Co-localization of Cell Lineage Markers and the Tomato Signal
Summary
We developed two sets of tracing combinations in Rosa26tdtomato (ubiquitously expressed in all cells)/Cre (specifically expressed in chondrocytes) mice: one with 2.3Col1a1-GFP (specific to osteoblasts) and one with immunofluorescence (specific to bone cells). The data demonstrate the direct transformation of chondrocytes into bone cells.
Abstract
The cell lineage tracing system has been used predominantly in developmental biology studies. The use of Cre recombinase allows for the activation of the reporter in a specific cell line and all progeny. Here, we used the cell lineage tracing technique to demonstrate that chondrocytes directly transform into osteoblasts and osteocytes during long bone and mandibular condyle development using two kinds of Cre, Col10a1-Cre and Aggrecan-CreERT2 (Agg-CreERT2), crossed with Rosa26tdTomato. Both Col10 and aggrecan are well-recognized markers for chondrocytes.
On this basis, we developed a new method-cell lineage tracing in conjunction with fluorescent immunohistochemistry-to define cell fate by analyzing the expression of specific cell markers. Runx2 (a marker for early-stage osteogenic cells) and Dentin matrix protein1 (DMP1; a marker for late-stage osteogenic cells) were used to identify chondrocyte-derived bone cells and their differentiation status. This combination not only broadens the application of cell lineage tracing, but also simplifies the generation of compound mice. More importantly, the number, location, and differentiation statuses of parent cell progeny are displayed simultaneously, providing more information than cell lineage tracing alone. In conclusion, the co-application of cell lineage tracing techniques and immunofluorescence is a powerful tool for investigating cell biology in vivo.
Introduction
During development, endochondral bone formation accounts for over 80% of the skeletal volume. It is widely believed that it begins with the apoptosis of hypertrophic chondrocytes. Subsequently, the cells from the underlying bone marrow invade and initiate angiogenesis, followed by new bone deposition by bone marrow- and periosteum-derived cells1,2. The cell fate of hypertrophic chondrocytes (HCs), however, has been an issue of debate for decades3. Initially, HCs were considered to be the end of the chondrocyte differentiation pathway, and apoptosis was generally thought to be the ultimate fate of HCs. Now, some researchers suggest that at least some HCs could survive and contribute to endochondral bone formation. Although they proposed that growth plate chondrocytes had the ability to transdifferentiate into osteoblasts based on ultrastructure, immunohistochemical staining, and in vitro studies46, none of these methods were definitive in demonstrating chondrocyte contribution to the osteoblast lineage.
The cell lineage tracing technique provides a more rigorous way to study cell fate. Briefly speaking, a recombinase enzyme, which is only expressed in a specific type of cell, stimulates the expression of the reporter gene. In this way, this type of cell and their descendants are permanently labeled7. The Cre-loxP system is commonly used in lineage tracing. Cre (the recombinase enzyme) will excise the STOP sequence between the two loxP sites and activate the reporter in a specific cell line (Figure 1A). In some cases, the investigator can choose a favorable time point to activate Cre by using a drug, such as tamoxifen, causing Cre to fuse to a modified form of the estrogen receptor (CreERT2)8. Fluorescent reporters have become the standard in lineage tracing experiments because they dramatically reduce the complexity and improve the accuracy and efficiency of cell fate tracing8,9. tdTomato is becoming the best choice among fluorescent reporters since it has the brightest fluorescent protein and the strongest epifluorescence, making it easily visualized7 (Figure 1A).
By using the Rosa26tdTomato lineage tracing system, our group and other investigators have shown that HCs can change their phenotype into bone cells during development10-14. To accomplish this, we developed two sets of tracing combinations with Rosa26tdtomato (ubiquitous expression in all cells)/Cre (specific to chondrocytes) mice: 2.3Col1a1-GFP (specific to osteoblasts) and immunofluorescence (specific to bone cells). The data demonstrate that both methods are viable ways to study cell fate in vivo.
Protocol
All protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Texas A&M University College of Dentistry.
1. Animal Breeding
- Use three animal models in this study. To investigate the fate of the embryonic chondrocytes in condyle formation, first use Col10a1-Cre15 mice and cross them with Rosa26tdTomato (B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) mice to obtain Col10a1-Cre and Rosa26tdTomato mice. Next, cross these mice with 2.3Col1a1-GFP mice16. Use Col10a1-Cre; Rosa26tdTomato; 2.3Col1a1-GFP mice to perform the cell lineage tracing experiments (Figure 1B).
- To study the fate of the postnatal chondrocytes, follow the same procedure as in step 1.1, but use the Aggrecan-creERT2 (Agg-CreERT2)17,18 line and keep the Agg-CreERT2; Rosa26tdTomato; 2.3Col1a1-GFP mice to perform the cell lineage tracing experiments (Figure 1C). Inject tamoxifen on postnatal day 14.
