Analysis of Chromosome Segregation, Histone Acetylation, and Spindle Morphology in Horse Oocytes
Summary
This manuscript describes an experimental approach to morphologically and biochemically characterize horse oocytes. Specifically, the present work illustrates how to collect immature and mature horse oocytes by ultrasound-guided ovum pick-up (OPU) and how to investigate chromosome segregation, spindle morphology, global histone acetylation, and mRNA expression.
Abstract
The field of assisted reproduction has been developed to treat infertility in women, companion animals, and endangered species. In the horse, assisted reproduction also allows for the production of embryos from high performers without interrupting their sports career and contributes to an increase in the number of foals from mares of high genetic value. The present manuscript describes the procedures used for collecting immature and mature oocytes from horse ovaries using ovum pick-up (OPU). These oocytes were then used to investigate the incidence of aneuploidy by adapting a protocol previously developed in mice. Specifically, the chromosomes and the centromeres of metaphase II (MII) oocytes were fluorescently labeled and counted on sequential focal plans after confocal laser microscope scanning. This analysis revealed a higher incidence in the aneuploidy rate when immature oocytes were collected from the follicles and matured in vitro compared to in vivo. Immunostaining for tubulin and the acetylated form of histone four at specific lysine residues also revealed differences in the morphology of the meiotic spindle and in the global pattern of histone acetylation. Finally, the expression of mRNAs coding for histone deacetylases (HDACs) and acetyl-transferases (HATs) was investigated by reverse transcription and quantitative-PCR (q-PCR). No differences in the relative expression of transcripts were observed between in vitro and in vivo matured oocytes. In agreement with a general silencing of the transcriptional activity during oocyte maturation, the analysis of the total transcript amount can only reveal mRNA stability or degradation. Therefore, these findings indicate that other translational and post-translational regulations might be affected.
Overall, the present study describes an experimental approach to morphologically and biochemically characterize the horse oocyte, a cell type that is extremely challenging to study due to low sample availability. However, it can expand our knowledge on the reproductive biology and infertility in monovulatory species.
Introduction
A vast array of assisted reproduction techniques has been developed to treat infertility in women, companion animals, and endangered species. One of the most common procedures in clinical settings is the retrieval of metaphase II (MII)-stage oocytes from the ovarian follicles by ultrasound-guided transvaginal aspiration, ovum pick-up (OPU)1. These oocytes are then fertilized in vitro (IVF), with the resulting embryo(s) implanted in a recipient uterus. The MII-stage (mature) oocytes are retrieved following the administration of exogenous gonadotropins. However, this treatment is associated, in some patients, with the development of ovarian hyperstimulation syndrome (OHSS)2.
Taking advantage of the intrinsic ability of fully-grown, immature oocytes (GV-stage) to spontaneously resume meiosis once isolated from their follicles, it is possible to obtain mature oocytes without administering gonadotropin3. This procedure is called oocyte in vitro maturation (IVM) and represents a less drug-oriented, less expensive, and more patient-friendly approach to assisted reproductive technology. However, the success of embryo development with in vitro-matured oocytes is generally lower than with in vivo matured oocytes4,5. A possible explanation is that in vitro matured oocytes are more affected by errors in chromosome segregation, and the resulting aneuploidy impairs normal embryonic development6.
Understanding the molecular basis of chromosomal mis-segregation during IVM would ultimately disclose the full potential of this technique. In this vein, the experimental approach used to investigate the morphological and biochemical features of in vitro-matured oocytes, compared to in vivo matured oocytes is here described7,8. Specifically, the procedures for the OPU of immature and mature oocytes and the IVM of immature oocytes are illustrated using adult and naturally-cycling horses as an experimental model. Then, immunofluorescence and image analysis are used to investigate chromosome segregation, spindle morphology, and the global pattern of histone acetylation on these gametes. Finally, a protocol of reverse-transcription and quantitative PCR is described for the analysis of mRNA expression.
Compared with rodent animal models, horses do not allow genetic manipulation, are less easy to manipulate, and require expensive maintenance. However, this model is gaining considerable interest for the study of oocyte maturation9,10 due to the similarity to human ovarian physiology11,12. Moreover, the development of reliable protocols of IVM-IVF in the horse has a substantial economic interest, as it would allow for an increase in the number of foals from mares of high genetic value.
