Generation of Null Mutants to Elucidate the Role of Bacterial Glycosyltransferases in Bacterial Motility
Summary
Here, we describe the construction of null mutants of Aeromonas in specific glycosyltransferases or regions containing glycosyltransferases, the motility assays, and flagella purification performed to establish the involvement and function of their encoded enzymes in the biosynthesis of a glycan, as well as the role of this glycan in bacterial pathogenesis.
Abstract
The study of glycosylation in prokaryotes is a rapidly growing area. Bacteria harbor different glycosylated structures on their surface whose glycans constitute a strain-specific barcode. The associated glycans show higher diversity in sugar composition and structure than those of eukaryotes and are important in bacterial-host recognition processes and interaction with the environment. In pathogenic bacteria, glycoproteins have been involved in different stages of the infectious process, and glycan modifications can interfere with specific functions of glycoproteins. However, despite the advances made in the understanding of glycan composition, structure, and biosynthesis pathways, understanding of the role of glycoproteins in pathogenicity or interaction with the environment remains very limited. Furthermore, in some bacteria, the enzymes required for protein glycosylation are shared with other polysaccharide biosynthetic pathways, such as lipopolysaccharide and capsule biosynthetic pathways. The functional importance of glycosylation has been elucidated in several bacteria through mutation of specific genes thought to be involved in the glycosylation process and the study of its impact on the expression of the target glycoprotein and the modifying glycan. Mesophilic Aeromonas have a single and O-glycosylated polar flagellum. Flagellar glycans show diversity in carbohydrate composition and chain length between Aeromonas strains. However, all strains analyzed to date show a pseudaminic acid derivative as the linking sugar that modifies serine or threonine residues. The pseudaminic acid derivative is required for polar flagella assembly, and its loss has an impact on adhesion, biofilm formation, and colonization. The protocol detailed in this article describes how the construction of null mutants can be used to understand the involvement of genes or genome regions containing putative glycosyltransferases in the biosynthesis of a flagellar glycan. This includes the potential to understand the function of the glycosyltransferases involved and the role of the glycan. This will be achieved by comparing the glycan deficient mutant to the wild-type strain.
Introduction
Protein glycosylation has been described in both Gram-positive and Gram-negative bacteria and consists of the covalent attachment of a glycan to an amino acid side chain1,2. In prokaryotes, this process usually occurs via two major enzymatic mechanisms: O- and N-glycosylation3. In O-glycosylation, the glycan is attached to the hydroxyl group of a serine (Ser) or threonine (Thr) residue. In N-glycosylation, the glycan is attached to the side chain amide nitrogen of an asparagine (Asn) residue within the tripeptide sequences Asn-X-Ser/Thr, where X could be any amino acid except proline.
Glycans can adopt linear or branched structures and are composed of monosaccharides or polysaccharides covalently linked by glycosidic bonds. In prokaryotes, glycans usually show diversity in sugar composition and structure in comparison to eukaryotic glycans4. Furthermore, two different bacterial glycosylation pathways that differ in how the glycan is assembled and transferred to the acceptor protein have been described: sequential and en bloc glycosylation5,6. For sequential glycosylation, the complex glycan is built up directly on the protein by successive addition of monosaccharides. In en bloc glycosylation, a pre-assembled glycan is transferred to the protein from a lipid-linked oligosaccharide by a specialized oligosaccharyltransferase (OTase). Both pathways have been shown to be involved in N- and O-glycosylation processes7.
Protein glycosylation has a role in modulating the physicochemical and biological properties of proteins. The presence of a glycan can influence how the protein interacts with its ligand, which affects the biological activity of the protein, but can also affect protein stability, solubility, susceptibility to proteolysis, immunogenicity, and microbe-host interactions8,9. However, several glycosylation parameters, such as the number of glycans, glycan composition, position, and attachment mechanism, could also affect protein function and structure.
Glycosyltransferases (GTs) are the key enzymes in the biosynthesis of complex glycans and glycoconjugates. These enzymes catalyze the glycosidic bond formation between a sugar moiety from an activated donor molecule and a specific substrate acceptor. GTs can use both nucleotides and non-nucleotides as donor molecules and target different substrate acceptors, such as proteins, saccharides, nucleic acids, and lipids10. Therefore, understanding GTs at the molecular level is important to identify their mechanisms of action and specificity, and also enables understanding how sugar composition of glycans that modify relevant molecules are related to pathogenicity. The Carbohydrate Active enzyme database (CAZy)11 classifies GTs according to their sequence homology, which provides a predictive tool since, in most of the GT families, the structural fold and mechanisms of action are invariant. However, four reasons make it difficult to predict substrate specificity of many GTs: 1) no clear sequence motif determining substrate specificity has been determined in prokaryotes12, 2) many GTs and OTases show substrate promiscuity13,14, 3) functional GTs are difficult to produce in high yield in recombinant form and 4) the identification of both donor and acceptor substrates is complex. Despite this, recent mutagenesis studies have made it possible to obtain significant advances in the understanding of catalytic mechanisms and subtract binding of GTs.
In bacteria, O-glycosylation seems to be more prevalent than N-glycosylation. The O-glycosylation sites do not show a consensus sequence, and many of the O-glycosylated proteins are secreted or cell-surface proteins, such as flagellins, pili, or autotransporters1. Flagellin glycosylation shows variability in the number of acceptor sites, glycan composition, and structure. For example, Burkholderia spp flagellins have only one acceptor site, while in Campylobacter jejuni, flagellins have as many as 19 acceptor sites15,16. Furthermore, for some bacteria, the glycan is a single monosaccharide, while other bacteria possess heterogeneous glycans compromised of different monosaccharides to form oligosaccharides. This heterogenicity occurs even among strains of the same species. Helicobacter flagellins are only modified by pseudaminic acid (PseAc)17, and Campylobacter flagellins can be modified by PseAc, the acetamidino form of the pseudaminic acid (PseAm) or legionaminic acid (LegAm), and glycans derived from these sugars with acetyl, N-acetylglucosamine, or propionic substitutions18,19. In Aeromonas, flagellins are modified by glycans whose composition ranges from a single PseAc acid derivative to a heteropolysaccharide20, and the attachment of glycans to the flagellin monomers is always via a PseAc derivative.
In general, glycosylation of flagellins is essential for flagellar filament assembly, motility, virulence, and host specificity. However, while flagellins of C. jejuni16, H. pylori17, and Aeromonas sp.21 cannot assemble into filament unless the protein monomers are glycosylated, Pseudomonas spp. and Burkholderia spp.15 do not require glycosylation for flagella assembly. Furthermore, in some C. jejuni strains, changes in sugar composition of the flagella glycan affect bacterial-host interaction and may play a role in evading certain immune responses16. Autoagglutination is another phenotypic characteristic affected by modifications in the composition of glycans associated with flagellins. A lower autoagglutination leads to a reduction in the ability to form microcolonies and biofilm22. In some bacteria, the ability of flagella to trigger a pro-inflammatory response was linked to flagellin glycosylation. Thus, in P. aeruginosa, glycosylated flagellin induces a higher pro-inflammatory response than unglycosylated23.