- To combine the cell lineage tracing technique with immunofluorescence, cross Agg-CreERT2 mice and Rosa26tdTomato mice. Use mice that carry both genes in the experiment and inject the tamoxifen on postnatal day 3 (Figure 1D).
2. Material Preparation
- Dissolve the tamoxifen powder in 10% ethanol and 90% corn oil at a concentration of 10 mg/ml. The dosage for injection is 75 mg/kg.
- Dissolve the paraformaldehyde (PFA) powder in PBS to a concentration of 4%, using 2 M sodium hydrate to adjust the pH value to 7.4. Use 4% PFA solution to fix the tissue (the mandibular condyle and hind leg are used here) after sacrifice. Because of its toxicity, handle PFA in the hood with gloves and a facemask.
- Dissolve the EDTA powder in distilled water to a concentration of 10%, using 2 M sodium hydrate to adjust the pH value to 7.4. Use 10% EDTA to decalcify the mandibular condyle and hind leg.
- Dissolve sucrose powder in PBS at concentrations of 15% and 30%. Use the sucrose solution to dehydrate the tissue after decalcification.
- Dissolve the hyaluronidase powder in PBS at a concentration of 2 mg/ml, pH 5.0. This solution is for immunofluorescence antigen retrieval.
- Use a 1.5-ml tube to prepare a blocking solution that contains 3% bovine serum albumin (BSA) and 20% goat serum in PBS for Runx2 or DMP1 immunofluorescent staining.
- Use a 1.5-ml tube to prepare a primary antibody solution that contains 2% goat serum in PBS for Runx2 or DMP1 immunofluorescent staining. The concentration of the primary antibody is 1:400 for Runx2 and 1:100 for DMP1.
- Use a 1.5-ml tube to prepare the rabbit IgG solution as the control for immunofluorescent staining to avoid false positive results. The solution contains 2% goat serum in PBS. The concentration of the rabbit IgG for Runx2 control is 1:400; for DMP1, 1:100. Perform the experiment and control staining simultaneously.
- Use a 1.5 ml tube to prepare a secondary antibody solution that contains 2% goat serum in PBS for Runx2 or DMP1 immunofluorescent staining. Use the secondary antibody at a dilution of 1:500.
3. Slide Preparation for Confocal Microscopy (Cell Lineage Tracing Only, No Combination with Immunofluorescence)
- Inject tamoxifen in the Agg-creERT2 mice at a favorable time point.
- First, remove a mouse from its cage. Then, use the left thumb and index finger to grab the skin on the back of the mouse and turn it over, exposing the abdomen. Use the right hand to hold the syringe. The optimal entry point for injection is on the left or right side of hypogastrium, avoiding the liver and bladder.
- Keep the syringe parallel to the hind legs of the mouse and inject intraperitoneally. The dosage for injection is 75 mg/kg. The weights of the mice at the ages of 2 weeks, 3 weeks, and 4 weeks old are approximately 7 – 9 g, 11 – 13 g, and 16 – 18 g, respectively.
- Anesthetize the mice with a Xylazine/Ketaset combination.
- To prepare the working solution, first dilute the Xylazine and Ketaset with distilled water to a concentration of 1 mg/ml and 5 mg/ml, respectively. Next, mix the Xylazine and Ketaset in a 1:2 ratio. The dosage for injection is 30 µl/g.
- Inject as in step 3.1. Confirm the anesthetization by pinching the mouse's ankle. The mouse is unconscious if it has no reaction.
- Perfuse the mice with 4% PFA after anesthetization.
- After the mouse loses consciousness, fix the four legs of the mouse on a board to entirely expose the abdomen.
- Saturate the abdomen with 70% ethanol and make an excision from the lower abdomen to the neck along the middle line. Pinch and simultaneously pull the skin to the lateral sides to reveal the peritoneal membrane. Use dissection scissors to make a longitudinal excision.
- Cut off and remove the front ribs to expose the heart. Puncture the heart from the left ventricle with a 22 G syringe, hold the syringe, and simultaneously cut a slot in the right auricle.
- Slowly inject the 4% PFA, which is perfused along the cardiovascular system while the blood flushes out of the cut from the right auricle. The volume of the PFA for perfusion is 1 ml/gram. Perform this step in a Class I biosafety cabinet that is hard-ducted to the building exhaust system.
- Peel off the mouse's skin and put the whole body into a 50-ml polypropylene centrifuge tube that contains 40 ml of 4% PFA to fix overnight at 4 °C.
- Use dissection scissors and # 3 and # 5 forceps to carefully remove the mandible and hind leg from the body and to remove the muscles and tendons on the surface. Perform this step in a Class I biosafety cabinet that is hard-ducted to the building exhaust system.
