T4 Bacteriophage and E. coli Interaction in the Murine Intestine: A Prototypical Model for Studying Host-Bacteriophage Dynamics In Vivo
Summary
Bacteriophages (phages), viruses that infect bacteria, are an integral component of the gut microbiome. Though these symbiotic inhabitants drive bacterial fitness and population dynamics, little is understood about how they impact gut homeostasis and disease. This protocol studies isolated T4 phages within a mouse model, adaptable to other phage-bacterial pairs.
Abstract
Bacteriophages (phages) are viruses that infect bacteria with species- and strain-level specificity and are the most abundant biological entities across all known ecosystems. Within bacterial communities, such as those found in the gut microbiota, phages are implicated in regulating microbiota population dynamics and driving bacterial evolution. There has been renewed interest in phage research in the last decade, in part due to the host-specific killing capabilities of lytic phages, which offer a promising tool to counter the increasing threat of antimicrobial resistant bacteria. Furthermore, recent studies demonstrating that phages adhere to intestinal mucus suggest they may have a protective role in preventing bacterial invasion into the underlying epithelium. Importantly, like bacterial microbiomes, disrupted phageomes have been associated with worsened outcomes in diseases such as inflammatory bowel disease. Previous studies have demonstrated that phages can modulate the microbiome of animals and humans through fecal filtrate transplants, benefiting the host's health. With this recent wave of research comes the necessity to establish and standardize protocols for studying phages in the context of the gut microbiome. This protocol provides a set of procedures to study isolated T4 phages and their bacterial host, Escherichia coli, in the context of the murine gastrointestinal tract. The methods described here outline how to start from a phage lysate, administer it to mice and assess effects on bacterial host and phage levels. This protocol can be modified and applied to other phage-bacterial pairs and provides a starting point for studying host-phage dynamics in vivo.
Introduction
Bacteriophages, or phages, are viruses that infect and kill bacteria with species and strain-level specificity1. Phages play important roles within complex bacterial communities such as the gut microbiota, where they have been implicated in regulating population dynamics and driving bacterial fitness2. Throughout the last decade, there has been renewed interest in phage research owing to the rise of antimicrobial resistant pathogens3, and the potential for phage therapy as an alternative treatment strategy. In recent years, lytic phage cocktails have been used intravenously with some success in serious, antibiotic-resistant bacterial septic infections in humans3,4. Oral phage therapy has also been proposed as a potential alternative to antibiotics to treat intestinal infections and inflammation. Furthermore, phages have been implicated in the success of fecal filtrate transplants (FFT), which are fecal microbiota preparations that have been filtered to remove bacteria, in the treatment of recurrent Clostridioides difficile infection (rCDI)5,6, inflammatory bowel disorders (IBD)7,8 and necrotizing enterocolitis in pre-term pigs9. Given these results, it is important to consider interactions both between phages and the gut microbiota, and phages and the mammalian host, as the addition of novel phages into a preexisting community may have indirect effects on the community as a whole, and not only its target bacteria2,10.
The study of phage interactions with their target bacteria in vitro has proven useful for understanding the mechanisms and impacts of phage and bacteria interactions in the gut. In this setting, it has been shown that Escherichia coli-specific T4 phages of the order Caudovirales require immunoglobulin (Ig)-like domains located within highly antigenic outer capsid (Hoc) proteins on the virion surface to adhere to intestinal mucus11. Additionally, transwell assays have shown that T4 phages are capable of interacting with epithelial cell cultures and translocating through cell layers by macropinocytosis12,13. These results support the hypothesis that phages can interact with their metazoan host, even though they are incapable of infecting eukaryotic cells. These models, while useful, lack the full range of complex interactions that occur in a gut ecosystem that are required for a comprehensive exploration of the tripartite interaction between phages, bacteria and the metazoan host.
Mouse models are an important tool for investigating phages within complex environments. A desirable application of phage administration is as an alternative strategy to treat antimicrobial resistant infections or pathobionts associated with chronic inflammatory diseases, including IBD. However, emerging literature suggests that phage behavior in vitro does not fully represent in vivo functions. Buttimer et al.14 demonstrated that a phage cocktail was able to deplete the targeted bacteria in a simplified human microbiota consortium in vitro, but could not be replicated in vivo in gnotobiotic mice colonized with the same bacteria-phage consortium. Furthermore, in a conventional mouse microbiome, T7 phage led to selective depletion of its target gut bacteria, although gradual recovery was observed over time, indicative of evolved resistance15. Other studies have demonstrated co-existence of orally administered phages and their target bacterial strains in vivo2,16. Indeed, beyond phage/bacteria co-existence, phage administration led to widespread changes in overall microbiota community composition and function2,16. This is relevant in disease settings as several studies have found associations between increased relative abundance of Caudovirales and IBD7,8,17 that were independent of changes in bacterial abundance7. It remains unknown whether this is a driver or consequence of disease pathogenesis.
The historic focus of phage investigation has been around the relationship between a phage and its target bacterium. However, it is also important to consider potential interactions between phage and the mucosa, epithelium, and immune system of the metazoan host. These interactions all play an important role in the overall response to intestinal phage infection. To demonstrate this, phages have been studied using germ-free (GF) mice to elucidate their impact on the immune system without interference by the microbiota8. In this system, phage nucleic acids were detected by Toll-like receptors (TLRs) located within endosomes of phagocytic immune cells (macrophages and dendritic cells). This activated downstream signaling and stimulated T cell dependent production of interferon (IFN)-γ8 or type I IFNs18. Moreover, Fluckiger et al.19 implicated memory CD8+ T cells in the recognition of phage-encoded (prophage) antigens, which resulted in T cell cross-reactivity with tumor antigens, resulting in reduced tumor burden. Finally, phage-specific antibody production has been documented in mouse studies where phages were delivered to animal models in a continual manner through drinking water8,20, or by repeated oral gavage over several months20, demonstrating the capacity for phage proteins to promote humoral immune responses. Although these modes of phage inoculation allow for optimal and continual priming of the immune system, they may not represent the naturally occurring interactions between phages and the intestinal environment, nor the kinetics of orally applied phage therapy. Thus far, a limited number of studies have examined the interactions of phage with a single bacterial species in monocolonized mouse models21. However, monocolonized mice proved critical in deciphering microbe-specific effects of individual species on gastrointestinal (GI) tract and immune development22,23,24, and they may yet prove useful in understanding tripartite interactions between phages, their target bacteria, and the metazoan host.
Excitingly, there is still much to learn about the interactions between intestinal phage and gut commensal bacteria, as well as the interactions that occur between the metazoan host and the phages that reside within it. This protocol provides a set of procedures to study isolated T4 phage and its bacterial counterpart, E. coli (K-12, BW25113), using a gnotobiotic mouse model. These standardized procedures also provide a foundation for optimizing other phage/bacteria dyads by adapting the growth parameters to the pairs of interest. The methods described here outline: (1) Preparation of T4 phage and vehicle lysates for oral gavage of mice; (2) Oral administration of T4 phage to E. coli monocolonized gnotobiotic mice; (3) Monitoring T4 phage levels in mouse feces and tissues over time.
For the representative results presented here, purified T4 phage lysates were propagated from phage bank stocks maintained by the Rohwer Lab. The Phage-on-Tap method for propagating T4 phage was adapted25, as referenced in this protocol. The method yields high titer, endotoxin-low phage stocks within three days. Utilizing this approach, 10 mL of ≥ 1010 plaque forming units (pfu)/mL of T4 phage with < 0.5 endotoxin units (EU)/mL were routinely collected. The recommended endotoxin levels for oral or intravenous administration into mice are ≤ 20 EU/mL and ≤ 5 EU/kg/h (or 0.1 EU administered over 1 h for a 20 g mouse), respectively, making this a suitable method of phage preparation for in vivo inoculation. All phage stocks were stored at 4 °C in saline magnesium (SM) phage buffer (recipe provided in step 1.1.5.1). E. coli was cultivated in LB media. For various phage-bacteria pairs, diverse culture media and growth conditions may be adapted from this protocol. Phages can also be sourced from the environment, such as wastewater, marine water, soil and intestinal contents and can be isolated and purified as per Sambrook and Russell26 prior to preparation using the appropriate growth and propagation conditions for each phage-host pair of interest25. Alternatively, phages can be obtained from commercial sources (see Table of Materials) or from phage banks.
Protocol
All experiments were conducted in accordance with the guidelines established by the UBC Animal Care Committee and Biosafety Committee-approved protocols (A23-0113, B19-0038). Mice were housed at the University of British Columbia under pathogen-free conditions at the Center for Disease Modelling. C57BL/6 mice were bred within the facility in a sterile flexible film isolator, provided with sterile mouse diet, water, bedding, and nesting material. The mice were maintained on a 12 h day/night cycle. Experimental mice, both male and female, were age-matched within each experiment, ranging between 6 to 12 weeks of age and weighing 15-30 g for all experiments.