Aeromonas are Gram-negative bacteria ubiquitous in the environment, which allows them to be at the interface of all One Health components24. Mesophilic Aeromonas have a single polar flagellum, which is constitutively produced. More than half of clinical isolates also express lateral flagellin, inducible in high viscosity media or plates. Different studies have related both flagella types with the early stages of bacterial pathogenesis25. While polar flagellins reported to date are O-glycosylated at 5-8 Ser or Thr residues of its central immunogenic domains, lateral flagellins are not O-glycosylated in all the strains. Although polar flagella glycans from different strains show diversity in their carbohydrate composition and chain length20, the linking sugar has been shown to be a pseudaminic acid derivative.
The goal of this manuscript is to describe a method to obtain null mutants in specific GTs or chromosomal regions containing GTs to analyze their involvement in the biosynthesis of relevant polysaccharides and in bacterial pathogenicity, as well as the role of the glycan itself. As an example, we identify and delete a chromosomal region containing GTs of Aeromonas to establish its involvement in polar flagellin glycosylation and analyze the role of the flagellin glycan. We show how to delete a specific GT to establish its function in the biosynthesis of this glycan and the role of modified glycan. Although using Aeromonas as an example, the principle can be used to identify and study flagella glycosylation islands of other Gram-negative bacteria and analyze the function of GTs involved in the biosynthesis of other glycans such as the O-antigen lipopolysaccharide.
Protocol
The schematic representation of the procedure is shown in Figure 1.
1. Bioinformatic identification of flagella glycosylation island (FGIs) in Aeromonas
- To identify the PseAc biosynthetic clusters in the Aeromonas genomes, use the tblastn tool from the NCBI database26. First, retrieve orthologous proteins to PseC and PseI of A. piscicola AH-3 (identification codes are OCA61126.1 and OCA61113.1) from databases of sequenced Aeromonas strains, and then use the tblastn tool to search their translated nucleotides sequences.
NOTE: The pse genes flank the Aeromonas FGIs, with pseB and pseC located at one end of the cluster and pseI at the other end. The location of the rest of the pse genes can be different from one FGI group to another. - Given that those polar flagellin genes of many strains are adjacent to the FGIs, use the tblastn from the NCBI database to find orthologous proteins to the polar flagellins FlaA and FlaB of A. piscicola AH-3 (identification codes are OCA61104.1 and OCA61105.1) and the genes that encode them.
- In addition, Aeromonas FGIs involved in the biosynthesis of heteropolysaccharidic glycans (group II)27 contain several genes encoding enzymes involved in the fatty acid synthesis, such as luxC and luxE. Use the tblastn from the NCBI database to find orthologous proteins to LuxC of A. piscicola AH-3 (identification code is OCA61121.1) and the gene that encodes it.
2. Generation of null mutants in flagella glycosylation island genes
NOTE: This method of mutagenesis is based on the allelic exchange of polymerase chain reaction (PCR) in-frame deletion products using the suicide vector pDM428 (GenBank: KC795686.1). Replication of pDM4 vector is lambda pir dependent, and the complete allelic exchange is coerced by utilizing the sacB gene located on the vector.
- Construction of PCR in-frame deletion products.
- Design two pairs of primers that amplify DNA regions upstream (A and B primers) and downstream (C and D primers) of the selected gene or region to be deleted (Figure 2B).
- Ensure that Primers A and D have more than 600 bp upstream from the start and downstream from the stop, respectively, of the gene or genes to be deleted. These two primers need to include at the 5' end a restriction site for an endonuclease that allows cloning in pDM4.
NOTE: The restriction site included at the 5' end of A and D primers should not be the same as sites within AB or CD amplicons. - Ensure that Primers B and C are within the gene to be deleted or inside the first and last gene, respectively, of the region to be deleted. Ensure that both primers (B and C) are in-frame and located between 5-6 codons inside the gene. Furthermore, ensure that both primers include at the 5' end a 21 bp complementary sequence (Table 1) that allows joining the DNA amplicons AB and CD.
- Grow the selected Aeromonas strain in 5 mL of tryptic soy broth (TSB) overnight (O/N) at 30 °C. Use a commercially available kit for genomic DNA purification and follow the manufacturer's instructions (Table of Materials). Quantify 2 µL of the purified DNA using a spectrophotometer.
NOTE: Use double distilled water as a blank, with the instrument set to record absorbance at 260 nm. Record absorbance with 2 µL of purified DNA 3-5 times and calculate an average absorbance reading. Use the Beer-Lambert law to calculate the DNA concentration, where A = Ɛ.c.l. wherein "A" is absorbance, "Ɛ" is the molar average extinction coefficient for DNA, "c" is DNA concentration, and "l" is the path length (refer to the manufacturer's instruction for the path length of each instrument). - Use 100 ng of purified chromosomal DNA as the template in two sets of asymmetric PCRs with A-B and C-D primers (Table of Materials). Use a 10:1 molar ratio of A:B and D:C primer pairs (Supplementary Material).
NOTE: Asymmetric PCRs allow the preferential amplification of one strand of the template more than the other. - Analyze the PCR products by electrophoresis in a 1% agarose gel. Use TAE (40 mM Tris-acetate, 1 mM EDTA) as gel running buffer and a power setting of 8 V/cm. To visualize DNA, add ethidium bromide (0.5 µg/mL) to the gel and visualize the PCR products using a bio-imaging station (Table of Materials).
- Excise the amplicons from the gel using a scalpel, purify following the manufacturer's protocol (Table of Materials) and quantify them.
- Join AB and CD amplicons by their 21 bp overlapping sequences in the 3' end of PCR products (Figure 3). Mix 100 ng of each amplicon (AB and CD) in a PCR tube with PCR reagents (pre-AD PCR), but without primers, and extend using the thermocycler program (Supplementary Material). Then, add 100 µM of A and D primers to the reaction (AD PCR) and amplify as a single fragment (Supplementary Material).
- Analyze the PCR product (AD amplicon) by electrophoresis in a 1% agarose gel using the conditions described in step 2.1.6, excise the amplicon from the gel, purify following the manufacturer's protocol and quantify the amplicon (Table of Materials).
- Construction of pDM4 recombinant plasmid with the in-frame deletion.
- Grow an O/N culture of Escherichia coli cc18λ pir containing pDM4 in Luria-Bertani (LB) Miller broth (10 mL) with chloramphenicol (25 µg/mL) at 30 °C.