One of the limitations of performing experiments on oocytes, especially in monovulatory species, is the restricted sample availability. This limit has been overcome here by adjusting an approach, previously developed in mice, to horse oocytes13,14 in order to conduct chromosome counting that minimizes the sample loss (see the discussion for a comparison with other available techniques). Moreover, a triple-fluorescence staining protocol has been optimized to conduct multiple analyses on the same sample, and q-PCR analyses were performed on pools of 2 oocytes only.
Overall, the present study describes an experimental approach aimed to morphologically and biochemically characterize the horse oocyte, a cell type that is extremely challenging to study due to the low sample availability. However, it can expand our knowledge of the reproductive biology and infertility of monovulatory species.
Protocol
All the procedures were approved by the Animal Care and Use Committee CEEA Val de Loire Number 19 and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
1. Oocyte Collection and In Vitro Maturation
- Ovum pick-up
- Daily assess by ultrasound the diameter of ovarian follicles in a cohort of adult mares. At the emergence of a follicle ≥33 mm (dominant follicle), inject the mare with 1,500 IU of human chorionic gonadotrophin (hCG) in the upper part of the jugular vein (i.v.).
- Before performing the injection, swab the area with alcohol, locate the jugular furrow, and place the thumb 2-3 cm under the site of injection to induce the vein to rise. Insert the needle almost parallel to the neck and aspirate to make sure that the needle is in the vein (the blood will enter the syringe). At this point, inject; hCG will induce the maturation of the oocyte in the dominant follicle.
- 35 h after the hCG injection, sedate the mare with detomidine (15 µg/kg, i.v.), butorphanol tartrate (10 µg/kg, i.v.) and butylscopolamine bromure (0.2-0.3 mg/kg, 15 mL/mare, i.v.).
- Connect the needle to the ultrasound probe. Insert the ultrasound probe vaginally and hold the ovary transrectally to face the ultrasound probe. Instruct one operator to maneuver the needle and the other to hold the ovary in place, with the follicle facing the ultrasound probe. Start from the ovary with the dominant follicle.
- Puncture the vaginal wall and the wall of the dominant follicle with the needle (ultrasound probes for OPU are equipped with a channel for the needle connected to a suction system); theneedle will be visible in the echographic image.
- Aspirate the follicular fluid in a 50-mL conical tube connected to the suction system. Pour the content of the conical tube into a large petri dish and search for the cumulus-oocyte complex (COC) using a stereomicroscope.
- Since the COC is not always retrieved with the follicular fluid, flush the follicle with Dulbecco's modified phosphate-buffered saline (DPBS) containing 5 IU/mL heparinat 37 °C. Examine DPBS for COC presence, as previously described for the follicular fluid in step 1.1.5. Flush several times until the COC is retrieved.
- Once the COC from the dominant follicle is retrieved, puncture and flush the follicles ≥5 and ≤25 mm, as described for the dominant follicle in steps 1.1.3-1.1.4; follicles ≥5 and ≤25 do not express luteinizing hormone/choriogonadotropin receptor (LHCGR), and therefore, their oocytes will not mature in response to hCG and will be collected at GV stage (immature). Only keep COCs with several complete layers of cumulus cells and brown, finely-granulated ooplasm. Discard the others.
- At the end of the OPU, inject the mare with benzyl-penicillin 15,000 IU/animal to prevent infection.
- House the mare in a quiet, clean stable for at least 2 h. Check signs of awareness by determining the position of the ears, reaction to stimuli (e.g., noise), and gait before returning the mare to the company of other animals.
- Daily assess by ultrasound the diameter of ovarian follicles in a cohort of adult mares. At the emergence of a follicle ≥33 mm (dominant follicle), inject the mare with 1,500 IU of human chorionic gonadotrophin (hCG) in the upper part of the jugular vein (i.v.).
- In vitro maturation
- Warm HEPES-buffered TCM199 (20 mM Hepes) supplemented with 1,790 IU/L heparin, 0.4% bovine serum albumin (BSA), penicillin, and streptomycin to 37 °C. Prepare this medium in advance. Filter-sterilize and keep it at 4 °C for up to 6 months.
- Prepare the IVM medium by supplementing NaHCO3-buffered TCM199 (25 mM NaHCO3) with 50 ng/mL epidermal growth factor (EGF) and 20% calf serum7. Use NaHCO3-buffered TCM199 within 3 weeks of opening a new bottle. Prepare the EGF in advance as 100x stocks, freeze it at -20 °C, and use it within 6 months. Dispense 500 µL in the wells of a 4-well dish and incubate in humidified air with 5% CO2 at 38.5 °C for at least 2 h.