1. Preparation of phage and vehicle lysates for oral gavage into mice
- Phage purification and stock generation
NOTE: In this study, T4 phages are grown and titered by plating onto a lawn of bacteria using the single agar layer method, described below. The double agar layer method has been described previously25,27 and can also be used with similar efficacy. The single agar layer method was selected for this protocol as it provides improved plaque visibility28.- Grow E. coli in 5 mL of sterile LB media in sterile polystyrene or glass culture tubes (with caps). Incubate tubes at 37 °C with shaking at 200 rpm overnight, until stationary phase is reached.
- Prepare soft agar in a glass media bottle by autoclaving LB with 0.5% agar and cooling to 50 °C or below (while remaining a liquid) prior to supplementation. Prepare enough soft agar for 15 mL per plate.
- Supplement soft agar with MgSO4 and CaCl2 to a final concentration of 1 mM for each. Add 100 µL of overnight E. coli culture for every 3 mL of soft agar. Stir gently using a magnetic stir bar to homogenize the supplements and culture in the soft agar.
NOTE: Consistency in the volume and density of E. coli that is added to the soft agar preparation is critical (outlined in the representative results section). - Add 15 mL of soft agar + E. coli per Petri dish (15 cm diameter) using a serological pipette. Allow plates to set at room temperature for same-day use.
NOTE: Plates will set in approximately 20 min with lids open. The agar will remain soft but will not shift when plates are inverted. - Prepare 8-10 serial dilutions of the source T4 phage in factors of 10 by diluting in SM buffer. Spot 5 µL of each dilution onto plates. Allow spots to dry, invert and incubate plates at 37 °C overnight.
- To prepare Saline Magnesium (SM) phage buffer, in 1 L of deionized H2O, dissolve 100 mM NaCl, 8 mM MgSO4·7H2O, and 50 mM Tris-HCl (pH 7.4). Autoclave and store at room temperature.
NOTE: The next day, E. coli should have grown into a lawn throughout the soft LB agar. Plaques, or cleared areas of growth visible in the E. coli lawn, will appear where phage killing of infected E. coli hosts occurred. Each plaque represents a plaque forming unit (pfu).
- To prepare Saline Magnesium (SM) phage buffer, in 1 L of deionized H2O, dissolve 100 mM NaCl, 8 mM MgSO4·7H2O, and 50 mM Tris-HCl (pH 7.4). Autoclave and store at room temperature.
- Pick a single plaque from the soft agar plate by pushing a sterile pipette tip into the center of the plaque, creating an agar plug at the end of the tip. Resuspend the plaque plug into 1 mL of SM buffer in a 1.7 mL microcentrifuge tube. Vortex the tube at max speed for 1 min to mix.
- Centrifuge the tube at 4000 x g for 5 min at room temperature to remove any debris from the lysate. Transfer the supernatant into a new microcentrifuge tube.
- To propagate the isolated phage, repeat steps 1.1.1-1.1.3. Add 100 µL of the isolated phage to a 15 mL aliquot of soft agar containing E. coli in a conical tube. Invert the tube three times to mix and pour the agar into a petri dish.
- Allow plates to dry before inverting and incubating overnight at 37 °C. Prepare a lawn control plate without phage to confirm E. coli viability.
NOTE: After overnight incubation, the control plate should have a lawn of E.coli while the phage-containing plate should be completely lysed. - To extract the phage from the plate, add 5 mL of SM buffer to the surface of the phage-cleared plate and shake at 70 rpm on a rocker for 15 min at room temperature.
- Collect the buffer from the plate into a 50 mL conical centrifuge tube and centrifuge at 4000 x g for 5 min at room temperature to pellet any debris from the agar plate.
- Collect the T4 phage stock lysate into a new tube and store at 4 °C until titration and propagation.
NOTE: T4 phages are stable for over 10 years25; however, stability will depend on the type of phage and storage conditions. Phage titer will also vary with storage time. Phages are sensitive to freezing. Therefore, cryopreservation is recommended in liquid nitrogen and avoiding freeze-thaw cycles as this may damage phages, reducing phage titer25. - Titer the T4 phage stock lysate to determine the concentration in pfu/mL, as previously described25,29,30. A high titer T4 phage stock will contain 108 pfu/mL or more.
- Preparation of experimental phage lysates
- Culture E. coli in 5 mL of LB media in sterile polystyrene or glass culture tubes (with caps). Incubate tubes overnight at 37 °C with shaking at 200 rpm, until stationary phase is reached.
- Subculture the overnight E. coli culture 1:50 in 100 mL LB media in a glass conical flask. Incubate at 37 °C with shaking at 200 rpm until the bacteria have reached early to mid-exponential phase (approx. 1.5 h).
NOTE: An initial growth curve can be performed to determine the optical density at 600 nm (OD600) range at which the bacterial culture is in exponential phase. Culture in a glass conical tube is suggested as recent evidence has found that phages are able to adhere to plastics such as polypropylene31. - Add 100 µL of the high titer T4 phage stock from step 1.1 into the E. coli subculture and incubate at 37 °C with shaking for 3 h, or until the new lysate is no longer cloudy. Collect the T4 phage lysate into 50 mL conical tubes and either proceed directly to clean-up steps or, if required, store at 4 °C until the next day.
NOTE: A larger phage burst size (indicating a higher average number of virions released per replication cycle) necessitates a longer incubation period for the bacterial subculture in step 1.2.2. This provides increased bacteria density prior to spiking with phage stocks. - Centrifuge lysates at 4000 x g for 20 min at room temperature to pellet any remaining bacteria and cellular debris. Filter-sterilize the resulting supernatant using a 0.22 µm nylon filter and transfer the filtrate to new 50 mL conical centrifuge tubes.
- Add 0.1 volumes of chloroform to each volume of filtered T4 phage lysate to kill any remaining bacteria and prevent bacterial growth. Vortex briefly to mix and incubate at room temperature for 10 min.
NOTE: Lipid enveloped phages are sensitive to chloroform, which may reduce phage titer25. Skip this step if required.
CAUTION: Chloroform is a toxic organic solvent that is dangerous when inhaled, ingested, or absorbed through the skin. Use a fume hood and appropriate personal protective equipment (PPE) when working with chloroform. Use glass instead of polystyrene serological pipettes when working with chloroform, as it is not compatible with most plastics. Chloroform can be placed in polypropylene conical centrifuge tubes for steps 1.2.5-1.2.6 but should not be stored long term in plastic tubes. - Centrifuge the lysate at 4000 x g for 5 min at room temperature to separate the chloroform from the lysate. Use a serological pipette to carefully transfer the top lysate layer into a new 50 mL conical tube without disturbing the underlying chloroform layer. Discard the chloroform waste into the appropriate hazardous liquid waste container. Store lysates at 4 °C until concentration and buffer exchange the following day.
- Concentrate phage lysates in a 100 kDa centrifugal filter device (see Table of Materials) by adding 13 mL of phage lysate to the upper reservoir of the device and centrifuging at 4000 x g for 5 min, or until most of the lysate has passed through the filter into the lower reservoir.
NOTE: Aim to have approximately 2 mL of lysate at the end of the spin time. As the phage concentration increases in the filter with the addition of lysate, spin times will likely increase. Centrifugal filter devices have a physical dead stop to prevent them from spinning dry32. Do not let the filter membrane dry out if continued use is intended (i.e., by removing all liquid from the upper reservoir). Spin times may vary based on phage type and titer. - Using a P200 or P1000 pipette, gently pipette the remaining ~2 mL of lysate up and down within the upper reservoir to unclog the filter membrane after each spin. Discard the filtrate from the lower reservoir into a waste container, leaving the concentrated phage in the upper reservoir.
- Repeat steps 1.2.7-1.2.8 until the entire volume of phage lysate has been passed through the filter device, with ~2 mL of concentrated phage retained in the upper reservoir after each spin.
- Unclog the filter membrane after the last spin by pipetting the remaining lysate (~2 mL) in the upper reservoir up and down. Wash the phage (buffer exchange) by adding 12 mL of SM buffer to the upper reservoir and centrifuge at 4000 x g for 10 min, or until most of the buffer has passed through the filter.
- Discard the filtrate and repeat the wash step (step 1.2.10). Resuspend the remaining 2 mL of lysate in SM buffer to a final volume of 10 mL (or less), for long-term storage. Transfer the lysate to a 50 mL conical centrifuge tube and store at 4 °C until endotoxin removal.
NOTE: It is optional to titer the phage at this stage to ensure that the phage lysate has been concentrated (> 108 pfu/mL), and to determine phage loss during the endotoxin removal process. - Remove contaminating endotoxins from the T4 phage lysate by adding 0.4 volumes of 1-octanol (see Table of Materials) to the total volume of lysate.