- Spin down the culture at 5,000 x g at 4 °C and suspend the pellet in 600 µL of sterile distilled water. Perform extraction and purification of plasmid pDM4 using a commercially available plasmid purification kit, following the provider's instructions (Table of Materials).
- Digest 1 µg of pDM4 and 400 ng of AD amplicon for 2 h with an endonuclease able to digest both ends of the AD amplicon and the cloning site of pDM4 (Figure 3). Follow the enzyme manufacturer's protocol for reaction conditions (Table of Materials).
- Clean up the digested plasmid and amplicon using a DNA purification kit and quantify them following the manufacturer's instructions (Table of Materials).
- To prevent religation of the linearized pDM4, remove the phosphate group from both 5' ends using the alkaline phosphatase enzyme following the manufacturer's instructions (Table of Materials). Then, clean up the treated plasmid and quantify it using a spectrophotometer as described in step 2.1.4 (Table of Materials).
- Combine 150 ng of the digested and phosphatase alkaline treated pDM4 with 95-100 ng of digested AD amplicon at a molar vector:insert ratio of 1:4. Prepare the ligation reaction in a volume of 15 µL, following the manufacturer's instructions (Table of Materials).
- Incubate O/N at 20 °C or over the weekend at 4 °C. After incubation, inactivate the T4 ligase at 70 °C for 20 min.
- Use ligation to introduce the plasmid into Escherichia coli MC1061λpir strain by electroporation. Use electrocompetent bacteria and 2 mm-gap electroporation cuvettes.
- Preparation of electrocompetent E. coli and electroporation of pDM4 recombinant plasmid
- Inoculate a single colony of E. coli MC1061λpir strain into Luria-Bertani (LB) Miller broth and grow O/N at 30 °C with 200 rpm shaking.
- Dilute 2 mL of the bacterial culture in 18 mL of LB broth and incubate at 30 °C with shaking (200 rpm) until an optical density (OD600) between 0.4-0.6 is attained.
- Pellet the bacterial culture by centrifugation at 5,000 x g and 4 °C for 15 min. Discard the supernatant and suspend the pellet in 40 mL of chilled distilled H2O.
- Repeat this cleaning step two more times to remove all culture salts. Finally, suspend the pellet in 4 mL of chilled distilled H2O and transfer 1 mL of the suspension to each of four conical 1.5 mL microfuge tubes.
- Pellet the bacterial culture by centrifugation at 14,000 x g and 4 °C for 5 min. Remove the supernatant without disturbing the pellet and suspend each pellet in 100 µL of chilled distilled H2O.
- Add 3-3.5 µL of the ligation mixture to each tube and incubate on ice for 5 min. Then, transfer the contents of each tube to a chilled 2 mm-gap electroporation cuvette and apply 2 kV, 129 Ω, and time constant around 5 ms.
NOTE: Pre-cool the electroporation cuvettes on ice. Maintain the bacteria on ice during the entire procedure. - After electroporation, add 250 µL of SOC medium to each of the four cuvettes to recover their contents, transfer them to a culture tube (about 1 mL), and incubate for 1 h at 30 °C with shaking (200 rpm).
- Plate the transformed cells on Luria-Bertani (LB) agar plates supplemented with chloramphenicol (25 µg/mL) to ensure all colonies growing in the plate have the plasmid backbone.
- Pick some colonies from the LB agar plates with chloramphenicol and incubate O/N at 30 °C. When colonies grow, check insertion of the deletion construct into pDM4 by PCR using primers that flank the pDM4 cloning side: pDM4for and pDM4rev (Table 1), and lysed colonies as the template (Supplementary Material).
- Pick one colony with a sterile toothpick and dip it into a 1.5 mL tube containing 15 µL of 1% Triton X-100, 20 mM Tris-HCl pH 8.0, 2 mM EDTA pH 8.0. Incubate the tubes at 100 °C for 5 min, and then 1 min on ice.
- Spin the tubes in a microfuge at 12,000 x g for 10 min to pellet bacteria and debris. Use 1 µL of the supernatant as the template in the PCR reaction (Table of Materials). The annealing temperature of the primer pair is 50 °C, and the PCR reaction requires 10% DMSO (Supplementary Material).
- Inoculate the colonies that amplified the product of interest in 10 mL of LB broth with chloramphenicol (25 µg/mL) and grow O/N at 30 °C. Extract the plasmid from cultures as described in steps 2.2.1-2.2.2. Sequence using the pDM4 primers following the manufacturer's instructions (Table of Materials).
- Transfer of in-frame deletion to the Aeromonas strain
NOTE: The pDM4 recombinant plasmid has to be introduced into the selected Aeromonas strain by triparental mating (Figure 4). Aeromonas strain has to be rifampicin-resistant to be selected after the triparental mating. Therefore, grow the selected strains O/N on TSB at 30 °C, centrifuge 2 mL, 4 mL, and 6 mL of cultures at 5,000 x g for 15 min, suspend each pellet in 100 µL of TSB, and plate in TSA with rifampicin (100 µg/mL).- Prepare O/N cultures of E. coli MC1061λpir with the pDM4 recombinant plasmid on LB agar with 25 µg/mL chloramphenicol (donor strain), the Aeromonas strain rifampicin-resistant on TSA with 100 µg/mL rifampicin (recipient strain), and the E. coli HB101 with the pRK2073 plasmid on LB agar with 80 µg/mL spectinomycin (helper strain) at 30 °C.
- When the colonies have grown, suspend the same number of colonies (10-15 colonies) of the donor, recipient, and helper strains gently with a sterile toothpick in three LB tubes with 150 µL of LB.
- Then, on an LB agar plate, without antibiotics, mix the three bacterial suspensions in two drops at a recipient: helper: donor ratio of 5:1:1 (50 µL of recipient, 10 µL of the donor, and 10 µL of helper). Incubate the plate face up O/N at 30 °C. Perform negative control of conjugation using the donor strain without recombinant plasmid.
NOTE: The helper strain carries the conjugative plasmid pRK2073 and assists the transfer of the pDM4 into the Aeromonas. Do not use vortex to suspend the donor, recipient, and helper strains to avoid breaking the pili. - Harvest bacteria from the LB agar plate by adding 1 mL of LB broth and spreading it with a sterile glass spreader to obtain a bacterial suspension. Use a pipette to transfer the bacterial suspension to a conical 1.5 mL microfuge tube and vortex vigorously.
- To select the recipient colonies containing the pDM4 recombinant plasmid, plate 100 µL of the harvested bacterial suspension on LB agar plates with rifampicin (100 µg/mL) and chloramphenicol (25 µg/mL).