- Cut the mandible into two pieces at the distal region of the third molar. Similarly, cut the femur and tibia in the midshaft to expose the bone marrow cavity in order to accelerate decalcification. Put the part that includes the condyle and condylar process and along with the hind leg into 40 ml of 10% EDTA to decalcify at 4 °C for 2 – 4 days in a 50-ml polypropylene centrifuge tube.
- Use 50 ml of 15% sucrose to dehydrate the condyle and hind leg overnight at 4 °C in a 50-ml polypropylene centrifuge tube.
- Use 30% sucrose to dehydrate the condyle and hind leg overnight at 4 °C in a 50-ml polypropylene centrifuge tube.
- Along the sagittal plane, embed the sample with OCT on the cutting plate in the cryosection machine.
- Horizontally lay the condyle or the hind leg in the mounting mold. Submerge the tissue in OCT and leave it in the cryosection machine until the OCT freezes.
- Mount the OCT block on the cutting plate. Wait for approximately 15 min before cutting to ensure that the OCT is completely frozen.
- Cut the condyle and hind leg into 10-µm sections. Collect the sections on slides and store at -20 °C.
- Incubate the slide in a 37 °C chamber to remove the water before staining.
- Wash the slides twice with distilled water for 5 min.
- Wipe off the water around each section. Use a hydrophobic barrier pen to circle the sections and drop DAPI or non-fluorescing anti-fade mounting solution into the circle. Carefully lay down the cover slip.
4. Immunohistochemical Staining for Runx2 and DMP1
NOTE: The IgG control is necessary for immunohistochemical staining to avoid false-positive signals. The staining for the experimental and control groups needs to be performed simultaneously.
- Incubate the slides in a 37 °C chamber to remove the water before staining.
- Wash the slides twice with distilled water.
- Use the hydrophobic barrier pen to circle all the sections on the slide. From this step, add all of the prepared solution into the circle to completely cover the sections.
- Treat the sections with hyaluronidase in a humid chamber at 37 °C for 30 min. The volume of the solution (in steps 4.5 – 4.8) depends on the size of the section. Use 50 µl of solution for the condyle and 100 µl for the long bone. Wash with PBST (PBS that contains 0.1% Tween 20) three times.
- Prepare and apply the blocking solution to each section and incubate them in a humid chamber for 1 hr at room temperature.
- Incubate the sections with primary antibody solution (rabbit anti-mouse Runx2, or rabbit anti-mouse DMP1) at 4 °C overnight. Wash with PBS three times.
- Incubate the sections with secondary antibody solution (goat anti-rabbit, Alexa Fluor 488) for 2 hr at room temperature. Wash with PBS three times.
- Wipe off the water around the section and drop DAPI into the circle to cover the section on the slide. Carefully lay down the cover slip.
5. Confocal Microscopy
- Capture fluorescent cell images using a confocal microscope at wavelengths ranging from 488 µm (green) to 561 µm (red). Take multiple stacked images at 200 Hz (dimensions of 1,024 × 1,024)19 using 10X, 20X, and 63X lenses.
Representative Results
Chondrocytes Directly Transform into Bone Cells (Osteoblasts and Osteocytes) in the Mandibular Condyle and Long Bone.
Aggrecan, a critical gene for chondrogenesis, is mainly expressed in early and mature chondrocytes18. As a result, the injection of tamoxifen at 2 weeks of age in Agg-CreERT2; Rosa26tdTomato mice activated the red-tomato reporter in all chondrocytes and their daughter cells. The 2.3Col1a1-GFP line gave rise to a green-fluorescing color in collagen 1-expressing bone cells, specifically osteoblasts and pre-osteocytes. The yellow color (red combined with green) indicated the presence of chondrocyte-derived bone cells that expressed the collagen 1 gene. In Figure 2A, most of the bone cells in the condylar process beneath the cartilage were red or yellow (a red cell that had begun to secrete green-fluorescing Col1a1, thereby appearing yellow when the two fluorophores were superimposed). These results provided strong evidence that these bone cells were derived from chondrocytes. Similar results were also observed during long bone development, where the majority of bone cells were derived from chondrocytes in both the epiphysis (Figure 2B) and the metaphysis (Figure 2C).