NOTE: Endotoxins are removed as they are highly immunostimulatory, and therefore their presence may lead to the induction of a phage-independent innate immune response25.
CAUTION: 1-Octanol is an aromatic, organic, flammable compound with a strong odor and is an eye irritant. Wear appropriate PPE when working with 1-octanol. Perform all work in a fume hood to avoid vapor inhalation. Use sealing film to prevent the leaking of tubes when working outside of the fume hood. - Seal the lid of the conical centrifuge tube with sealing film to prevent leaking. Shake at 120 rpm on a platform or rocking shaker at room temperature for 1 h, followed by incubation at 4 °C for 1.5 h, without shaking.
- Centrifuge at 4000 x g for 10 min to separate the endotoxin-cleared lysate from the 1-octanol. The 1-octanol layer will float on top of the lysate. Use a P1000 pipette to carefully remove as much 1-octanol as possible and discard into the appropriate hazardous/flammable liquid waste container.
- Use an 18 G needle and 10 mL syringe to collect the phage lysate beneath the remaining 1-octanol layer, taking care not to collect the 1-octanol layer. Store lysate at 4 °C until speed vacuum.
- Transfer 1 mL aliquots of T4 phage lysate into sterile 1.5 mL microcentrifuge tubes. Speed vacuum with lids open at 4000 x g at room temperature to evaporate residual 1-octanol from the lysate. Speed vacuum for 3 h or until the phage lysate volume has been reduced by 30%25. Store at 4 °C until titration.
- Titer the phage lysate to determine the concentration in pfu/mL. Dilute the resulting T4 phage lysate in SM buffer to the desired concentration and re-titer to confirm.
- Quantify endotoxins present in the T4 phage lysate using a chromogenic endotoxin quantitation kit as per manufacturer instructions (see Table of Materials). Compare endotoxin levels in the final phage lysate with a pre-endotoxin-removal sample, vehicle controls, buffer, and mouse drinking water.
NOTE: The chromogenic endotoxin quantification kit is inactivated by 1-octanol25. Perform speed vacuum steps to remove 1-octanol (step 1.2.16) before quantifying endotoxins. The endotoxin content for distilled water is estimated at 20 EU/mL25; however, the endotoxin content of mouse drinking water may vary between facilities. If the endotoxin levels in the phage lysate exceed the amount present in drinking water, consider repeating endotoxin removal steps (1.2.12-1.2.16). - Store concentrated, endotoxin-cleared T4 phage lysates in SM buffer at 4 °C.
- Preparation of phage-free bacterial lysate for vehicle controls
NOTE: A phage-free bacterial lysate can be prepared as a vehicle to control for any effects due to bacterial contaminants (e.g., endotoxin) generated in the production of phage lysate in step 1.2, which could impact experimental outcomes when inoculated into mice.- From an overnight culture of E. coli, subculture the E. coli 1:50 into 100 mL of LB media. Without adding phage, continue to culture the bacteria while shaking at 37 °C for 3 h, or matching the incubation time for the phage lysate produced in step 1.2.
- Transfer the E. coli culture to 50 mL conical tubes and lyse cells using a sonicator probe (see Table of Materials) on ice at 30 kHz for 30 s pulses (3x) to manually lyse bacterial cells.
- Follow the rest of the protocol as per step 1.2, starting at step 1.2.4, including all clean up, wash and endotoxin removal steps.
NOTE: During concentration using the centrifugal filter device, the vehicle lysate will flow through the filter more quickly than the phage lysate, due to the absence of phage. Therefore, shorten centrifugation times from 5 mins to 2-5 min to ensure that the ultrafilter membrane does not dry out during prolonged centrifugation. - Titer the vehicle lysate to confirm that it does not contain phage.
- Use a chromogenic endotoxin quantitation kit as per manufacturer instructions to measure endotoxin levels in the vehicle lysate. Dilute the vehicle lysate in SM buffer to match the endotoxin levels in the phage lysate.
- Alternative experimental controls: preparation of heat-inactivated phage lysate
NOTE: An alternative to the phage-free bacterial lysate is a heat-inactivated phage lysate. Heat-inactivation dissociates phage virions, while residual endotoxins such as lipopolysaccharides (LPS) are heat stable8,33. This method was used by Gogokhia et al.8 to determine whether intact, viable phage virions were required for immune activation. Researchers are encouraged to test both methods (phage-free vs. heat-inactivated) and determine which control is most appropriate for their experimental needs. Whichever is chosen, it is important to acknowledge that a buffer control is unlikely to be appropriate due to the significant manipulation that is required to concentrate and clean-up a phage stock.- Transfer the required volume of cleaned, purified, and diluted T4 phage lysate from step 1.2 (100 µL per mouse) to sterile microcentrifuge tubes fitted with lid locks.
- Heat lysates on a heat block at 95 °C for 15 min16.
- OPTIONAL: If the removal of phage/bacterial nucleic acids is required, perform DNase I and RNase A treatment as per Jakočiūnė and Moodley34. Briefly:
- Add 50 µL DNase I 10x buffer, 1µL DNase I (1 U/µL) and 1 µL RNase A (10 mg/mL) (see Table of Materials) to 450 µL of heat-inactivated phage lysate.
- Incubate lysates at 37 °C for 1.5 h in a heat block, without shaking.
- Inactivate the DNase I and RNase A by adding 20 µL 0.5 M ethylenediaminetetraacetic acid (EDTA)34.
- Hold lysates at room temperature for 10 min to cool and combine lysates if multiple tubes are used. Store at 4 °C until titration.
- Perform a phage titer to confirm that no viable T4 phage is present in the lysate. Store heat-inactivated lysates at 4 °C.
- Use a chromogenic endotoxin quantitation kit as per manufacturer instructions to measure endotoxin levels in the heat-inactivated lysate. Dilute the heat-inactivated lysate in SM buffer according to the endotoxin levels present in the matched phage lysate.
2. Administration and monitoring of T4 phage in E. coli monocolonized mice
- Monocolonization of mice with E. coli
- Prepare E. coli for administration into GF mice8 by culturing E. coli picked from a single colony in LB media overnight.
- Under strict, sterile conditions35, administer 200 µL of E. coli culture to GF mice housed in either sterile vinyl isolators or bioexclusion airtight cages by oral gavage. If using aerotolerant bacteria, mice may also be colonized by application of 200 µL of bacterial culture to the backs of each mouse.
NOTE: The inoculation dose will be strain dependent, as different bacteria demonstrate different pH survival ability36,37. Zucoloto et al.35 recommend inoculation with 1 x 108 cfu per animal. Representative results shown here were generated from mice inoculated with an overnight culture (cfu was not determined). Pilot experiments can be performed to determine a dose that results in reliable colonization of the mouse strain of interest. The volume of bacteria to be administered by oral gavage may be impacted by the age and weight of the mice. Consult individual lab or institutional animal ethics protocol for maximum allowable doses. The volume suggested here is based on a gavage allowance of 10% of the mouse body weight (e.g., 200 µL maximum for a 20 g mouse). Some bacterial species will not tolerate the acidity of the stomach and fail to colonize the gut. Consider first gavaging with 100 µL of 1 M NaHCO3 to neutralize stomach acids2. If using a strict anaerobe to colonize, the microbe will need to be grown under anaerobic conditions and transferred to the gnotobiotic animal facility in individually prepared air-tight containers (1 for each mouse). Perform each gavage quickly once the container is opened to limit loss of microbial viability due to oxygen exposure. - Monitor mice for adverse health effects.
NOTE: In the event of adverse health effects, refer to the standards set by the relevant animal care facility. Possible adverse health effects include, but are not limited to: (1) Aspiration of gavage fluid: symptoms include expulsion of bubbles via the nose, "open mouth" breathing/gasping. (2) Perforation of the esophagus: symptoms include rapidly ailing health, hunching, lethargy, leading to death of the animal within 24 h. (3) Inflammation of the esophagus due to repeated gavages: symptoms include difficulty inserting the gavage needle. (4) Diarrhea due to alterations to the microbiome. - Confirm bacterial colonization in fecal pellets by culturing at least once per week and/or by 16S rRNA sequencing35.
NOTE: If colonizing breeders within a gnotobiotic isolator for production of experimental mice, plan at least 9 weeks in advance for generation of 6-week-old first generation of offspring (F1) experimental mice. Alternatively, GF adult mice can be monocolonized. In this approach, waiting 7 weeks post-colonization prior to phage inoculation is recommended, as this is the time needed for the intestinal mucosa to stabilize after introduction of a complex microbiota into GF mice38.
- Oral inoculation of T4 phage into E. coli monocolonized mice
- Dilute T4 phage lysates and vehicle controls to a predetermined concentration in SM buffer. As per Hsu et al.2, dilute phage lysates to allow for administration of 2 x 106 pfu per mouse2.