- Spin down 200 µL and 600 µL of the samples in a microfuge at 5,000 x g for 15 min, suspend the pellet in 100 µL of LB broth and plate on LB agar with rifampicin (100 µg/mL) and chloramphenicol (25 µg/mL). Incubate all plates for 24-48 h at 30 °C.
- Pick up the grown colonies, transfer to LB plates with rifampicin (100 µg/mL) and chloramphenicol (25 µg/mL), and incubate them O/N at 30 °C. Then, confirm that the colonies are Aeromonas by the oxidase test29 and that pDM4 recombinant plasmid was inserted (first recombination) into the specific chromosomal region by PCR using the E and F primers (Supplementary Material), which bind outside the region amplified with the A and D primers (Table of Materials). As described in step 2.3.11, use 1 µL of lysed colonies as the template.
- To complete the allelic exchange for the in-frame deletion, coerce the colonies with the integrated suicide plasmid to a second recombination. Grow the recombinant colonies in 2 mL of LB with 10% sucrose and without antibiotics, O/N at 30 °C.
- Plate 100 µL of the bacterial cultures from step 2.4.8 on LB agar plate with sucrose (10%). Also, plate 100 µL of different culture dilutions (10-2-10-4) and incubate O/N at 30 °C.
- To confirm the loss of pDM4, pick up the colonies, transfer to LB plates with and without chloramphenicol (25 µg/mL), incubate them O/N at 30 °C, and then select the chloramphenicol sensitive colonies.
- Analyze the sensitive colonies by PCR using the E-F primers pair and 1 µL of lysed colonies as the template (Table of Materials and Supplementary Material). Select the colonies whose PCR product correlated with the size of the deleted construct.
3. Motility assays
NOTE: In some bacterial species with glycosylated flagella, modifications in the levels of glycosylation or glycan composition affect the assembly of flagellins, which is usually reflected as a motility reduction or absence of motility. Therefore, two motility assays were performed with the null mutants.
- Motility assay in liquid media
- Culture Aeromonas strains O/N in 1 mL of TSB at 25-30 °C. To adhere a microscope cover slide to an excavated slide, pick up a small amount of silicone with the tip of a toothpick and put a small drop on the four corners of the cover slide. Then, deposit 1 µL of the Aeromonas culture in the middle of the cover slide and mix in a drop of fresh TSB media (2 µL).
- Place an excavated slide on the cover slide. Match the excavation with the deposited drop, press gently, and turn the slide upside down.
- Analyze swimming motility by light microscopy using an optic microscope (Table of Materials) with a 100x objective using immersion oil.
NOTE: The wild-type strain of Aeromonas must be used as the positive control. As a negative control, use a non-flagellated bacterium such as Klebsiella.
- Motility assay in semi-solid media
- Culture Aeromonas strains O/N in TSB (1 mL) at 25-30 °C. Then, transfer 3 µL of the culture onto the center of a soft agar plate (1% tryptone, 0.5% NaCl, 0.25% bacto agar) and incubate the plate face up O/N at 25-30 °C.
- Using a ruler, measure the bacterial migration through the agar from the plate's center toward the periphery.
4. Flagella purification
- Isolation of flagella anchored to the bacterial surface
- Culture Aeromonas strains O/N in TSB (10 mL) at 25-30 °C. Then, expand the culture into a flask with TSB (900 mL) and let it grow O/N at 25-30 °C with shaking (200 rpm).
- Centrifuge at 5,000 x g for 20 min at 4 °C. Suspend the pellet in 20 mL of 100 mM Tris-HCl buffer (pH 7.8).
- To remove the anchored flagella, shear the suspension in a vortex with a glass bar (2-3 mm diameter) for 3-4 min, and then pass repetitively (minimum six times) through a 21-G syringe.
- Pellet bacteria by two consecutive centrifugations at 4 °C: first, at 8,000 x g for 30 min. Remove the supernatant and centrifuge at 18,000 x g for 20 min. Collect the supernatant, which contains free flagella.
- Ultracentrifuge the supernatant at 100,000 x g for 1 h at 4 °C and suspend the pellet in 150 µL of 100 mM Tris-HCl (pH 7.8) plus 2 mM EDTA buffer. The pellet contains flagella filaments.
- Analyze 5-10 µL of the resuspended pellet from step 4.1.5 by electrophoresis in a 12% SDS-Polyacrylamide gel and visualize the proteins using Coomassie blue staining.
NOTE: Follow the manufacturer's instructions for protein electrophoresis and gel staining. As a positive control of glycosylated flagellin, use the purified flagella of the wild-type strain. Purify the enriched fraction of flagella in a cesium chloride (ClCs) gradient.
- Cesium chloride gradient to purify flagella.
- Mix 12.1 g of CsCl with 27 mL of distilled H2O.
- Transfer the enriched fraction of flagella (150 µL) to a thin-walled ultra-clear tube (14 mm x 89 mm) (Table of Materials) and fill it with 12 mL of CsCl solution.
- Ultracentrifuge the tube at 60,000 x g for 48 h at 4 °C in a swinging bucket rotor (Table of Materials).
- Collect the flagella band into the CsCl gradient with a syringe and dialyze against 100 mM Tris-HCl (pH 7.8) plus 2 mM EDTA. Then, analyze the dialyzed flagella by electrophoresis in a 12% SDS-Polyacrylamide gel and Coomassie blue staining (step 4.1.6) or by mass spectrometry.
Representative Results
This methodology provides an effective system to generate null mutants in genes or chromosomal regions of Aeromonas that can affect flagella glycosylation and the role of flagella filament (Figure 1).
The protocol starts with the bioinformatic identification of putative FGIs and the genes encoding GTs presents in this region. In Aeromonas, the chromosomal location of FGIs is based on the detection of three types of genes: genes involved in the biosynthesis of the pseudaminic acid (pseI and pseC), polar flagellin genes, and luxC. Strains whose polar flagella are glycosylated by a single pseudaminic acid derivative, such as A. hydrophila AH-130, do not have genes encoding GTs between the pse genes located in FGIs group I27. Most of the strains belonging to this FGI group show polar flagellin genes adjacent to this region. In contrast, strains whose polar flagella is glycosylated with a heteropolysaccharide glycan, such as A. piscicola AH-319, show different genes encoding GTs downstream of luxC, localized between pse genes of FGIs group II27, and they are not always adjacent to polar flagellin genes (Figure 2A).