- After the retrieval of the COCs from the ≥5 and ≤25 mm follicles, put them in a petri dish (3.5-cm diameter) filled with 2 mL of HEPES-TCM199. Move the COCs to different areas of the dish in order to wash away the cellular debris.
- Transfer the COCs to the IVM medium and incubate for 28 h in humidified air with 5% CO2 at 38.5 °C.
2. Immunofluorescence and Image Analysis
- Chromosome count
- After collection from the dominant follicle or at the end of IVM, incubate the COCs with 100 µM monastrol for 1 h in humidified air with 5% CO2 at 38.5 °C. Prepare monastrol in advance as 100x stocks and store it at -20 °C for up to 1 year.
- Remove the cumulus cells by treating them with 0.5% hyaluronidase or by gently pipetting; remove the zona pellucida by treating it in 0.2% pronase. Prepare hyaluronidase and pronase in advance as 10x stocks and store them at -20 °C for up to 1 year.
- Fix the oocytes in 4% paraformaldehyde in DPBS for 15 min at 38.5 °C followed by a further 45 min at 4 °C.
CAUTION! Wear personal protective equipment when handling paraformaldehyde, and dispose of contaminated materials in accordance with hazardous waste disposal guidelines. - Wash the oocytes by sequentially transferring in 3 wells filled with 500 µL of DPBS supplemented with 0.1% polyvinyl alcohol (PVA). Permeabilize in 0.3% Triton X-100 for 10 min at room temperature (RT).
- Block non-specific bindingby incubation in DPBS supplemented with 1% bovine serum albumin (DPBS-1%BSA) and 10% normal donkey serum for at least 30 min at RT. Samples can be kept in blocking solution at 4 °C for 3-4 days.
- For primary staining, incubate the oocytes in rabbit anti-Aurora B phospho-Thr232 in DPBS supplemented with 1% BSA (dilution 1:50) at 4 °C overnight.
- Wash the oocytes as described in step 2.1.4. Incubate the oocytes in a solution of tetra-methylrhodamine isothiocyanate (TRITC)-conjugated donkey anti-rabbit IgG (1:100 in DPBS-1% BSA) for 1 h at RT in the dark.
- Wash the oocytes as described in step 2.1.4, and then place 3-4 oocytes in a drop of non-hardening, anti-fade mounting medium supplemented with 20 µM YOPRO1 on a glass slide. Repeat the procedure until all the oocytes are mounted (3-4 oocytes per slide).
- Allow the oocytes to settle for about 10 min in the mounting medium, and then cover them with a coverslip. To avoid crushing the oocytes, apply two strips of double-sided tape before laying the coverslip on the glass slide.
NOTE: This precaution will also ensure that all the samples are equally compressed between the glass slide and the coverslip. Glass slides can be frozen at -20 °C and imaged during the following 2-3 days. - Image the samples on a confocal laser scanning microscope with a 60X objective using the red (centromeres) and green (chromosomes) channels7,13,14,20,21. Scan the samples spanning the whole metaphasic plate on the z-axis, with steps every 0.35 µm. Save the digitalized images of every single plan.
- Count the centromeres in serial confocal sections using the cell count function of the NIH ImageJ software (available at: https://imagej.nih.gov/ij/plugins/cell-counter.html).
NOTE: Avoid a repeated count of centromeres that appear on adjacent sections by identifying the centromeres that appear, stay, and disappear in sequential sections with a numeric code. - Repeat the chromosomal counting in step 2.1.11 with an independent operator.
NOTE: Classify the oocytes as euploid (32 chromosomes), hyperploid (>32), or hypoploid (<32).
- Spindle morphology and histone acetylation
- After collection from the dominant follicle or at the end of the IVM, remove the cumulus cells by treatment in 0.5% hyaluronidase or by gentle pipetting, as described in step 2.1.2.
- Wash the oocytes in DPBS-PVA, as in step 2.1.4, and fix them in 2.5% paraformaldehyde for 20 min at 38 °C. Extensively wash, as described in step 2.1.4, and permeabilize in 0.1% Triton X-100 for 5 min at RT.
NOTE: Wear personal protective equipment when handling paraformaldehyde and dispose of contaminated materials in accordance with hazardous waste disposal guidelines. - Block non-specific bindingby incubation in DPBS supplemented with 2% bovine serum albumin (DPBS – 2%BSA), 0.05% saponin, and 10% normal donkey serum for 2 h at RT.
NOTE: Samples can be kept in blocking solution at 4 °C for 3 – 4 days. - For primary staining, incubate the oocytes in mouse anti-alpha-tubulin (1:150) and rabbit anti-acH4K16 (1:250) in DPBS-2% BSA with 0.05% saponin at 4 °C overnight.