NOTE: A higher or lower concentration of phage may be used depending on the stability of the phage in vivo. Doses of 2 x 102, 2 x 104 and 2 x 106 pfu of T4 phage per mouse resulted in stable and long-term colonization in the gut that did not appear to be dose-dependent (shown in representative results section). Phage-bacteria kinetics should therefore be piloted in vivo prior to large scale experiments. - Under sterile, gnotobiotic conditions, gavage each mouse with 100 µL of autoclaved 1 M NaHCO3 to neutralize stomach acids. Wait 10 min, then gavage with 100 µL of T4 phage lysate or vehicle control.
- Monitor mice for adverse health effects.
- Dilute T4 phage lysates and vehicle controls to a predetermined concentration in SM buffer. As per Hsu et al.2, dilute phage lysates to allow for administration of 2 x 106 pfu per mouse2.
3. Monitoring T4 phage levels in vivo
NOTE: Once mice have been inoculated with phage, the concentration of both phages and target bacteria can be measured in fecal or tissue samples. This provides information on the kinetics of the phage infection and colonization dynamics of both organisms.
- Spot plating T4 phage to determine concentration in fecal pellets
- Collect fecal pellets from each mouse into sterile, pre-weighed, microcentrifuge tubes to measure T4 phage and E. coli levels. Store tubes on ice until plating.
NOTE: Place samples on ice in the interim between collection and plating to slow the growth of aerotolerant bacteria. Over several hours, there may still be growth of aerobic bacteria, or some death of anaerobic bacteria due to oxygen exposure which could lead to skewed bacteria counts. Therefore, bacteria and phage plating should be performed as soon as possible after sample collection. Obligate anaerobic bacteria will not tolerate oxygen exposure. To preserve the sample, collect samples into sealed tubes and transfer tubes to an anaerobic chamber as soon as possible after collection. If no growth is detected in fecal samples, consider alternatives such as 16S rRNA qPCR for bacterial quantification. - Record final weights of each tube and calculate the sample weight by subtracting the initial tube weight. This will be used for normalizing T4 phage and E. coli concentration to sample weight (pfu/g or cfu/g, respectively).
- Add 1 mL of sterile SM buffer to each tube and vortex thoroughly at maximum speed (>1 min) to homogenize fecal pellets. If the sample is less than 15 mg, a smaller volume of SM buffer may be added. Record the volume of SM buffer added to each sample for calculations of cfu/g or pfu/g of sample.
- Prepare a series of 8 (or more depending on the expected phage concentration) serial dilutions of 20 µL of each sample in 180 µL SM buffer, in factors of 10. Vortex each homogenized sample briefly to mix before adding to the first tube/well. Pipette to mix in between each addition and change tips between each dilution to prevent inflating phage or bacteria counts via sample carryover.
NOTE: If fibers and debris present in the sample hinder pipetting, add additional buffer to the fecal slurry to further dilute the stock sample. - Spot 5 µL of each dilution onto LB soft agar plates containing E. coli (for phage plaque assays) or LB agar plates (1.5% agar, for bacterial colony assays) to determine T4 phage and E. coli concentration in each sample. For accuracy, spot each sample in triplicate.
NOTE: If preparing serial dilutions in a 96-well plate, an 8-channel P20 multichannel pipette may be used to pipette each column of serially diluted sample onto plates. Change tips between each dilution, even if moving from most dilute to most concentrated, as phage may adhere to the walls of the pipette tip and alter the amount of phage added to each new well31. - Allow each spot to dry before inverting the plate and placing it in the incubator. Incubate overnight at 37 °C.
- For each sample, select the dilution at which there are 3-30 countable plaques per spot. Count and record the number of plaques in the spot and the dilution used.
- Calculate pfu/g of sample by dividing the number of plaques by the volume plated in each spot to give pfu/µL. Multiply this by the dilution factor and by the volume of SM buffer added to each sample to give pfu/sample. Lastly, divide by the sample weight to give pfu/g30.

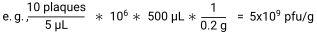
- Collect fecal pellets from each mouse into sterile, pre-weighed, microcentrifuge tubes to measure T4 phage and E. coli levels. Store tubes on ice until plating.
- Tissue sampling at experimental endpoints
- At each selected time point, euthanize mice as per approved institutional animal ethics protocol and collect cecal contents, small and large intestinal contents, and any tissues of interest into sterile, pre-weighed 2 mL round-bottom microcentrifuge tubes.
- Record final weights of each tube to calculate the sample weight. Store tissues and cecal contents on ice until same day plating.
- Add sterile SM buffer to each tube and record the volumes added.
- For cecal and intestinal contents, vortex samples thoroughly (>1 min) as per step 3.1.3.
- For tissue samples, add one sterile metal bead to each tube. Homogenize tissues using a tissue lyser (see Table of Materials) at 20 Hz for 5 min or until tissues are dissociated and the suspension is homogenous.
NOTE: If samples do not homogenize well, consider increasing the volume of SM buffer added, increasing the homogenization time, or increasing the homogenization frequency to 30 Hz. A tissue homogenizer may also be used to dissociate tissues while allowing for bacterial and phage recovery39,40. Homogenizers are operated at higher frequencies than tissue lysers while maintaining bacteria integrity. - Prepare serial dilutions of each sample in factors of 10 and perform spot plating to determine E. coli and T4 phage concentrations in each sample, as described in step 3.1.4-3.1.841.
Representative Results
To investigate the interactions between the T4 phage/E. coli dyad in the murine intestine, T4 phage and vehicle lysates were prepared, cleaned, and purified (Figure 1A). T4 phage lysates were titered by plaque assay and diluted to 2 x 107 pfu/mL (2 x 106 pfu/mouse) in SM buffer. Vehicle lysates were also titered to confirm no viable phage presence and diluted in the same volume of SM buffer as the T4 phage lysate. Endotoxin levels were quantified in diluted lysates using a chromogenic endotoxin quantification kit. T4 phage, heat-inactivated phage, and vehicle lysates had endotoxin levels below acceptable levels for drinking water (<20 EU/mL)25 (Figure 1B). As the vehicle lysate had a similar amount of endotoxin as the T4 phage lysate, it was deemed to be a suitable control. The heat-inactivated phage lysate (2 x 107 pfu/mL prior to heat treatment) had less endotoxin than both the vehicle and T4 phage lysate. As per the kit instructions, samples that had been 1-octanol-treated were tested for assay inhibition by addition of an endotoxin standard to the vehicle lysate (spiked vehicle), to give a final concentration of 0.5 EU/mL. The spiked vehicle endotoxin levels minus vehicle endotoxin levels measured 0.5 EU/mL ±25%, as per manufacturer recommendations (Figure 1B). This demonstrated that 1-octanol was successfully removed and that there was no assay inhibition caused by the lysates that could interfere with the results of the assay.
E. coli K-12 (BW25113) monocolonized C57BL/6 mice were bred within a sterile flexible film vinyl isolator under gnotobiotic conditions (Figure 2A). F0 generation mice were colonized with E. coli through its addition to their environment and co-housing. Only the progeny (F1 and F2 generation) that were colonized with E. coli from birth through vertical transmission and co-housing were used for experiments. This strategy controlled for the changes to the immune system, mucus production, and GI tract development that occurs in response to bacterial inoculation22,37, therefore allowing the measurement of changes that occur only because of phage introduction. Once matured, experimental mice were transferred to sterile isocages, where they each received 2 x 106 pfu of T4 phage or an equal volume of vehicle lysate by oral gavage at 6-8 weeks of age. T4 phage and E. coli abundance was monitored over four weeks by collection of fecal pellets and spot plating assays on either 0.5% (soft) agar or 1.5% agar, respectively. Over the course of this experiment, T4 phage and E. coli were able to co-exist without either population being depleted (Figure 2B, C). Oral administration of T4 phage at lower doses of 2 x 102 and 2 x 104 pfu/mouse did not reduce the ability of T4 phage to colonize the gut compared to 2 x 106 pfu (Figure 2D). Similarly, a dose-dependent effect of T4 inoculation on E. coli levels was not observed (Figure 2E).
Importantly, consistency in plating between samples was found to be crucial to the plaque count. For instance, when adding bacteria into the soft agar, it was determined that the density of bacteria greatly influenced the outcome of the assay. Addition of E. coli that was subcultured for 1 h, 1.5 h, or 2 h, resulted in lower plaque counts than with the addition of E. coli subcultured for 2.8 h or more. Furthermore, plaque quantification stabilized at ~1 x 109 pfu/mL when E. coli was subcultured for 2.8 h to 16 h (overnight), which was approximately a 5-fold increase compared to the value calculated from a 1 h sub-culture (~2 x 108 pfu/ml) (Figure 3A). Rather than sub-culturing, target bacteria can be inoculated into soft agar from overnight cultures. Here, the volume of the 16 h (overnight) E. coli culture that was inoculated into the soft agar impacted plaque counts (Figure 3B). These results indicate that the density of E. coli within the agar can influence plaque assay counts. In both approaches, phage quantification results plateaued at the same value (109 pfu/ml), suggesting that target bacteria must be of sufficiently high density in the soft agar to ensure plaque count accuracy.