The region involved in the biosynthesis of the flagella heteropolysaccharide glycan and the function of putative GTs contained there was confirmed by the construction of null mutants. The generation of each mutant requires four primers: A, B, C, and D (Table 1). Primer B anneals 5-6 codons downstream of the start of the gene (fgi-4) or first gene of the cluster (fgi-1) to be deleted and primer A anneals 600-800 bp upstream from the start of this gene (fgi-4 or fgi-1). Primer C anneals upstream of the last 5-6 codons of the gene (fgi-4) or last gene of the cluster (fgi-12) to be deleted and primer D anneals 600-800 bp downstream from the stop of this gene (fgi-4 or fgi-12) (Figure 2B). Primers A and D for the deletion of fgi cluster and fgi-4 of A. piscicola AH-3 contain a restriction site for the endonuclease BamHI at their 5' ends (Table 1). This endonuclease has no internal targets in the AB nor CD PCR fragments. One important step is the design of B and C primers. Both have to be in-frame to not break the open reading frame of gene or fragment to be deleted. The 21 bp complementary sequences at the 5' end of B and C primers allow the generation of AB and CD amplicons with 3' end complementary sequences, which allow the joining and extending of these amplicons. The PCR program to join and extend the amplicons consists of an initial denaturalization step and five cycles with an annealing temperature of 54 °C (Supplementary Material). Then, the addition of A and D primers allows the amplification of the extended fragment (Figure 3). The enzymatic action of BamHI gives rise to an amplicon with sticky ends compatible for ligation with the suicide vector pDM4 digested with BglII.
Given that pDM4 is a λpir dependent plasmid, the ligation product was electroporated into the E. coli strain MC1061λpir (thi thr1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5 supE44 λ pir) and selected in chloramphenicol plates. pDM4 does not have a direct system to select the recombinant clones, and colonies were analyzed by PCR with a pDM4 primers pair (pDM4for and pDM4rev) (Table 1 and Supplementary Material) that flank the cloning region. The E. coli strain MC1061λpir without pDM4 was used as the negative control in the PCR reaction.
To perform the allelic exchange, the recombinant pDM4 plasmid was transferred to the recipient strain A. piscicola AH-3. Given that many Aeromonas are not transformable by electroporation, the recombinant pDM4 was transferred by conjugation using triparental mating (Figure 4). To select the transconjugant colonies, we use a rifampicin-resistant recipient strain, which allows selecting the Aeromonas strain from the conjugation mating. Furthermore, the selected colonies were submitted to the oxidase test because while Aeromonas is oxidase-positive, E. coli is oxidase negative. The first and second recombinations were confirmed by PCR with external primers (E and F primers) of amplified regions (AB and CD fragments) (Table 1, Supplementary Material, and Figure 5A).
In Aeromonas, the glycosylation of polar flagellins is essential for the assembly of the flagellar filament. The presence of functional polar flagella was analyzed using motility assays of the bacterial colonies with deleted GTs genes or region. Light microscopy assays showed that swimming motility in liquid medium was reduced in both mutants in comparison to the wild-type strain. Furthermore, both showed a decreased radial expansion in relation to the wild-type strain when motility was analyzed on soft agar (Figure 5B). Given that both mutants were able to swim but with reduced motility in relation to the wild-type strain, their polar flagella were purified, and the flagellin molecular weight was analyzed in a 12% SDS-polyacrylamide gel. This analysis showed that both mutants have polar flagellins with lower molecular weight than the wild-type strain (Figure 5C), which suggests alterations in the flagella glycan. Flagella purified by CsCl gradients (Figure 5C) will be used in mass-spectrometry assays to identify glycan composition and null mutants used in biofilm, adhesion, or other assays to identify the role of the glycan and glycan composition.
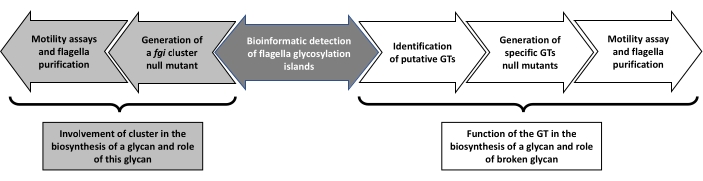
Figure 1: Overview of the steps used in this procedure. Scheme of the process described in the protocol to identify flagella glycosylation island of Aeromonas, and GTs involved in glycan biosynthesis. Please click here to view a larger version of this figure.
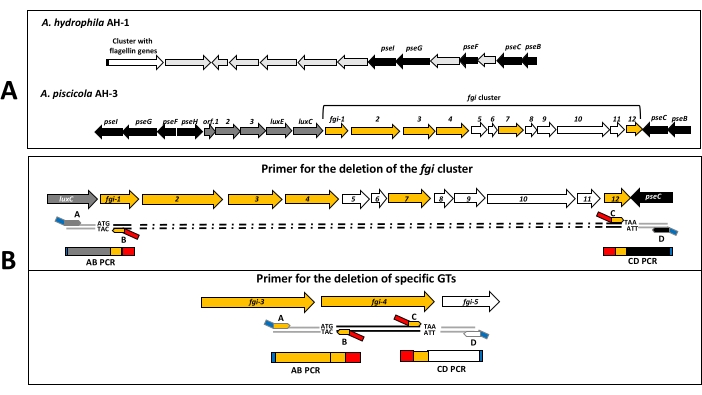
Figure 2: Bioinformatic detection of chromosomal regions and primer design. (A) Scheme of chromosomal regions identified in A. hydrophila AH-1 and A. piscicola AH-3. A. hydrophila AH-1 flagella glycosylation island is representative of strains whose flagella glycan only have a pseudaminic acid derivative, and A. piscicola AH-3 flagella glycosylation island is representative of strains whose flagella glycan is a heteropolysaccharide. Genes denoted in black are involved in the biosynthesis of pseudaminic acid, and those denoted yellow are putative GTs. (B) Scheme of the chromosomal location of primer pairs designed for the PCR in-frame deletions. Blue boxes in A and D primers contain the restriction binding site for an endonuclease. Red boxes in B and C primers contain 21 bp complementary sequences. Please click here to view a larger version of this figure.
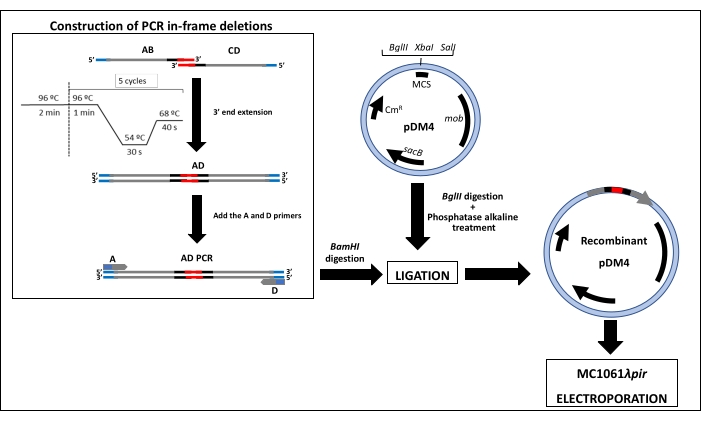
Figure 3: Scheme depicting the method to construct PCR in-frame deletions and ligation to the suicide plasmid pDM4. MCS: multi cloning site, CmR: chloramphenicol resistant genes, sacB: encodes the Bacillus subtilis levansucrase, whose expression is induced by sucrose and is lethal for Gram-negative bacteria, and mob: mobilization genes. Please click here to view a larger version of this figure.
