (2010).
- Wright, G. J. Signal initiation in biological systems: the properties and detection of transient extracellular protein interactions. Molecular BioSystems. 5 (12), 1405-1412 (2009).
- Ramani, S. R., et al. A secreted protein microarray platform for extracellular protein interaction discovery. Analytical Biochemistry. 420 (2), 127-138 (2012).
- Tom, I., Lewin-Koh, N., Ramani, S. R., Gonzalez, L. C. Protein microarrays for identification of novel extracellular protein-protein interactions. Current Protocols in Protein Science. 27, 1-27 (2013).
- Kaushansky, A., et al. Quantifying protein-protein interactions in high throughput using protein domain microarrays. Nature Protocols. 5 (4), 773-790 (2010).
- Popescu, S. C., et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proceedings of the National Academy of Sciences of the United States of America. 104 (11), 4730-4735 (2007).
- Zhu, H., et al. Global analysis of protein activities using proteome chips. Science. 293 (5537), 2101-2105 (2001).
- Bushell, K. M., Sollner, C., Schuster-Boeckler, B., Bateman, A., Wright, G. J. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Research. 18 (4), 622-630 (2008).
- Voulgaraki, D., et al. Multivalent recombinant proteins for probing functions of leucocyte surface proteins such as the CD200 receptor. Immunology. 115 (3), 337-346 (2005).
- Jiang, L., Barclay, A. N. Identification of leucocyte surface protein interactions by high-throughput screening with multivalent reagents. Immunology. 129 (1), 55-61 (2010).
- Yeh, F. L., Wang, Y., Tom, I., Gonzalez, L. C., Sheng, M. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 91 (2), 328-340 (2016).
- Jaworski, A., et al. Operational redundancy in axon guidance through the multifunctional receptor Robo3 and its ligand NELL2. Science. 350 (6263), 961-965 (2015).
- Martinez-Martin, N., et al. The extracellular interactome of the human adenovirus family reveals diverse strategies for immunomodulation. Nature Communications. 7, 11473 (2016).
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods in Molecular Biology. 1611, 59-73 (2017).
- Krogh, A., Larsson, B., von Heijne, G., Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 305 (3), 567-580 (2001).
- Kall, L., Krogh, A., Sonnhammer, E. L. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Research. 35 (Web Server issue), W429-W432 (2007).
- Bernsel, A., Viklund, H., Hennerdal, A., Elofsson, A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Research. 27 (Web Server issue), W465-W468 (2009).
- Gonzalez, R., et al. Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 107 (8), 3552-3557 (2010).
- Durocher, Y., Perret, S., Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Research. 30 (2), E9 (2002).
- Battle, T., Antonsson, B., Feger, G., Besson, D. A high-throughput mammalian protein expression, purification, aliquoting and storage pipeline to assemble a library of the human secretome. Combinatorial Chemistry & High Throughput Screening. 9 (9), 639-649 (2006).
- Clark, H. F., et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Research. 13 (10), 2265-2270 (2003).