Taken together, these results highlight the importance of developing an understanding of phage-bacteria kinetics through in vitro and in vivo pilot experiments prior to performing experimental manipulations.
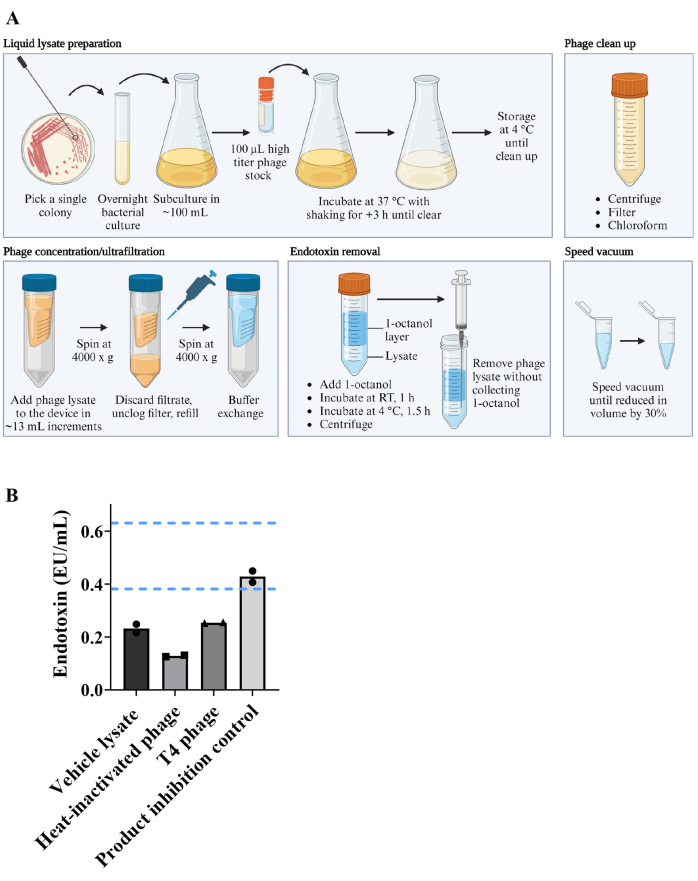
Figure 1: Preparation of phage lysates. (A) Workflow for starting a phage lysate from a high titer phage stock. Lysates were propagated, cleaned, concentrated, and endotoxins were removed. Contaminating 1-octanol was removed by speed vacuum. (B) Endotoxins were quantified in phage and vehicle lysates using a chromogenic endotoxin quantification kit. Dashed blue lines indicate the upper and lower boundaries to disprove product inhibition in the product inhibition control (0.5 EU/mL ± 25%). Each point represents one of two technical replicates. Bar height represents the mean across replicates. Please click here to view a larger version of this figure.
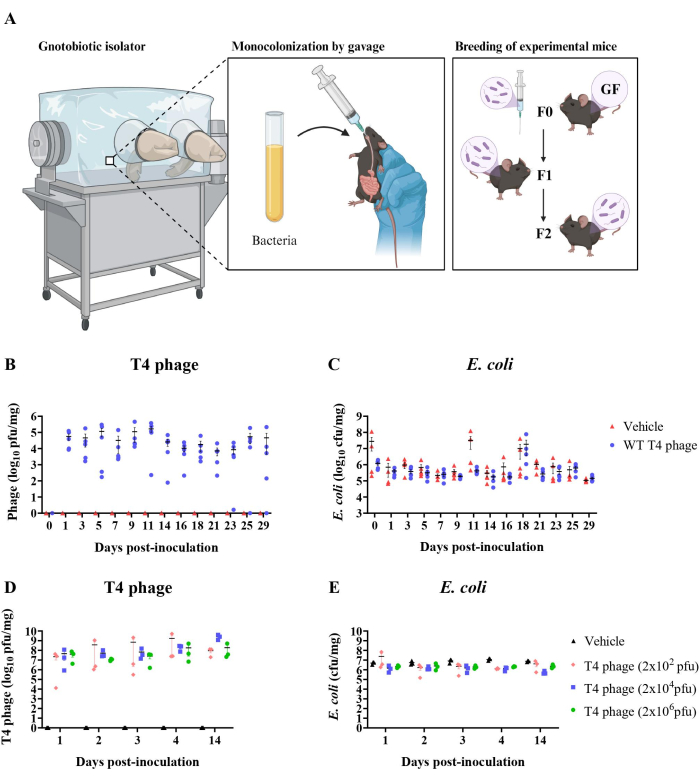
Figure 2: Generation of monocolonized mice and monitoring of phage and bacteria levels in fecal pellets. (A) Workflow for colonizing germ-free mice with a single bacteria species and generation of F1 and F2 monocolonized mice. (B–E) Levels of T4 phage and E. coli detected in fecal pellets of C57BL/6 mice that were born monocolonized with E. coli and were inoculated with T4 phage at 6-8 weeks old. At day 0 (pre-colonization) and the indicated days post-colonization, T4 phage levels were measured by spot plating and plaque assays using the single agar layer method. E. coli levels were measured by spot plating on 1.5% LB agar. (B, C) Kinetics of (B) T4 phage and (C) E. coli levels in recovered fecal pellets following inoculation with 2 x 106 pfu of T4 phage or vehicle lysate (normalized for volume and endotoxin levels). (D, E) (D) T4 phage and (E) E. coli levels in recovered fecal pellets following inoculation with 2 x 102 pfu, 2 x 104 pfu or 2 x 106 pfu of T4 phage. Error bars represent mean and standard error of the mean (SEM). Please click here to view a larger version of this figure.
Figure 3: Troubleshooting accuracy in plaque assays. (A) T4 phage lysate titer using E. coli overnight cultures or subcultures for soft agar preparation (0.5% agar, LB). Subcultures were made using a 1:50 dilution of overnight E. coli culture into fresh LB. 5 mL of soft agar preparations were inoculated with 100 µL of E. coli and 20 µL of T4 phage. CaCl2 and MgSO4 were not added to soft agar for these tests but can be added if desired. E. coli subculture times below 2.8 h resulted in a lower apparent phage titer. (B) T4 phage lysate titer using different E. coli densities in soft agar preparation. Adding different volumes of E. coli from an overnight culture (not sub-cultured, as in A) into the soft agar resulted in different apparent phage titers. Error bars represent calculated phage titer uncertainty. Please click here to view a larger version of this figure.
Discussion
The study of phages in the microbiome presents a significant challenge compared to their bacterial counterparts. Specifically, phages do not contain a conserved phylogenetic marker common to all phages akin to the 16S and 18S ribosomal subunits that allow for the ease in sequencing and identification of prokaryotic and eukaryotic species, respectively42. However, with advances in next generation sequencing approaches, including increasing read lengths, throughput and decreasing costs, comes the rapid expansion of bacteriophage genome databases42,43,44. With much groundwork in phage discovery well underway, phage research is now more accessible than ever. As key members of the gut microbiome, and the most genetically diverse organisms on Earth, phages present an exciting angle for new research investigating the complexity of the microbiome. The protocols described here focus on investigating known phage species in mice colonized with their target bacteria. It should be noted that these protocols provide a guideline for studying phage in vivo and can be expanded upon with the growth of the field.
Bacteriophage research for therapeutic treatment of bacterial infections largely fell out of fashion with the advent of antibiotics in the 1940s. However, over-prescription and misuse of antibiotics has accelerated the emergence of multi-drug resistant pathogens as a major public health concern3. Phages present an attractive potential alternative to antibiotics, due to their high degree of host specificity3. Importantly, as phages naturally take up residence as members of the human gut microbiome, it is necessary to first understand the roles that phages play in these complex environments. While many studies have investigated the relationship between a phage and its target bacteria in vitro, it is important to consider how these relationships might hold up within the landscape of the GI tract. As in exploratory studies investigating the bacterial components of the microbiota, simplified mouse models such as GF and monocolonized mice allow us to isolate effects of phage on its target species, and how this relationship contributes to the immune response. This is an important step towards phage therapy, as some phages may not provide benefit when introduced to the gut. For example, the Pseudomonas aeruginosa phage Pf4 exacerbates the disease caused by its host, by inhibiting both anti-bacterial immune responses and keratinocyte migration, resulting in impaired wound healing18,45. The procedures described here aim to standardize techniques for studying phages as members of the murine microbiota. Echoing pioneering studies investigating the impact of the microbiota on the metazoan host, continued research in phages within the context of the microbiota may prove exciting and significant in the broader understanding of the multibiome46.