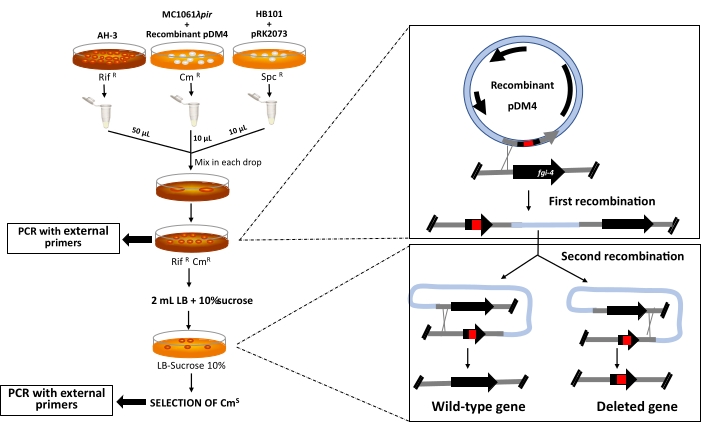
Figure 4: Triparental conjugation and procedure used for the allelic exchange. First recombination occurs due to the absence of λpir into the Aeromonas strains and gives rise to the integration of the recombinant plasmid into the selected gene. Second recombination is induced by growth in LB with 10% sucrose, which leads to the expression of sacB gene, and the plasmid is excised from the chromosome. After the cross-over, the wild-type or the mutated gene can remain on the bacterial chromosome. Null mutants are selected by PCR with external primers. RifR: rifampicin-resistant; CmR: chloramphenicol resistant; SpcR: spectinomycin resistant. Please click here to view a larger version of this figure.
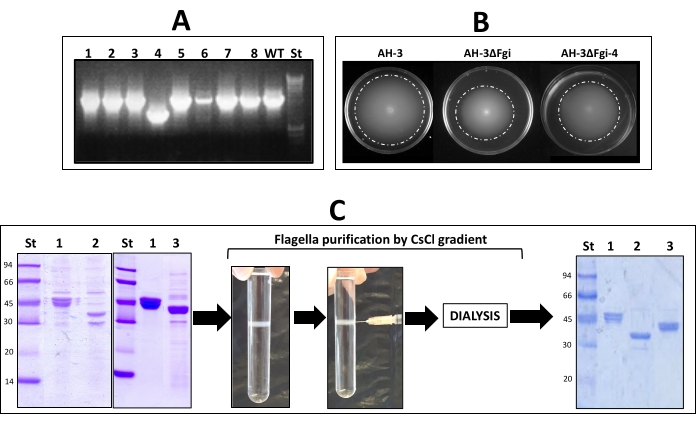
Figure 5: Confirmation of null mutants and phenotypic analysis of polar flagella. (A) PCR with fgi-4 external primers (EF primers) of A. piscicola AH-3 to confirm the allelic exchange after second recombination in chloramphenicol sensitive colonies. Lane WT: A. piscicola AH-3; lanes 1-8: chloramphenicol-sensitive colonies of the second recombination; St: HyperLadder 1 Kb marker. Lanes 1-3 and 5-8 show the colonies with wild-type fgi-4 gene as lane WT. Lane 4 shows a colony with the deleted fgi-4 gene. (B) Motility on soft agar of A. piscicola AH-3 and null mutants in the fgi region (AH-3ΔFgi) and fgi-4 gene (AH-3ΔFgi-4) at 25 °C. (C) Polar flagellum from AH-3 (lane1) and the null mutants in the fgi region (lane 2) and fgi-4 gene (lane 3), isolated, purified in CsCl gradients, analyzed in 12% SDS-PAGE, and stained using Coomassie blue. Size standard (St). Please click here to view a larger version of this figure.
| Primer name | Sequence in 5’ to 3’ direction | Used for | ||
| A. piscicola AH-3 | ||||
| A-Flgi1 | CGCGGATCCGACTGTACCCGTTTCAATCA | fgi mutant | ||
| B-Flgi1 | CCCATCCACTAAACTTAAACAGATCACCTCGAACTCGAAA | |||
| C-Flgi12 | TGTTTAAGTTTAGTGGATGGGGGAACCTTAAATGCCATGA | |||
| D-Flgi12 | CGCGGATCCCAGTCTTCAGCTTCCATCC | |||
| E-Flgi1 | ACCCGCTTCATTCGCTAT | |||
| F-Flgi12 | TCCGATTTTCTGACTCAGGG | |||
| A-Fgi4 | CGCGGATCCGATGCGTACGCTAATATGAA | fgi-4 mutant | ||
| B-Fgi4 | CCCATCCACTAAACTTAAACACATATTATCTTGCCCCTGAT | |||
| C-Fgi4 | TGTTTAAGTTTAGTGGATGGGATGGAGCTAATCACTCGTTT | |||
| D-Fgi4 | CGCGGATCCACATATCAACCCCCAAC | |||
| E-Fgi4 | ATTTCCCTGCCAAATACG | |||
| F-Fgi4 | CCTGCCAACAGGATGTAAG | |||
| pDM4 vector | ||||
| pDM4for | AGTGATCTTCCGTCACAGG | Insertions into the pDM4 vector | ||
| pDM4rev | AAGGTTTAACGG TTGTGGA | |||
Table 1: Primers used for the construction of null mutants. Overlapping regions in primers B and C are underlined. BamHI site is bolded in primers A and D.
Supplementary Material. Reagents and reactions used in asymmetric PCRs to obtain the AB and CD amplicons, Pre-AD and AD PCRs to extend and obtain the AD amplicons, pDM4 PCRs to verify the insertion of deleted construct in pDM4 vector, and EF PCRs to verify the first and second recombination. Please click here to download this File.
Discussion
The critical early step of this method is the identification of regions involved in the glycosylation of flagella and putative GTs because these enzymes show high homology and are involved in many processes. Bioinformatic analysis of Aeromonas genomes in public databases shows that this region is adjacent to the polar flagella region 2, which contains the flagellin genes in many strains and contains genes involved in the biosynthesis of pseudaminic acid27. This has made it possible to develop a guideline for detecting flagella glycosylation islands in Aeromonas that could be used to identify this region in other bacteria whose flagellar glycan contain pseudaminic acid derivatives. In addition, although this method describes how to analyze a gene cluster involved in flagella glycosylation, it can also be used to study genes encoding proteins that might be involved in flagella formation, rotation, or regulation in different Gram-negative bacteria.