To further investigate these results, we crossed Col10al-Cre mice with Rosa26tdTomato; 2.3Col1a1-GFP mice. Col10a1 is a highly specific marker for HCs, expressed only in HCs and their descendants. Moreover, Col10a1-Cre is non-inducible, which reflects cell differentiation from the very beginning of Col10a1 expression (at E14.5). In the trabeculae near the cartilage-bone interface (superior level) of 3-week-old mice, red and some yellow fluorescing cells were predominant, while green fluorescing cells were scarce. In a slightly more inferior area (middle level), the majority of the cells were fluorescing yellow, with slightly fewer fluorescing red. Green fluorescing cells appeared to dominate only in the most inferior area of the condylar process (inferior level) (Figure 3).This trend indicates that condylar growth is mostly contributed to by transformed bone cells from HCs. The distribution of the red and green fluorescing cells in the condylar process is also consistent with the inferior-to-superior direction of condylar growth.
The cell lineage tracing technique clearly demonstrates that apoptosis is not the only fate for HCs. Both the Agg-CreERT2 and Col10a1-Cre lines indicate that chondrocyte-derived bone cells are the major source for bone development in the condylar process and in the epiphysis and metaphysis in long bone.
Co-application of Immunofluorescent Staining and Cell Lineage Tracing Enables the Tracking of Cell Differentiation.
The cell lineage tracing technique can also be combined with fluorescent immunohistochemistry to determine the cell type by the detection of a cell-specific marker. This method has several advantages for the study of cell fate. First, the researcher can still define the cell characteristics by selecting appropriate markers, even without the specific GFP mouse line, which broadens the application for the cell lineage tracing technique. Secondly, it simplifies the generation of compound mice. For instance, we only need to generate Agg-CreERT2; Rosa26tdTomato compound mice to produce the red color after activating Cre (at postnatal day 3), instead of creating Agg-CreERT2; 2.3Col1a1-GFP; Rosa26tdTomato mice, which requires more crosses.
In this study, we chose two antibodies for immunofluorescent staining. One was against Runx2, a critical transcriptional factor during osteoblast maturation (early stage of osteogenic cells); the other was against DMP1, expressed in mature osteocytes (late stage of osteogenic cells). In Figure 4, the green fluorescence represents the Runx2 or DMP1 antibody expression in the 2-week-old condylar process. In Figure 4A, three types of colored cells in the subchondral bone are present. The yellow color (superimposition of red and green fluorescence) in the nucleus represents the chondrocyte-derived bone cells expressing Runx2, indicating that these cells were immature osteoblasts on the bone surface. In addition, there were red-fluorescing bone cells that did not express Runx2 (red fluorescence without superimposed green). These cells represent mature chondrocyte-derived bone cells in the bone matrix. The least common cells were those with only green-fluorescing nuclei, indicating non-chondrocyte-derived bone cells with Runx2 expression or a transformation that occurred before Cre activation. The image suggests that the red bone cells and those with yellow nuclei were the majority cell populations contributing to the condylar subchondral bone formation.
On the other hand, almost every red, chondrocyte-derived bone cell in the bone matrix had DMP1 staining around their cell bodies. Few osteocytes were positive for DMP1 but lacked red cell bodies (Figure 4B). There are also two possible explanations for these cells: either they are non-chondrocyte-derived bone cells, or they are chondrocyte-derived bone cells that transformed prior to the tamoxifen injection. In addition, no red bone cells on the surface of the bone expressed DMP1, indicating that they were not mature osteocytes.
Taken together, the data from the expression patterns for both early- (Runx2) and late-stage markers (DMP1) of osteogenic cells were consistent with the previous outcome using Cre combined with 2.3Col1a1-GFP. These data demonstrate that the combination of immunofluorescence and Rosa26tdTomato tracing is a good way to study cell fate. More importantly, investigators can distinguish the cell type, identify the differentiation stage, and observe the transformed cell numbers simultaneously by using immunofluorescence with different markers, which provides more information than Rosa26tdTomato tracing alone.