- Wash the oocytes as in step 2.1.4. Incubate the oocytes in a solution of green fluorescent dye-conjugated donkey anti-mouse IgG (1:500) and TRITC-conjugated donkey anti-rabbit IgG (1:100) in DPBS-2% BSA with 0.05% saponin for 1 h at RT in the dark.
- Wash the oocytes as in step 2.1.4, and then place 3-4 oocytes in a drop of the non-hardening, anti-fade mounting medium supplemented with 1 µg/mL 4',6-diamidino-2-phenylindole (DAPI) on a glass slide.
- Mount the oocytes on glass slides as described in step 2.1.8.
- Image the samples on a confocal laser scanning microscope with a 60x objective using the red (acH4K16), green (alpha-tubulin), and blue (DNA) channels.
NOTE: Keep the laser settings for the red and blue channels constant throughout the acquisition of all the samples. Scan the samples spanning the whole meiotic spindle and metaphasic plate on the z-axis, with steps every 0.35 µm. Save the digitalized images (single plans and projected 3D image). - Measure the pole-to-pole spindle length and the spindle diameter at the maximum width on the projected image using the measurement function of the NIH ImageJ software (https://imagej.nih.gov/ij/docs/guide/146-30.html). Repeat each 3 times and calculate the mean.
- Quantify the relative fluorescence of acH4K16 using the integrated density function of the NIH ImageJ software (https://imagej.nih.gov/ij/docs/guide/146-30.html). Normalize it for the integrated density of the DAPI staining.
3. Analysis of mRNA Expression
- After the COCs have been retrieved from the 5 to 25 mm follicles, collect the granulosa cell suspension that remained in the Petri dish and wash it twice in cold DPBS by centrifuging at 10,600 x g for 2 min. Snap freeze in liquid nitrogen. Store the samples at -80 °C for up to 6 months; the cells will serve for the standard curve.
- Carefully remove all the cumulus cells, as described in step 2.1.2, from immature (GV), in vitro matured, and in vivo matured oocytes.
- Wash the oocytes, as described in 2.1.4, in DPBS-PVA, collect them singly in 1 µL volumes, and lay 2 µL RNALater on top. Snap-freeze in liquid nitrogen. Store the samples at -80 °C for up to 6 months.
- Add 2 pg of Luciferase RNA to each sample as a spike-in. Pool the oocytes 2 x 2 and extract the total RNA using a silica membrane filter-based kit suitable to recover RNA from small samples.
- Reverse-transcribe the RNA in 10 µL with 0.25 µg random hexamers and mouse Moloney leukaemia virus reverse transcriptase for 1 h at 37 °C. Store the samples at -20 °C for up to 6 months.
- Dilute the cDNA from the granulosa cells in RNAse-free water to 50 ng/µL. Serially dilute this solution 1:10 to obtain 5 ng/µL, 0.5 ng/µL, 0.05 ng/µL, and 0.005 ng/µL solutions.
- Assemble reactions in triplicate in a total volume of 20 µL per reaction using cDNA equivalent to 0.05 oocytes as the substrate, or 5 µL of the serial dilutions of granulosa cell cDNA, 10 µL of SYBR green supermix, and 0.3 µM of each specific primer (Table 1).
- Run the reaction in a thermal cycler using a 3-step protocol: 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 20 s, repeated 40 times. Finally, acquire the melting curve, starting from 60 °C, and increase the temperature by 0.5 °C every 20 s up to 95 °C.
- Assess the starting quantity (SQ) of the specific mRNA in the oocyte samples using the standard curve15 calculated based on the granulosa cell sample. Express the data as the ratio between the SQ of the gene of interest and the SQ of the housekeeping genes.
Representative Results
The original findings of these experiments were described in depth previously7 and are reported herein as an example of results that can be obtained using the described protocols.
Maturation rate
Of the 32 COCs retrieved by OPU from dominant follicles, 28 (88%) were at the MII stage. Fourteen of the 58 COCs collected from follicles 5-25 mm in size were immediately snap-frozen as immature oocytes for the mRNA expression study. The remaining 44 were in vitro matured, and 37 reached the MII stage (85%). The efficiency of maturation was not significantly different in the two groups (Fisher's exact test P >0.05).