Limitations
The protocols described here have been optimized for studying the competition between T4 phage and its target bacteria, E. coli, when T4 phage are administered by oral gavage to E. coli monocolonized mice. Like bacteria, distinct phage species behave differently from one another, having varying replication times, burst sizes, and ranges of bacterial targets47. Therefore, when investigating other phage-bacterial pairs in similar animal models, one should take care to optimize these protocols at all steps. For example, phages with a longer replication time and/or a smaller burst size may require a longer incubation with their bacterial target for production of a high titer lysate. Similarly, plate incubation times may need to be extended for plaque assays. Furthermore, phages with a short replication time and/or high burst size may require shorter plate incubations, as overnight incubation may result in plaque overgrowth. If the propagation conditions are unknown for a specific phage of interest, the groundwork for determining growth conditions should be established before beginning in vivo work48.
Ig-like domains found within the Hoc capsid proteins of T4 phage facilitate adherence to intestinal mucus11. Depending on the mucus-binding capacity of the phage of choice, the kinetics of phage-bacteria levels in fecal samples may differ from results shown in Figure 2B–E. For instance, it was shown in gut-on-a-chip systems that phage containing intact Hoc proteins had increased E. coli killing capacity compared to Hoc-deficient phage11. While it is suspected that T4 phages are retained through mucus-binding in vivo11,12,13, the impact of re-inoculation through the coprophagic behavior of mice cannot be ruled out. These effects could be reduced by using wire cage bottoms, or by frequently changing cages. Since Ig-like domains have not been identified in ssDNA or RNA phages49, it will be interesting to tease apart the other strategies used by phage to retain residence within the gut mucosa.
Finally, these exploratory studies have examined phage-bacteria-host interactions at homeostasis only. In animal models of human disease, it is unknown how changes in intestinal barrier integrity, such as in IBD, might impact these interactions. Recent studies have shown that Caudovirales phages have increased richness and diversity in patients with IBD7,8. In mouse models, it was shown that continuous phage treatment exacerbated dextran sodium sulfate (DSS)-induced experimental colitis8. It remains to be determined how phage-bacteria-host interactions play out in inflammatory environments, and whether impaired barrier integrity facilitates phage-induced inflammation.
Troubleshooting and alternative methods
Mouse models
Monocolonized mouse models allow for the interrogation of a single species of bacteria on host physiology16. Research involving monocolonized mice has played a crucial role in elucidating how the microbiota influences the immune system22. As a model system, monocolonized mice do not recapitulate the physiology of their conventional microbiota counterparts. More similar to GF mice, E. coli monocolonized mice have reduced mucus production (similar to GF mice50), and an immature immune system23. However, monocolonized mice have been invaluable for unravelling the microbe-specific effects of individual species on GI tract and immune development. A classic example is the discovery that segmented filamentous bacteria (SFB) are potent inducers of CD4+ T helper 17 cells in mice24. With reference to mucus, there are microbe-specific effects on mucus-gene transcription51and mucus thickness50. While limited, monocolonized mice provide opportunities to investigate the impact of one phage-bacteria pair on the mammalian host in a controlled model. Importantly, phages can, and should, be investigated in the context of a complex microbiota. At the time of writing, few studies have addressed the impacts of phage predation of commensal bacterial species within a conventional microbiome. This is a future application of the protocols presented in this paper, which could be adapted to facilitate these studies.
Use of appropriate vehicle controls
Appropriate controls are essential in any experiment. Here, a suitable vehicle for oral administration to mice was defined that controls for the multi-step cleaning and purification of T4 phage lysates. An important requirement is that the vehicle control contains an equal level of bacterial endotoxins as the phage lysate, to control for any endotoxin-mediated immune response. Chloroform and 1-octanol are added to the lysate as part of the purification process and subsequently removed. To control for potential immune responses that may be generated to trace levels of these chemicals, a phage-free bacterial lysate can be produced as a vehicle control. T4 phage and vehicle lysates contained a very low concentration of bacterial endotoxins, well below the levels allowed in drinking water25 (Figure 1B). Alternative controls may be used and can be modified to suit the intended research question. Most simply, phage buffer containing an equivalent amount of endotoxin can be used, although this does not account for the multi-step cleaning and purification processes. Gogokhia et al.8reported the use of heat-inactivated phage as a control for whether phage protein without DNA was sufficient to elicit an immune response. In our hands, heat-inactivated phage had lower levels of endotoxin than the T4 phage lysate (Figure 1B), and therefore was not used to control for endotoxin levels in this study. However, we encourage the testing of both protocols to determine which method is most suitable for individual experimental purposes. If the vehicle and T4 phage lysates differ significantly in the amount of endotoxin present, the lysate containing the lower levels can be supplemented with purified endotoxin. 1-Octanol is added to lysates during the purification process but inactivates the chromogenic endotoxin quantification kit test. It is important to test product inhibition by potentially interfering substances in the sample to ensure that endotoxin measurements are accurate. If product inhibition is suspected, it is possible that the sample was not speed vacuumed for long enough. If problems persist, dialysis can be used to remove remaining 1-octanol25.
Administration route and dose
Due to the mucus-binding ability of T4 phage, mouse models were inoculated in a single oral gavage dose. The purpose of this was to monitor the duration of phage and E. coli co-habitation of the GI tract; however, other approaches to studying phage in in vivo models have been reported. For example, phage-enriched drinking water has been used to provide a continual supply of phage to mice8,20. The benefit of this method of administration is that phages are continually replenished, even in the absence of a bacterial host, as demonstrated by Gogokhia et al.8. This experimental design allowed for evaluation of the immune response to phages in the absence of bacteria and provided continual immune stimulation over the course of the experiment. Physiologically, this method does not represent a natural course of phage infection or phage colonization of an intestinal environment, but it does provide important insights into how repeated phage exposure may prime the immune system. This has significance in the context of phage therapy development, as phage cocktails may be delivered as a course of treatment to treat intestinal bacterial infections, similar to antibiotic regimens. In the context of the T4 phage-E. coli pair, it was determined that decreasing the dose of T4 phage applied orally to monocolonized mice did not alter fecal phage or bacteria levels (Figure 2D, E). Therefore, in this case the inoculation dose of T4 phage is not crucial for maintenance of stable T4 phage-E. coli colonization in the gut of bicolonized mice. It is noted that the lowest dose administered was 200 pfu/mouse, and the highest dose was 2 x 106 pfu/mouse (as per Hsu et al.)2. Therefore, data supporting T4 phage-E. coli kinetics outside this range is unavailable in this study.
Spot and whole plate phage titers
For measuring T4 phage levels in feces and tissues, the protocols outlined here rely upon spot plating techniques rather than whole plate plaque assays, which may contribute to the variability in phage levels between mice (Figure 2B). In whole plate techniques, plaques are counted over a larger area, allowing for more accurate quantification. However, an appropriate dilution of each sample must usually be predetermined by spot plating. Since samples were assayed on the day of sample collection, spot plating was deemed to be the most appropriate method for a more high-throughput approach. Therefore, the resolution of this data is represented most accurately by the order of magnitude of phage in each sample. For precise phage measurements, there are additional considerations for each phage. More accurate measurements can be obtained by whole plate assays, or by using smaller serial dilution ranges for each sample. If these methods still result in variable phage or target bacterial counts, it would be prudent to assess whether unique interactions between the phage and bacteria could account for differences among individual mice via metagenomic sequencing or other methods to experimentally assess co-evolution and resistance. As per Bonilla et al.25, when preparing soft agar for plaque assays, it is important to be consistent with the amount of bacterial host added25. As shown in Figure 3, even relatively small changes in bacteria culture times (Figure 3A) and density (Figure 3B) can affect the accuracy of plaque assays. These findings may be different for other phage-bacteria pairs, and similar experiments are recommended when starting out with new organisms. Additionally, for new phage-bacteria pairs, exploratory experiments should be performed to determine the growth characteristics of each. For example, growth curves can be performed to determine the lag, exponential and stationary phases, and growth rate of the bacteria. The one-step growth experiment can be used to determine the latent period (time to lysis) and burst size (number of phages released upon bacterium lysis) of phages52,53.
Alternative T4 phage measurement by qPCR
Quantifying phage can be performed with plaque assays, as outlined above, or via quantitative polymerase chain reaction (qPCR). These two approaches have an important distinction: plaque assays determine the number of viable phage capable of infecting and killing their bacterial hosts, while qPCR quantifies phage-specific genetic material (as a proxy of the number of phage present) but does not provide information about the viability and infectivity of the phage. qPCRs were performed to compare the quantification of phage gene copies with plaques formed in plaque assays using primers described by Hsu et al.2 (Figure 4). T4 phage DNA was extracted and amplified from the cecal contents of T4 phage/E. coli bicolonized mice. In most mice, qPCR and plaque assays detected similar levels of T4 phage in cecal contents (Figure 4A, B). While some amplification was detected in vehicle-inoculated cecal contents, the calculated gene copies/g were below the limit of detection (LOD) (Figure 4A). The absence of quantifiable phage in these samples was confirmed by gel electrophoresis. T4 phage gene products (96 bp) were readily visualized in DNA isolated from cecal contents of T4 inoculated mice but were absent from vehicle controls (Figure 4C).