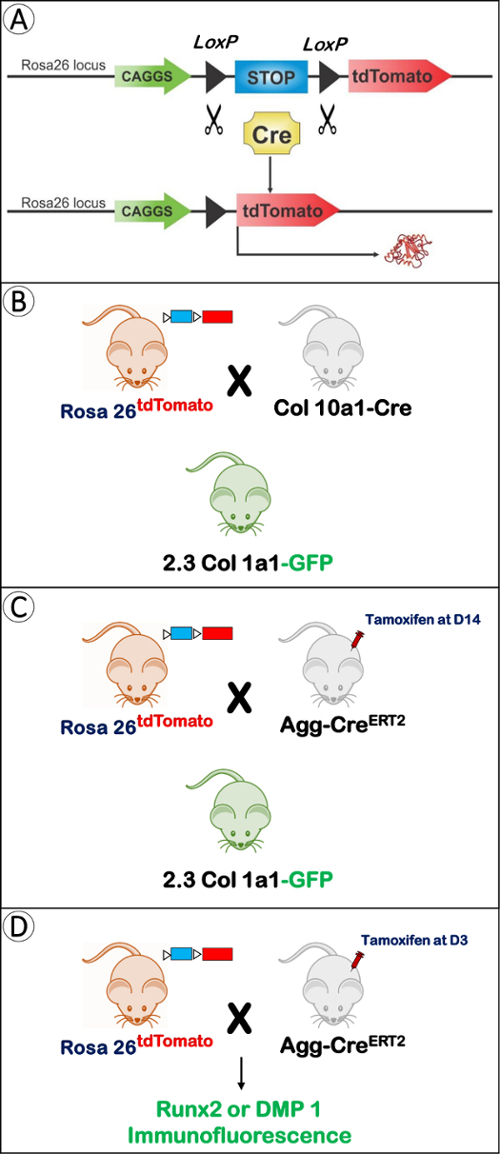
Figure 1. The Mechanism for Cell Lineage Tracing and the Generation of the Compound Mice. A)The Cre-loxP system is commonly used in lineage tracing. Cre excises the STOP sequence between the two loxP sites, and the tdTomato protein (fluorescing red) is permanently expressed in the specific cell line. B) Three animal models are mentioned in this paper. We used 2.3Col1a1-GFP; Rosa26tdTomato mice and crossed them with Col10a1-Cre mice. Col10a1-Cre is non-inducible, which reflects the cell differentiation from the very beginning of Col10a1 expression (at E14.5). C) Aggrecan-Cre ERT2(Agg-CreERT2) is another Cre line also crossed with 2.3Col1a1-GFP; Rosa26tdTomato mice. Cre was activated at 2 weeks of age by a tamoxifen injection (Tm: tamoxifen). D) In order to combine the immunofluorescence with cell lineage tracing, we generated Agg-CreERT2; Rosa26tdTomato mice, activated Cre at postnatal day 3, and performed the Runx2 and DMP1 immunofluorescence (Tm: tamoxifen). Please click here to view a larger version of this figure.
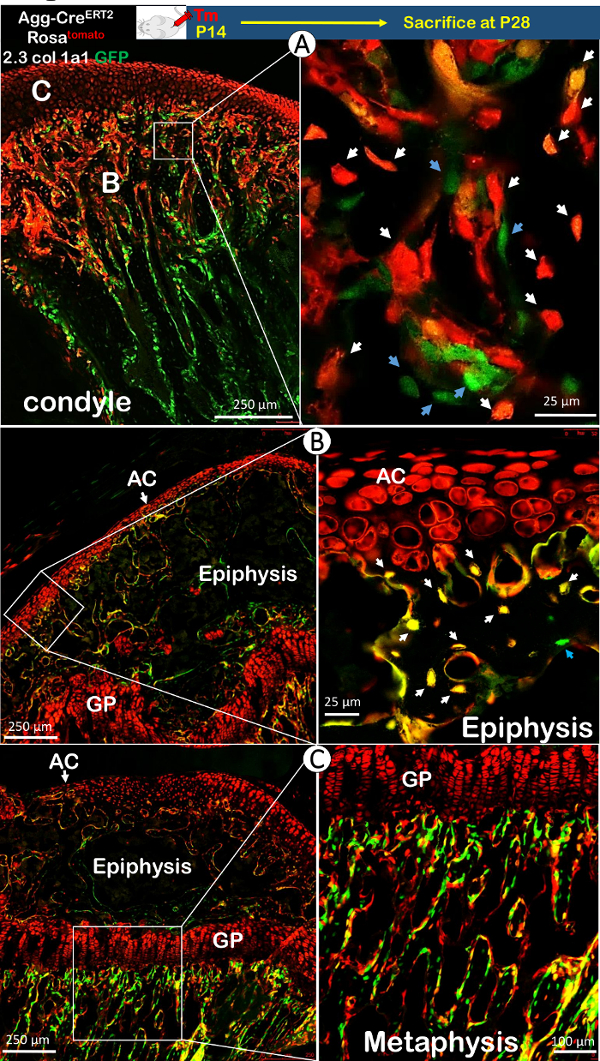
Figure 2. The Transformation from Chondrocytes to Bone Cells (Osteoblasts and Osteocytes) in the Mandibular Condyle and Long Bone Using Agg-CreERT2; 2.3Col1a1-GFP; Rosa26tdTomato Compound Mice. A) Cre was activated by tamoxifen on postnatal day 14, and the mice were sacrificed at 4 weeks old. There were three colored cells in the condylar process: pure red (chondrocyte-derived bone cells), yellow (red combined with green, indicating chondrocyte-derived bone cells that expressed the collagen 1 gene), and pure green (non-chondrocyte derived bone cells with the collagen 1 gene). Most of the bone cells in the condylar process beneath the cartilage were yellow or pure red (white arrows), while few bone cells were green (blue arrows). These data provide strong evidence that chondrocytes directly transform into bone cells and contribute to the formation of the condylar process during development (Tm: tamoxifen; C: cartilage; B: bone). B, C) The majority of bone cells in the epiphysis and metaphysis were chondrocyte-derived (white arrow: chondrocyte-transformed bone cells in yellow or pure red; blue arrow: non-chondrocyte-derived bone cells in pure green; AC: articular cartilage; GP: growth plate). Please click here to view a larger version of this figure.