Aneuploidy rate
In order to study whether IVM can perturb the faithful chromosomal partitioning between the secondary oocyte and the first polar body, the number of chromosomes at the MII stage was counted, as it represents the result of chromosome segregation during meiosis I. As shown in Figure 1A, the anti-Aurora B phospho-Thr232 antibody specifically stained the centromeric region in horse oocytes, allowing one to count the number of chromosomes. In vitro-matured oocytes were significantly more affected by aneuploidy (5/11) compared to in vivo matured oocytes (0/12; Fisher's exact test, P <0.05; Figure 1B). Most of the aneuploid oocytes (4/5) were hyperploid; therefore, the aneuploidy rate does not result from artifacts in the preparation of the sample determining chromosomal loss, as happens with chromosomal spreads. Immunostaining of the centromeric region was also attempted with the anti-CENPA and anti-AURKB antibodies, but no signal was visualized in both cases (data not shown).
Spindle morphology and acetylation of H4K16
The pole-to-pole spindle length and the diameter of the spindle at the maximum width were measured, as illustrated in Figure 2. According to these measurements, in vitro-matured oocytes were significantly longer (19.89 ± 1.15 µm) and wider (17.28 ± 0.81 µm) compared to in vivo matured oocytes (14.57 ± 1.7 µm in length and 13.56 ± 0.91 µm in diameter; unpaired t-test, P <0.05). On the same samples, the global acetylation level of H4K16 was also measured, confirming that acetylation at H4K16 was significantly lower in IVM oocytes compared to their in vivo counterparts (Figure 2, red).
Expression of transcripts coding for histone acetyltransferases and deacetylases
Whether the expression of genes involved in the histone acetylation-deacetylation process was altered in in vitro-matured oocytes was investigated by q-PCR. Histone acetyl-transferase 1 (HAT1), K-acetyltransferase 8(KAT8), histone deacetylase 1 (HDAC1), and NAD-dependent protein deacetylase sirtuin 1 (SIRT1) were identified as putative targets. Significant differences in the expression of these transcripts in immature, in vitro matured, and in vivo matured oocytes were not observed, independent of the normalization used. Specifically, the results of the transcript expression analysis reported in Figure 3 were calculated against a standard curve using the SQ method and normalized for housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Similarly, no differences were observed when the results where normalized for the spike in luciferase or when the ΔΔct method was used (not shown).
| Target | Fw primer | Rv primer | Amplicon size |
| GAPDH | ATCACCATCTTCCAGGAGCGAGA | GTCTTCTGGGTGGCAGTGATGG | 241 bp |
| HAT1 | CTGACATGAGTGATGCTGAACA | TAACGCGTCGGTAATCTTCC | 219 bp |
| HDAC1 | GAAGGCGGTCGCAAGAAT | CCAACTTGACCTCCTCCTTGA | 166 bp |
| KAT8 | ACTGGTCTTGGGTCCTGCTG | GGGCACTTTTGAGGTGTTCC | 198 bp |
| Luciferase | TCATTCTTCGCCAAAAGCACTCTG | AGCCCATATCCTTGTCGTATCCC | 148 bp |
| SIRT1 | GACTCGCAAAGGAGCAGATT | GGACTCTGGCATGTTCCACT | 169 bp |
Table 1: Primer Setails. The 5' > 3' sequence of the forward (Fw) and (Rv) primers and the expected amplicon size are given for each transcript analyzed: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone acetyl-transferase 1 (HAT1), histone deacetylase 1 (HDAC1), K-acetyltransferase 8 (KAT8), luciferase, and NAD-dependent protein deacetylase sirtuin 1 (SIRT1). Reprinted with permission from reference7.
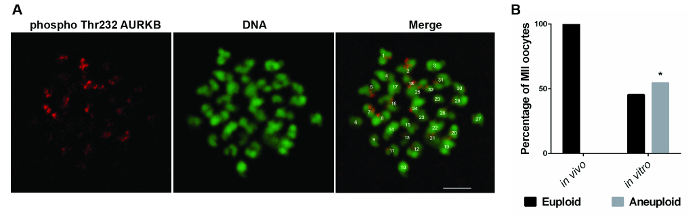
Figure 1: Aneuploidy Rate in In Vivo- and In Vitro-matured Horse Oocytes. (A) Representative images showing aurora B phospho-Thr232 (red) and DNA (green) staining of a MII-stage horse oocyte treated with monastrol. A euploid oocyte (32 chromosomes) is shown. Scale bar = 5 µm. (B) The bar graph represents the distribution of euploid and aneuploid MII-stage oocytes in in vivo (n = 12) and in vitro (n = 11) matured oocytes. *Indicates a significant difference in the aneuploidy rate (Fisher's exact test, P <0.05). Reprinted with permission from Reference 7. Please click here to view a larger version of this figure.