In these tests, T4 phage gene copies were detected in the cecal contents of one T4 phage-colonized mouse by qPCR (Figure 4A, arrow) despite the inability to detect T4 in plaque assays from the same sample (Figure 4B, arrow). These results suggest that viral particles that do not form plaques may arise in vivo, either due to production of defective viral particles or co-evolution of phage and/or bacteria. Bacterial plating techniques on hard agar can be used to determine fecal bacteria susceptibility to phage14. We suggest that qPCR-mediated detection may be a valuable addition to in vivo phage workflows as it may be more robust to phage evolution within the intestinal environment given that it targets short, conserved gene sequences. Non-biased approaches such as metagenomics are valuable for examining phage-bacterial co-evolution and relative abundances but may ultimately be more costly.
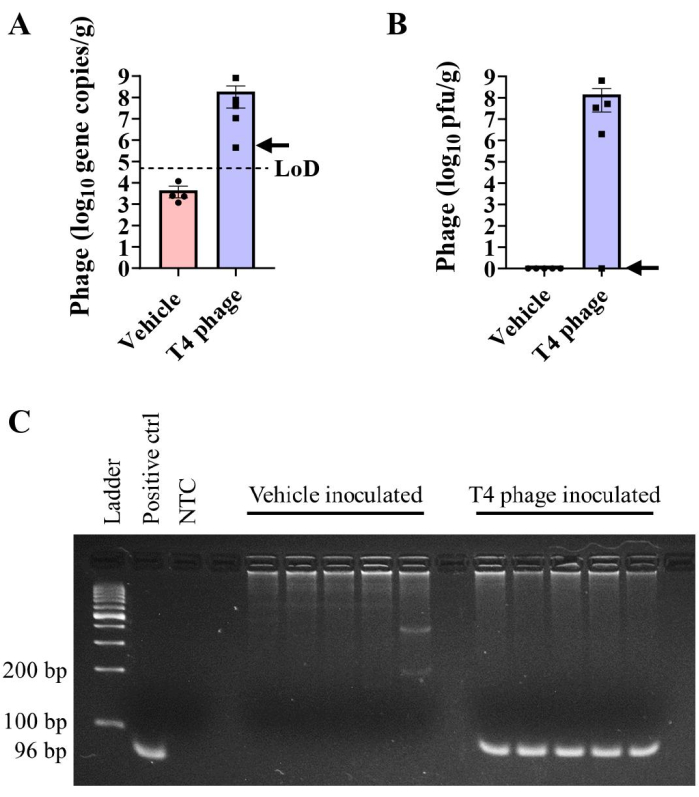
Figure 4: Detection of T4 phage by qPCR. (A,B) T4 phage burdens were measured via (A) absolute qPCR or (B) plaque assay in the cecal contents of E. coli colonized C57BL/6 mice at day 29 post-inoculation with T4 phage or vehicle. The arrow indicates results from an individual mouse that had detectable T4 genome copies but not plaquable virus in cecal contents. (C) Gel electrophoresis of PCR products showing the presence of the 96 bp band in T4 phage-inoculated cecal samples, but not in vehicle inoculated samples. A 100 bp DNA ladder was used. LOD = limit of detection. Error bars represent mean and SEM. Please click here to view a larger version of this figure.
Applications
Bacteriophages represent upwards of 90% of the virus-like particles present in the microbiome44; however, the impact of phages on the gut microbiome is poorly understood. Initial studies investigating the bacterial component of the microbiome did so by isolating particular species, and studying their impact on immune maturation22,24. Similar interrogations of the phageome present a daunting but necessary task, and a requirement to gain a comprehensive understanding of the multibiome46. While the focus of phage therapy development tends to be geared towards curing septic bacterial infections, it is important to understand how the addition of a phage cocktail to the GI tract may alter the intestinal ecosystem. Furthermore, the safety of therapeutic phage cocktails should be assessed by generating an immune profile. Phages have been investigated as potential treatments for intestinal bacterial infections such as C. difficile5 and Salmonella enterica serovar Typhimurium54. Recent studies have also demonstrated that FFTs are equally or more effective in treating necrotizing enterocolitis in pre-term pigs9, suggesting the viral component of fecal microbiota transplants (FMTs) may play an active role in disease reduction. Continued studies investigating the role of phages in the microbiome are warranted to further the development of phage therapies against intestinal pathogens, but also to better inform FMTs as treatment for disease. By standardizing methods for the study of phages in vivo, transparency and reproducibility in phage research will be increased, along with guidance for those extending their work into mouse models.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge that the land they performed this research on is the traditional, ancestral, and unceded territory of the xwməθkwəy̓əm (Musqueam) Nation. The land it is situated on has always been a place of learning for the Musqueam people, who for millennia have passed on in their culture, history, and traditions from one generation to the next on this site. We encourage others to learn more about the native lands in which they live and work at https://native-land.ca. The authors acknowledge support from the Natural Sciences and Engineering Council of Canada (NSERC) Canadian Graduate Scholarships – Master's (N.P.), Michael Smith Health Research BC Trainee Award (RT-2023-3174, to MH), Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grants Program (RGPIN-2019-04591 to C.T., RGPIN-2016-04282 to LCO), Canadian Institute for Advanced Research / Humans and the Microbiome (FL-001253 Appt 3362, to C.T.), Michael Smith Foundation for Health Research Scholar Award (18239, to C.T.), Canadian Institutes for Health Research (PJT-159458 to LCO) and the Canadian Foundation for Innovation (34673 to LCO and 38277 to CT). We are grateful for technical support from the UBC Centre for Disease Modelling and ubcFLOW, which is supported by the UBC GREx Biological Resilience Initiative, and to members of the Osborne and Tropini labs for critical discussions and evaluation of the manuscript. Figure 1A and Figure 2A were created using Biorender.com.
Materials
| 1-octanol (99%) | Thermofisher | CAAAA15977-AP | |
| 50 ml PES Steriflip Sterile Disposable Vacuum Filter Units | Millipore Sigma | SCGP00525 | |
| Agarose (Low-EEO/Multi-Purpose/Molecular Biology Grade) | Fisher BioReagents | BP160-500 | |
| Amicon® 100kDa Ultra-15 centrifugal filter device, Ultracel-100 | Millipore Sigma | UFC910008 | |
| BD Microtainer® Tubes, SST | BD Medical | 365967 | |
| Bioexclusion airtight cages (ISO cages) | Techiplast | 1245ISOCAGE | |
| C1000 Touch™ Thermal Cycler with 96-Well Fast Reaction Module | BioRad | 1851196 | |
| Calcium Chloride Dihydrate (White Crystals to Powder) | Fisher BioReagents | BP510-500 | |
| Cap Locks For 1.5ML Tube 100/pk | Andwin Scientific | 16812612 | |
| Chloroform (Ethanol as Preservative/Certified ACS) | Fisher | C298-500 | |
| Copper coated steel beads (4.5 mm) | Crosman Corporation | 0767 | |
| DNeasy Blood & Tissue Kit (50) | Thermo Scientific | 69504 | |
| DreamTaq Green PCR Master Mix (2X) | Thermo Scientific | K1081 | |
| Ethylenediaminetetraacetic acid (EDTA) disodium salt solution, for molecular biology, 0.5 M in H2O | Sigma Aldrich | E7889 | |
| Fisher BioReagents™ Agar, Powder / Flakes, Fisher BioReagents™ | Fisher Bioreagents | BP1423-500 | |
| Fisher BioReagents™ Microbiology Media: LB Broth (Powder) – Lennox | Fisher Bioreagents | BP1427-500 | |
| GeneRuler 100 bp DNA Ladder | Thermo Scientific | SM0241 | |
| Green FastMix® qPCR mix, 1250 rxns | QuantaBio | 95072-012 | |
| HEPA filters for isocage lids, AUTOCLAVABLE H14 FILTERS FOR ISO LINE- IRRADIATED | Techiplast | UISOHEPAXTBOX-300 | |
| Magnesium sulfate heptahydrate | Fisher BioReagents | BP213-1 | |
| MaxQ 6000 Incubated Shaker | Thermo Scientific | 8354-30-0009 | |
| Microbiology Media: LB Broth (Powder) – Lennox | Fisher BioReagents | BP1427-500 | |
| Microcentrifuge Tubes with Locking Snap Cap, 2ml | Fisher | 14-666-315 | |
| Parafilm sealing film | Bemis | PM-996 | |
| Phage stocks | Carolina Biological Supply | n/a | |
| PicoLab® Mouse Diet 20 EXT | LabDiet | 5R58 | |
| Pierce™ Chromogenic Endotoxin Quant Kit | Thermo Scientific | A39552S | |
| RNase A (17,500 U) | Qiagen | 19101 | |
| RNase-free DNase Set | Qiagen | 79254 | |
| Sodium Bicarbonate (Fine White Powder) | Fisher Chemical | BP328-500 | |
| Sodium Chloride (Crystalline/Certified ACS) | Fisher Chemical | S271 | |
| Sonicator (probe model CL-18; power source model FB50) | Fisher scentific | n/a | |
| Sterile flexible film isolator | Class Biologically Clean | n/a | |
| SYBR™ Safe DNA Gel Stain | Invitrogen | S33102 | |
| T100 Thermal Cycler | BioRad | 1861096 | |
| T4 phage primer, forward (CCACACATAGCGCGAGTATAA) | IDT | n/a | |
| T4 phage primer, forward (GAAACTCGGTCAGGCTATCAA) | IDT | n/a | |
| TissueLyser II | Qiagen | 85300 | |
| Tris-HCl, 1M Solution, pH 8.0, Molecular Biology Grade, Ultrapure | Thermo Scientific | AAJ22638AE | |
| Water, (DNASE, RNASE free) | Fisher BioReagents | BP2484100 |
Referenzen
- Rohwer, F., Segall, A. M. A century of phage lessons. Nature. 528 (7580), 46-47 (2015).