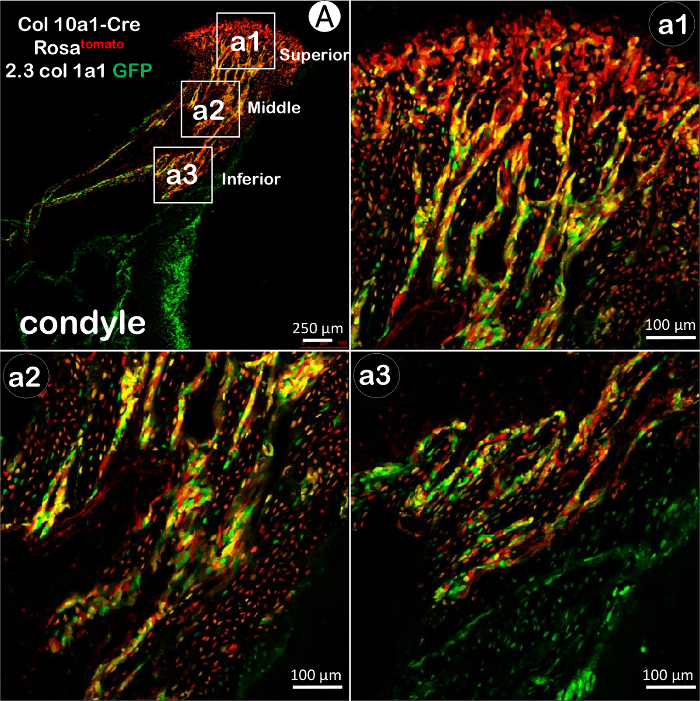
Figure 3. The Transformation from Chondrocytes to Bone Cells in the Mandibular Condyle Using Col10al-Cre; 2.3Col1a1-GFP; Rosa26tdTomato Compound Mice. A) In the condylar process from 3-week-old mice, green bone cells were a distinct minority in the trabeculae near the cartilage-bone interface (superior level, a1), whereas red and some yellow cells were predominant. In the middle level (a2), yellow cells were in the majority, with slightly fewer red cells. Green cells appeared to be in the majority only in the most inferior area (a3). Please click here to view a larger version of this figure.
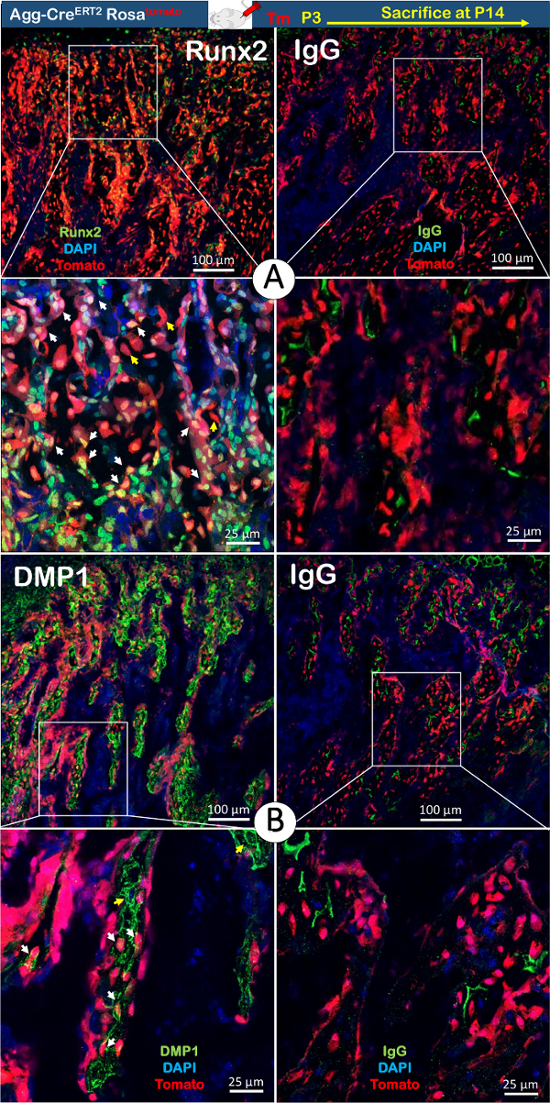
Figure 4. The Co-localization of two Markers for Osteogenic Cells with the Lineage-tracing Background in the Condylar Process Using 2-week-old Agg-CreERT2; Rosa26tdTomato Compound Mice. A) The yellow color in the nuclei represents chondrocyte-derived bone cells expressing Runx2 (white arrows) on the surface of the trabecular bone beneath the condyle cartilage. The pure red bone cells without Runx2 expression represent mature chondrocyte-derived bone cells in the bone matrix (yellow arrows). The fewest cells were those carrying only green nuclei, representing either non-chondrocyte-derived bone cells or transformed bone cells from chondrocytes before Cre activation (Tm: tamoxifen). B) In the trabecular bone beneath the condyle cartilage, almost every red chondrocyte-derived bone cell in the bone matrix carried DMP1 staining around their cell bodies (white arrows). Only a few of the osteocytes were positive for DMP1 but lacked red color in their cell bodies (yellow arrows). There are two possibilities for these cells: non-chondrocyte-derived bone cells or chondrocyte-derived bone cells arising before tamoxifen injection. Please click here to view a larger version of this figure.