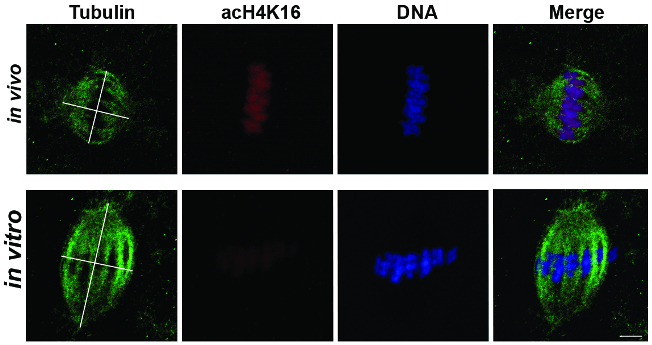
Figure 2: Spindle Measurement and H4K16 Acetylation in In Vivo- and In Vitro-matured Horse Oocytes. Representative images showing tubulin (green), acetylated H4K16 (red), and DNA (blue) in MII oocytes after in vivo (n = 4) or in vitro (n=14) maturation. The tubulin images show the axes used for spindle length and diameter measurements. Scale bar = 5 µm. Modified from reference7. Please click here to view a larger version of this figure.
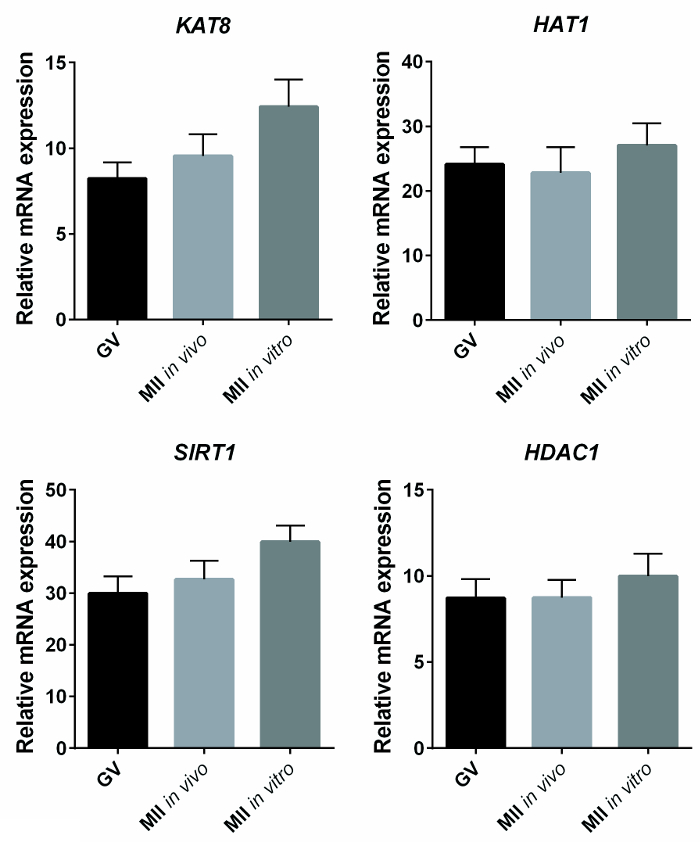
Figure 3: Expression of KAT8, SIRT1, HAT1, and HDAC1 in Immature, In Vivo, and In Vitro Matured Oocytes. The bar graphs represent the mean ± SEM of the relative mRNA expression of KAT8, SIRT1, HAT1, and HDAC1, expressed as the starting quantity (SQ) of the transcript of interest compared with the SQ of GAPDH, used as housekeeping. No statistical differences were observed between immature (GV, n = 14), in vivo (n = 14), and in vitro (n = 12) matured oocytes (one-way ANOVA, P >0.05). Reprinted with permission from reference7. Please click here to view a larger version of this figure.
Discussion
Even though IVM has been performed in horses for more than 20 years16, we do not yet know whether the oocyte can be the origin of embryonic aneuploidy, as has been proposed for humans17. The reason is probably that the preparation of oocyte spreads for chromosome counting results in considerable sample loss. With this in mind, a survey of the methods used for investigating errors in chromosome segregation was conducted to search for the most appropriate technique to apply to horse oocytes. Four main techniques were found in the scientific literature: 1) chromosomal spreads5,18,19, 2) fluorescent in situ hybridization (FISH)4,6,17,20, 3) centromere count on monastrol-collapsed spindles13,14,21,22, and 4) chromosome fluorescence staining without chromosome count23,24,25.