- Hsu, B. B., et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host & Microbe. 25 (6), 803-814.e5 (2019).
- Gordillo Altamirano, F. L., Barr, J. J. Phage Therapy in the postantibiotic era. Clinical Microbiology Reviews. 32 (2), (2019).
- Schooley, R. T., et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrobial Agents and Chemotherapy. 61 (10), (2017).
- Ott, S. J., et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology. 152 (4), 799-811.e7 (2017).
- Zuo, T., et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 67 (4), 634-643 (2018).
- Norman, J. M., et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 160 (3), 447-460 (2015).
- Gogokhia, L., et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host & Microbe. 25 (2), 285-299.e8 (2019).
- Brunse, A., et al. Fecal filtrate transplantation protects against necrotizing enterocolitis. The ISME Journal. 16 (3), 686-694 (2022).
- Duerkop, B. A., Clements, C. V., Rollins, D., Rodrigues, J. L. M., Hooper, L. V. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proceedings of the National Academy of Sciences of the United States of America. 109 (43), 17621-17626 (2012).
- Barr, J. J., et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 110 (26), 10771-10776 (2013).
- Nguyen, S., et al. Bacteriophage Transcytosis provides a mechanism to cross epithelial cell layers. mBio. 8 (6), (2017).
- Bichet, M. C., et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience. 24 (4), 102287 (2021).
- Buttimer, C., et al. Impact of a phage cocktail targeting Escherichia coli and Enterococcus faecalis as members of a gut bacterial consortium in vitro and in vivo. Frontiers in Microbiology. 13, 936083 (2022).
- Li, Y., et al. Bacteriophages allow selective depletion of gut bacteria to produce a targeted-bacterium-depleted mouse model. Cell Reports Methods. 2 (11), 100324 (2022).
- Reyes, A., Wu, M., McNulty, N. P., Rohwer, F. L., Gordon, J. I. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proceedings of the National Academy of Sciences of the United States of America. 110 (50), 20236-20241 (2013).
- Federici, S., et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 185 (16), 2879-2898.e4 (2022).
- Sweere, J. M., et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science (New York, N.Y.). 363 (6434), (2019).
- Fluckiger, A., et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science (New York, N.Y.). 369 (6506), 936-942 (2020).
- Majewska, J., et al. Induction of Phage-Specific Antibodies by Two Therapeutic Staphylococcal Bacteriophages Administered per os. Frontiers in Immunology. 10, 2607 (2019).
- Weiss, M., et al. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology. 393 (1), 16-23 (2009).
- Thomson, C. A., Morgan, S. C., Ohland, C., McCoy, K. D. From germ-free to wild: modulating microbiome complexity to understand mucosal immunology. Mucosal Immunology. 15 (6), 1085-1094 (2022).
- Al-Asmakh, M., Zadjali, F. Use of germ-free animal models in microbiota-related research. Journal of Microbiology and Biotechnology. 25 (10), 1583-1588 (2015).
- Ivanov, I. I., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139 (3), 485-498 (2009).
- Bonilla, N., et al. Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ. 4, e2261 (2016).
- Sambrook, J., Russell, D. W. . Molecular Cloning: A Laboratory Manual. 1, (2001).
- Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., Johnson, R. P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods in Molecular Biology (Clifton, N.J). 501, 69-76 (2009).
- Manikantha, B., Karthika, R., Murugadas, V., Vishnuvinayagam, S., Rao, B. M. Comparison of the single agar and double agar layer methods for enumeration of bacteriophages. Fishery Technology. 59, 60-63 (2022).
- Sanders, E. R. Aseptic laboratory techniques: plating methods. Journal of Visualized Experiments. 63, e3064 (2012).
- Louten, J. Chapter 7 – Detection and diagnosis of viral infections. Essential Human Virology. , 111-132 (2016).
- Richter, &. #. 3. 2. 1. ;., et al. Adsorption of bacteriophages on polypropylene labware affects the reproducibility of phage research. Scientific Reports. 11 (1), 7387 (2021).
- . Amicon Ultra-15 Centrifugal Filter Devices Available from: https://www.emdmillipore.com/CA/en/product/Amicon-Ultra-15-Centrifugal-Filter-Unit (2018)
- Hecker, W., Witthauer, D., Staerk, A. Validation of dry heat inactivation of bacterial endotoxins. PDA Journal of Pharmaceutical Science and Technology. 48 (4), 197-204 (1994).
- Jakočiūnė, D., Moodley, A. A Rapid bacteriophage DNA extraction method. Methods and Protocols. 1 (3), 27 (2018).
- Zucoloto, A. Z., Yu, I. L., McCoy, K. D., McDonald, B. Generation, maintenance, and monitoring of gnotobiotic mice. STAR Protocols. 2 (2), 100536 (2021).
- Ng, K. M., et al. Single-strain behavior predicts responses to environmental pH and osmolality in the gut microbiota. mBio. 14 (4), e0075323 (2023).
- McCallum, G., Tropini, C. The gut microbiota and its biogeography. Nature Reviews. Microbiology. , (2023).
- Bergstrom, K., Xia, L. The barrier and beyond: Roles of intestinal mucus and mucin-type O-glycosylation in resistance and tolerance defense strategies guiding host-microbe symbiosis. Gut Microbes. 14 (1), 2052699 (2022).
- Askar, M., Ashraf, W., Scammell, B., Bayston, R. Comparison of different human tissue processing methods for maximization of bacterial recovery. European Journal of Clinical Microbiology & Infectious Diseases. 38 (1), 149-155 (2019).
- Redanz, S., Podbielski, A., Warnke, P. Improved microbiological diagnostic due to utilization of a high-throughput homogenizer for routine tissue processing. Diagnostic Microbiology and Infectious Disease. 82 (3), 189-193 (2015).
- Bhinder, G., et al. The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. Journal of Visualized Experiments. 72, e50222 (2013).
- Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F., Gordon, J. I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nature Reviews Microbiology. 10 (9), 607-617 (2012).
- Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D., Lawley, T. D. Massive expansion of human gut bacteriophage diversity. Cell. 184 (4), 1098-1109.e9 (2021).
- Reyes, A., et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 466 (7304), 334-338 (2010).
- Bach, M. S., et al. Filamentous bacteriophage delays healing of Pseudomonas-infected wounds. Cell Reports. Medicine. 3 (6), 100656 (2022).
- Filyk, H. A., Osborne, L. C. The multibiome: The intestinal ecosystem’s influence on immune homeostasis, health, and disease. EBioMedicine. 13, 46-54 (2016).
- Rohwer, F., Merry, Y., Maughan, H., Hisakawa, N. Heather Life in Our Phage World: A Centennial Field Guide to the Earth’s Most Diverse Inhabitants. Wholon. , (2014).
- Glonti, T., Pirnay, J. P. In Vitro techniques and measurements of phage characteristics that are important for phage therapy success. Viruses. 14 (7), 1490 (2022).
- Fraser, J. S., Yu, Z., Maxwell, K. L., Davidson, A. R. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. Journal of Molecular Biology. 359 (2), 496-507 (2006).
- Li, H., et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nature Communications. 6, 8292 (2015).
- Bergström, A., et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Research Notes. 5, 402 (2012).
- Adams, M. H. . Bacteriophages. , (1959).
- Kutter, E., Sulakvelidze, A. . Bacteriophages: Biology and Applications. , (2004).
- Bao, H., et al. Dysbiosis and intestinal inflammation caused by Salmonella Typhimurium in mice can be alleviated by preadministration of a lytic phage. Microbiological Research. 260, 127020 (2022).