Discussion
Due to technological limitations, it is always difficult to investigate the behavior of cells in vivo. However, the cell lineage tracing technique is proving to be a powerful tool for studying cell biology7-9. In this study, we further improve this protocol by combining it with immunofluorescence. In this way, cell fate can be defined by multiple related markers, which broadens the application of lineage tracing. Moreover, this co-localization of immunofluorescence and tomato signal simultaneously displays the number of progeny of the founder cell, their location, and their differentiation status, providing more information than cell lineage tracing alone. In addition, the use of cell-specific markers can simplify the generation of compound mice, accelerating the investigation.
The specificity of Cre is crucial for the unequivocal attribution of lineage and the accuracy of the results20,21. It is very important to select the appropriate Cre lines to verify the cell fate. In this study, we chose Col10a1, because it is considered a specific marker for hypertrophic chondrocytes10,11, and aggrecan, because it is also a well-recognized marker for chondrocytes18. There are also good Cre models used in other fields. The Scleraxis-Cre (Scx-Cre) line, for example, is useful in tendon and ligament research since scleraxis is a basic helix-loop-helix transcription factor that is a marker for the tendon and ligament cell lineage22. Another Cre called DMP1-Cre is commonly used in skeleton- and tooth-related studies because DMP1 is highly expressed in odontoblasts and osteocytes23,24.
Compared with non-inducible Cre systems, the activation of inducible Cre can be restricted spatially and temporally20. However, some limitations should be considered when designing the experiment and interpreting the results of inducible Cre models. First, the dose of tamoxifen may change the efficiency of Cre activation and the number of labelled cells. Low doses will label the population of interest at a clonal density25; high doses may label the entire progenitor pool7,26. Thus, the dose must be chosen depending on the purpose of the experiment. Second, tamoxifen has potential toxicity, especially in high doses27,28. For this reason, it is better to decrease the dose to avoid late-term abortions when injecting during pregnancy29.
In conclusion, the co-application of cell lineage tracing techniques and immunofluorescence is a powerful tool for investigating cell biology in vivo. In the future, investigators can attempt to simultaneously perform immunofluorescence with two different antibodies over the tomato signal background. This method can show the expression pattern for two markers in one section, making it easier for the investigator to compare and analyze the results. Moreover, this co-application can be further improved by in situ immunofluorescence to decrease non-specific staining that may exist through conventional immunofluorescence.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This study was supported by NIH grant DE025014 to JQF.
Materials
| Tamoxifen | Sigma | T5648 | activate the Cre event |
| Paraformaldehyde | Sigma | P6148 | fix the sample |
| Ethylenediaminetetraacetic acid | Alfa Aesar | A10713 | decalcify the hard tissue |
| Sucrose | Sigma | S0389 | dehydrate the tissue |
| Hyaluronidase from bovine testes | Sigma | H4272 | retrieve antigen for immunochemical staining |
| Bovine serum albumin | Sigma | A3059 | blocking solution |
| primary antibody for Runx2 | Cell Signal | D1L7F | primary antibody for immunochemical staining |
| primary antibody for DMP1 | provided by Dr. Chunlin Qin | primary antibody for immunochemical staining | |
| anti-rabbit IgG | Sigma | 18140 | control for immunochemical staining |
| secondary antibody | Invitrogen | A11008 | second antibody for immunochemical staining |
| OCT | Tissue-Tek | 4583 | embed the sample for frozen section |
| Tween 20 | Fisher Scientific | BP337 | PBST |
| non-fluorescing antifade mountant | Life technologies | P36934 | mounting slides |
| DAPI | Life technologies | P36931 | nuclear staining |
| Hydrophobic Barrier Pen | Vector Laboratories | circle the section on the slide for for immunochemical staining | |
| Xylazine | AnaSed | anesthetization | |
| Ketaset | Zoetis | anesthetization | |
| cryosection machine | Leica | CM1860 UV | |
| confocal microscope | Leica | DM6000 CFS |
Referencias
- Gibson, G. Active role of chondrocyte apoptosis in endochondral ossification. Microsc Res Tech. 43 (2), 191-204 (1998).