As already mentioned, the preparation of chromosomal spreads followed by Giemsa staining is complicated by a high rate of sample loss. This method has been replaced in many cases by FISH, which uses specific probes to map different chromosomes. A drawback of FISH techniques is that when a small number of probes are used, the incidence of a false negative may increase (i.e., identifying as normal a cell that is aneuploid because the missing or supernumerary chromosome has not been probed). The reliable use of FISH is therefore largely based on the knowledge a priori of the chromosomes that are more frequently affected by aneuploidy (e.g., chromosome 21 in humans). FISH has been previously used to investigate chromosomal abnormalities in horse embryos using 2 probes, against chromosome 2 and chromosome 420. According to these experiments, the nuclei of in vitro produced embryos are more likely to be affected by chromosomal abnormalities than in vivo produced embryos. This finding is in agreement with the observation reported above using the monastrol-collapse method in IVM oocytes. However, the incidence of aneuploidy in IVM oocytes was even higher than that reported using FISH in in vitro-produced embryos20. This partial discrepancy can be explained by the fact that only two chromosome-specific probes were used for the FISH experiments. This approach might have underestimated the incidence of aneuploidy involving other chromosomes, especially when considering that horses have 32 chromosomes. However, it cannot be excluded that severe aneuploidies detectable in the oocytes are not maintained in the embryos because gametes affected by critical genetic abnormalities might not develop further.
The combination of monastrol treatment, chromosome and centromere staining, and confocal microscopy allowed for consistent counts of the chromosome number in MII-stage horse oocytes with minimal sample loss. The technical limitation encountered in developing this technique for horse oocytes was the availability of horse-specific antibodies. Before using Aurora B phospho-(Thr232), two other antibodies (anti-Aurora B and anti-CENPA) did not produce a centromeric stain. Notably, anti-Aurora B and anti-CENPA were already successfully used in bovine and human cells26,27,28; therefore, it was concluded that they were not specific to the horse. This technique can also be limited by the availability of a confocal laser scanning microscope and image analysis software. Also, blind counting by two independent operators is essential to exclude operator-induced artifacts. Even though the chromosomal count of a monastrol-collapsed spindle is an approach that better preserves the morphology of the cell and prevents sample loss, this technique only allows the observation of differences in the basic number of chromosomes; it is not informative on other chromosomal abnormalities (e.g., translocations, segmental interchanges, etc.) that can instead be revealed by karyotyping and, in some cases, by FISH.
Errors in chromosome segregation have also been investigated by spindle and chromosome immunofluorescence, without performing a chromosomal count23,24,25. In this case, spindles with severe anomalies and lagging or scattered chromosomes are considered signs of erratic chromosome segregation. Accordingly, chromosomes that were scattered out of the spindle were observed in in vitro matured oocytes7 (the class that is more likely affected by aneuploidy). However chromosomal counting has the advantage of conveying quantifiable information.
To summarize, compared to the other techniques described, the use of monastrol and centromere staining has the advantage of preventing sample loss, minimizing the incidence of false negatives, and being quantifiable.
The alteration of the epigenetic modifications has been proposed as a possible mechanism in the onset of oocyte aneuploidy18,24,25. Using a triple fluorescent stain, it was possible to visualize and measure the meiotic spindle and to observe lagging/scattered chromosomes. At the same time, thanks to the development of antibodies that recognize specific residue modifications, the acetylation level of H4K16 was carried out on the same sample.This approach served the double purpose of reducing the amount of oocytes necessary for the analysis and correlating H4 acetylation with the morphology of the spindle.
The quantification of covalent protein modifications can be performed by Western blot. However, this technique requires a bigger samples size and does not preserve the integrity of the sample for morphological studies. The triple-fluorescent staining approach can be applied to other histone modifications and, potentially, to any protein that can be recognized by different secondary antibodies8; the only limiting factor is the availability of horse-specific antibodies. In this context, the authors hope that raising attention to the horse model will increase interest in the investigation of basic biological mechanisms in this species, and therefore, in the development of dedicated tools.
Finally, the expression study of mRNAs coding for histone deacetylases (HDACs) and acetyl-transferases (HATs) conducted by reverse transcription and quantitative-PCR (q-PCR) is also described. q-PCR is a broadly-spread technique; however, its use for horse oocytes is not diffuse. It is essential to specify that, due to the general silencing of transcriptional activity during oocyte maturation29, the analysis of the total transcript amount can only reveal mRNA stability or degradation in this background30,31. Differences in the relative expression of transcripts between in vitro and in vivo maturation were not observed. These findings indicate that translational and post-translational regulations might be affected by IVM, pointing out the need for the development of other experimental approaches aimed to investigate translational and post-translational mechanisms at the single-cell level.