Also important in the generation of PCR in-frame deletions is the design of B and C primers. These primers should be localized 5-6 codons downstream of the start (B primer) and upstream of the stop (C primer) of the selected gene or region to be deleted and should not break the open reading frame. Furthermore, an important factor to consider is the length of the homology region amplified to generate the AB and CD amplicons. This protocol recommends that both amplicons contain a homolog region of 600-800 bp each. Shorter homolog regions make the first recombination more challenging.
Aeromonas strains do not carry the lambda pir gene, which is required to replicate the pDM4 suicide plasmid. Therefore, the pDM4 recombinant plasmid must integrate into the chromosomal DNA. After the triparental mating, it is important to identify the bacterial colonies in which the first recombination has occurred. These colonies contain the recombinant pDM4 inserted into the homologous chromosomal region. Identification of recombinant colonies is performed using a PCR reaction with primers (E and F primers) external to the region targeted to be deleted. If a standard DNA polymerase is used, the E-F primers will only lead to the generation of a PCR product in the wild-type. This primer pair will not produce any PCR product in bacterial colonies in which the pDM4 recombinant plasmid has been inserted into the chromosomal DNA. The lack of PCR product is due to the distance between the annealing sequences of E and F primers. This distance cannot be amplified by standard polymerases. However, other factors can also prevent the formation of PCR products. Therefore, first recombinant colonies can be identified by DNA polymerases for long fragment amplification or by PCR reactions using pairs of primers consisting of a pDM4 and an external primer. When large genetic clusters are deleted, amplification in the wild-type strain requires the use of DNA polymerases for long fragment amplification. Bacterial colonies with the first recombination can be identified by PCR with pDM4-external pairs of primers. However, first and second recombinations are usually produced at the same time, leaving the pDM4 with the wild-type or with the deleted gene after recombination. This leads to chloramphenicol resistant colonies whose PCR using external primers give amplicons with an identical size of the wild-type gene or small amplicons, which correlate to the deleted gene or fragment. Colonies shown to have the correct recombinant insert were incubated with sucrose to remove the non-inserted pDM4. Sucrose induces the expression of the sacB gene located on the pDM4, which encodes a lethal enzyme for gram-negative bacteria. Therefore, only colonies lacking the suicide plasmid will grow in LB with sucrose. To support the identity of colonies containing the deleted gene, the fragment amplified with the external primers pair can then be sequenced.
Implementing this in-frame mutation method in Aeromonas allows the generation of more stable null mutants without modifications in the expression levels of downstream genes. Other methods, such as inserting an antibiotic cassette, assure the transcription of downstream genes, but the expression level could be modified. Furthermore, this method could be extrapolated to other target genes and regions, including other Gram-negative bacteria29,31,32,33.
In some bacterial strains, the loss or modification of glycan linked to flagellins can lead to the inability of flagella assembly or the instability of flagella filament, which leads to a reduction of polar flagella motility. While it is easy to distinguish a non-motile phenotype from a motile one using motility assays in liquid media, differences in the degree of motility can be difficult to quantify. Motility plate assays are required to measure differences in the degree of motility. However, some bacterial species, including many mesophilic Aeromonas strains, express inducible lateral flagella when growing in high viscous media or plates, in addition to the constitutive polar flagella. Both flagella types contribute to the bacterial motility in semi-solid plates. Therefore, modifications of polar or lateral flagella only lead to reductions in the migration diameters. Thus, AH-3ΔFgi and AH-3ΔFgi-4 mutants show only reduced motility in relation to the wild-type strain. To improve the evaluation of motility produced by the rotation of polar flagella, the expansion diameter of null mutants should be compared in relation to not only the wild-type strain but also a mutant lacking the polar flagellum. Furthermore, a reduction in the number of glycan residues or changes in the glycan composition influences the electrophoretic motility of glycosylated flagellins. For example, the molecular weight of glycosylated flagellins observed using SDS-PAGE gels is higher than predicted from flagellin amino acid sequence, and disruptions to the glycan are observed as reductions in their molecular weight. In some cases, the changes in glycan modification to the flagellin protein may not be detectable using SDS-PAGE. In these cases, mass spectrometry analysis of either intact protein or flagellin peptides may be required to confirm the presence and mass of glycan.
Pathogenicity of Aeromonas, as well as other Gram-negative bacteria, can be affected by the absence of flagellum but also by changes in the composition of flagella glycans. These changes can affect bacterial interaction to biotic and abiotic surfaces, autoagglutination, play a role in evading immune response and/or induction of pro-inflammatory response. Therefore, adherence, biofilm formation, and pro-inflammatory response assays are required to evaluate the relation between flagella glycosylation and Aeromonas pathogenicity.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Research Council Canada, for the Plan Nacional de I + D (Ministerio de Economía y Competitividad, Spain) and for the Generalitat de Catalunya (Centre de Referència en Biotecnologia).
Materials
| ABI PRISM Big Dye Terminator v. 3.1 Cycle Sequencing Ready Reaction Kit | Applied Biosystems | 4337455 | Used for sequencing |
| AccuPrime Taq DNA Polymerase, high fidelity | Invitrogen | 12346-086 | Used for amplification of AB, CD and AD fragments |
| Agarose | Conda-Pronadise | 8008 | Used for DNA electrophoresis |
| Alkaline phosphatase, calf intestinal (CIAP) | Promega | M1821 | Used to remove phosphate at the 5’ end |
| Bacto agar | Becton Dickinson | 214010 | Use for motility analysis |
| BamHI | Promega | R6021 | Used for endonuclease restriction |
| BglII | Promega | R6081 | Used for endonuclease restriction |
| BioDoc-It Imagin System | UVP | Bio-imaging station used for DNA visualization | |
| Biotaq polymerase | Bioline | BIO-21040 | Used for colony screening |
| Cesium chloride | Applichem | A1126,0100 | Used for flagella purification |
| Chloramphenicol | Applichem | A1806,0025 | Used for triparental mating |
| Cytiva illustra GFX PCR DNA and Gel Band Purification Kit | Cytivia | 28-9034-71 | Used for purification of PCR amplicons and DNA fragments. |
| EDTA | Applichem | 131026.1211 | Used for DNA electrophoresis |
| Electroporation cuvettes 2 mm gap | VWR | 732-1133 | Used for transformation |
| Ethidium bromide | Applichem | A1152,0025 | Use for DNA visualization |
| HyperLadder 1 Kb marker | Bioline | BIO-33053 | DNA marker |
| Invitrogen Easy-DNA gDNA Purification Kit | Invitrogen | 10750204 | Used for bacterial chromosomal DNA purification |
| Luria-Bertani (LB) Miller agar | Condalab | 996 | Used for Escherichia coli culture |
| Luria-Bertani (LB) Miller broth | Condalab | 1551 | Used for Escherichia coli culture |
| Nanodrop ND-1000 | NanoDrop Techonologies Inc | Spectrophotometer used for DNA quantification | |
| Rifampicin | Applichem | A2220,0005 | Used for triparental mating |
| SOC Medium | Invitrogen | 15544034 | Used for electroporation recovery |
| Spectinomycin | Applichem | A3834,0005 | Used for triparental mating |
| SW 41 Ti Swinging-Bucket Rotor | Beckman | 331362 | Used for flagella purification |
| T4 DNA ligase | Invitrogen | 15224017 | Used for ligation reaction |
| Trypticasein soy agar | Condalab | 1068 | Used for Aeromonas grown |
| Trypticasein soy broth | Condalab | 1224 | Used for Aeromonas grown |
| Tryptone | Condalab | 1612 | Use for motility analysis |
| Tris | Applichem | A2264,0500 | Used for DNA electrophoresis and flagella purification |
| Triton X-100 | Applichem | A4975,0100 | Used for bacterial lysis |
| Ultra Clear tubes (14 mm x 89 mm) | Beckman | 344059 | Used for flagella purification |
| Veriti 96 well Thermal Cycler | Applied Biosystems | Used for PCR reactions | |
| Zyppy Plasmid Miniprep II Kit | Zymmo research | D4020 | Used for isolation of plasmid DNA |
Referenzen
- Schäffer, C., Messner, P. Emerging facets of prokaryotic glycosylation. FEMS Microbiology Review. 41 (1), 49-91 (2017).