- Kronenberg, H. M. Developmental regulation of the growth plate. Nature. 423 (6937), 332-336 (2003).
- Shapiro, I. M., Adams, C. S., Freeman, T., Srinivas, V. Fate of the hypertrophic chondrocyte: microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res C Embryo Today. 75 (4), 330-339 (2005).
- Kahn, A. J., Simmons, D. J. Chondrocyte-to-osteocyte transformation in grafts of perichondrium-free epiphyseal cartilage. Clin Orthop Relat Res. (129), 299-304 (1977).
- Roach, H. I. Trans-differentiation of hypertrophic chondrocytes into cells capable of producing a mineralized bone matrix. Bone Miner. 19 (1), 1-20 (1992).
- Park, J., et al. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 4 (5), 608-621 (2015).
- Kretzschmar, K., Watt, F. M. Lineage tracing. Cell. 148 (1-2), 33-35 (2012).
- Humphreys, B. D., DiRocco, D. P. Lineage-tracing methods and the kidney. Kidney Int. 86 (3), 481-488 (2014).
- Romagnani, P., Rinkevich, Y., Dekel, B. The use of lineage tracing to study kidney injury and regeneration. Nat Rev Nephrol. 11 (7), 420-431 (2015).
- Yang, G., et al. Osteogenic fate of hypertrophic chondrocytes. Cell Res. 24 (10), 1266-1269 (2014).
- Yang, L., Tsang, K. Y., Tang, H. C., Chan, D., Cheah, K. S. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. 111 (33), 12097-12102 (2014).
- Jing, Y., et al. Chondrocytes Directly Transform into Bone Cells in Mandibular Condyle Growth. J Dent Res. 94 (12), 1668-1675 (2015).
- Zhou, X., von der Mark, K., Henry, S., Norton, W., Adams, H., de Crombrugghe, B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 10 (12), (2014).
- Purcell, P., Trainor, P. A. The Mighty Chondrocyte: No Bones about It. J Dent Res. 94 (12), 1625-1627 (2015).
- Gebhard, S., et al. Specific expression of Cre recombinase in hypertrophic cartilage under the control of a BAC-Col10a1 promoter. Matrix Biol. 27 (8), 693-699 (2008).
- Kalajzic, Z., et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 31 (6), 654-660 (2002).
- Akiyama, H., et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 102 (41), 14665-14670 (2005).
- Henry, S. P., et al. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 47 (12), 805-814 (2009).
- Ren, Y., Lin, S., Jing, Y., Dechow, P. C., Feng, J. Q. A novel way to statistically analyze morphologic changes in Dmp1-null osteocytes. Connect Tissue Res. 55, 129-133 (2014).
- Pest, M. A., Beier, F. Developmental biology: Is there such a thing as a cartilage-specific knockout mouse?. Nat Rev Rheumatol. 10 (12), 702-704 (2014).
- Tsang, K. Y., Chan, D., Cheah, K. S. Fate of growth plate hypertrophic chondrocytes: death or lineage extension?. Dev Growth Differ. 57 (2), 179-192 (2015).
- Sugimoto, Y., Takimoto, A., Hiraki, Y., Shukunami, C. Generation and characterization of ScxCre transgenic mice. Genesis. 51 (4), 275-283 (2013).
- Feng, J. Q., et al. Generation of a conditional null allele for Dmp1 in mouse. Genesis. 46 (2), 87-91 (2008).
- Lu, Y., Xie, Y., Zhang, S., Dusevich, V., Bonewald, L. F., Feng, J. Q. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 86 (4), 320-325 (2007).
- Rios, A. C., Fu, N. Y., Lindeman, G. J., Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature. 506 (7488), 322-327 (2014).
- Blanpain, C., Simons, B. D. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 14 (8), 489-502 (2013).
- Huh, W. J., Khurana, S. S., Geahlen, J. H., Kohli, K., Waller, R. A., Mills, J. C. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 142 (1), 21-24 (2012).
- Lee, M. H., Kim, J. W., Kim, J. H., Kang, K. S., Kong, G., Lee, M. O. Gene expression profiling of murine hepatic steatosis induced by tamoxifen. Toxicol Lett. 199 (3), 416-424 (2010).
- Nakamura, E., Nguyen, M. T., Mackem, S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 235 (9), 2603-2612 (2006).