- De Maayer, P., Cowan, D. A. Flashy flagella: flagellin modification is relatively common and highly versatile among the Enterobacteriaceae. BMC Genomics. 17, 377 (2016).
- Nothaft, H., Szymanski, C. M. Protein glycosylation in bacteria: sweeter than ever. Nature Review Microbiology. 8 (11), 765-778 (2010).
- Benz, I., Schmidt, M. A. Never say never again: protein glycosylation in pathogenic bacteria. Molecular Microbiology. 45 (2), 267-276 (2002).
- Guerry, P., Szymanski, C. M. Campylobacter sugars sticking out. Trends in Microbiology. 16 (9), 428-435 (2008).
- Hug, I., Feldman, M. F. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 21 (2), 138-151 (2011).
- Iwashkiw, J. A., Vozza, N. F., Kinsella, R. L., Feldman, M. F. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Molecular Microbiology. 89 (1), 14-28 (2013).
- Szymanski, C. M., Wren, B. W. Protein glycosylation in bacterial mucosal pathogens. Nature Review Microbiology. 3, 225-237 (2005).
- Tan, F. Y. Y., Tang, C. M., Exley, R. M. Sugar coating: bacterial protein glycosylation and host-microbe interactions. Trends Biochemistry Sciences. 40 (7), 342-350 (2015).
- Lairson, L. L., Henrissat, B., Davies, G. J., Withers, S. G. Glycosyltransferases: structures, functions, and mechanisms. Annual Review Biochemistry. 77, 521-555 (2008).
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 42, 490-495 (2014).
- Weerapana, E., Imperiali, B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 16 (6), 91 (2006).
- Faridmoayer, A., et al. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. Journal of Biological Chemistry. 283 (50), 34596-34604 (2008).
- Wacker, M., et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proceedings of the National Academy of Sciences of the United States of America. 103 (18), 7088-7093 (2006).
- Scott, A. E., et al. Flagellar glycosylation in Burkholderia pseudomallei and Burkholderia thailandensis. Journal of Bacteriology. 193 (14), 3577-3587 (2011).
- Thibault, P., et al. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. Journal of Biological Chemistry. 276 (37), 34862-34870 (2001).
- Schirm, M., et al. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Molecular Microbiology. 48 (6), 1579-1592 (2003).
- McNally, D. J., et al. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. Journal of Biological Chemistry. 282 (19), 14463-14475 (2007).
- Logan, S. M., et al. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS Journal. 276 (4), 1014-1023 (2009).
- Wilhelms, M., Fulton, K. M., Twine, S. M., Tomás, J. M., Merino, S. Differential glycosylation of polar and lateral flagellins in Aeromonas hydrophila AH-3. Journal of Biological Chemistry. 287 (33), 27851-27862 (2012).
- Canals, R., et al. Non-structural flagella genes affecting both polar and lateral flagella-mediated motility in Aeromonas hydrophila. Microbiology. 153, 1165-1175 (2007).
- Howard, S. L., et al. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infection and Immunity. 77 (6), 2544-2556 (2009).
- Verma, A., Arora, S. K., Kuravi, S. K., Ramphal, R. Roles of specific amino acids in the N-terminus of Pseudomonas aeruginosa flagellin and of flagellin glycosylation in the innate immune response. Infection and Immunity. 73 (12), 8237-8246 (2005).
- Lamy, B., Baron, S., Barraud, O. Aeromonas: the multifaceted middleman in the One Health world. Current Opinion in Microbiology. 65, 24-32 (2022).
- Gavín, R., et al. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Molecular Microbiology. 43 (2), 383-397 (2002).
- Altschul, F. S., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 25 (17), 3389-3402 (1997).
- Forn-Cuní, G., et al. Polar flagella glycosylation in Aeromonas: Genomic characterization and Involvement of a specific glycosyltransferase (Fgi-1) in heterogeneous flagella glycosylation. Frontiers in Microbiology. 11, 595697 (2021).
- Milton, D. L., O’Toole, R., Horstedt, P., Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. Journal of Bacteriol.ogy. 178 (5), 1310-1319 (1996).
- Kovács, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 178 (4535), 703 (1956).
- Fulton, K. M., Mendoza-Barberá, E., Twine, S. M., Tomás, J. M., Merino, S. Polar glycosylated and lateral non-glycosylated flagella from Aeromonas hydrophila strain AH-1 (Serotype O11). International Journal of Molecular Sciences. 16 (12), 28255-28269 (2015).
- Khider, M., Willassen, N. P., Hansen, H. The alternative sigma factor RpoQ regulates colony morphology, biofilm formation and motility in the fish pathogen Aliivibrio salmonicida. BMC Microbiology. 18 (1), 116 (2018).
- Pawar, S. V., et al. Novel genetic tools to tackle c-di-GMP-dependent signalling in Pseudomonas aeruginosa. Journal of Applied Microbiology. 120 (1), 205-217 (2016).
- Vander Broek, W., Chalmers, K. J., Stevens, M. P., Stevens, J. M. Quantitative proteomic analysis of Burkholderia pseuomallei Bsa Type III secretion system effectors using hypersecreting mutants. The American Society for Biochemistry and Molecular Biology. 14 (4), 905-916 (2015).