Overall, the present study describes an experimental approach to morphologically and biochemically characterize horse oocytes. This approach can be applied to oocytes of other species that are similarly affected by a low recovery rate, including humans or endangered species. These studies will expand our knowledge of mammalian reproductive biology and will contribute to our understanding of infertility.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Fabrice Vincent for the support with laser scanning confocal microscopy (LSCM), and Philippe Barrière and Thierry Blard for performing daily ultrasound ovarian scanning and hCG injection. This work was supported in part by the "Regione Sardegna and Regione Lombardia" project "Ex Ovo Omnia" (Grant no. 26096200 to A. M. L.); the "L'Oreal Italia per le Donne e la Scienza 2012" fellowship (Contract 2012 to F. F.), FP7-PEOPLE-2011- CIG, Research Executive Agency (REA) "Pro-Ovum" (Grant no. 303640 to V. L.); and by the Postdoctoral School of Agriculture and Veterinary Medicine, co-financed by the European Social Fund, Sectorial Operational Program for Human Resource Development 2007-2013 (Contract no. POSDRU/89/1.5/S/62371 to I. M.). The in vivo oocyte collection was financed by the Institut Français du Cheval et de l'Equitation.
Materials
| ultrasound probe | Aloka | UST-5820-7,5 | |
| human chorionic gonadotrophin | centravet | CHO004 | 1500unit/animal IV |
| detomidine | centravet | MED010 | 9-15µg/kg IV |
| butylscopolamine bromure | centravet | EST001 | 0,2mg/kg IV |
| stereomicroscope | NIKON | SMZ-2B | |
| butorphanol | centravet | DOL003 | 10µg/kg IV |
| benzyl-penicillin | centravet | DEP203 IM | 15000UI/animal |
| TCM199 | Sigma-Aldrich | M3769 10X1L | powder for hepes-buffered TCM199 |
| Hepes sodium salt | Sigma-Aldrich | H3784-100G | |
| bovine serum albumin | Sigma-Aldrich | A8806 | |
| heparin | Sigma-Aldrich | H3149-10KU | |
| NaHCO3-buffered TCM199 | Sigma-Aldrich | M2154-500ML | liquid for IVM medium |
| epidermal growth factor | Sigma-Aldrich | E4127 | |
| newborn calf serum | Sigma-Aldrich | N4762-500ML | |
| 4-well dishes | NUNCLON | 144444 | |
| incubator | Heraeus | BB6060 | |
| monastrol | Sigma-Aldrich | M8515 | |
| hyaluronidase | Sigma-Aldrich | H3506 | |
| pronase | Sigma-Aldrich | P5147 | |
| paraformaldehyde | Sigma-Aldrich | P6148 | |
| polyvinyl alcohol | Sigma-Aldrich | 341584 | |
| triton-X 100 | Sigma-Aldrich | T8787 | |
| normal donkey serum | Sigma-Aldrich | D9663 | |
| rabbit anti-Aurora B phospho-Thr232 | BioLegend | 636102 | |
| TRITC-conjugated donkey anti-rabbit IgG | Vector Laboratories | 711-025-152 | |
| Vecta-Shield | Vector Laboratories | H-1000 | |
| YOPRO1 | Thermofisher Scientific | Y3603 | |
| confocal laser scanning microscope | LSM 780 | Zeiss | |
| confocal laser scanning microscope | LSM 700 | Zeiss | |
| ImageJ software | rsb.info. nih.gov/ij/download.html | free resource | |
| mouse anti-alpha-tubulin | Sigma-Aldrich | T8203 | |
| rabbit anti-acH4K16 | Upstate Biotechnology | 07-329 | |
| AlexaFluor 488-coniugated donkey anti-mouse IgG | Life Technologies | A21202 | |
| 4’,6-diamidino-2-phenylindole | Sigma-Aldrich | D8417 | DAPI |
| centrifuge | Eppendorf | 5417R | |
| RNALater | Invitrogen | AM7020 | |
| Luciferase RNA | Promega | L4561 | |
| PicoPure RNA Isolation Kit | Applied Biosystems | 12204-01 | |
| random hexamers | Thermofisher Scientific | N8080127 | |
| mouse Moloney leukaemia virus reverse transcriptase | Thermofisher Scientific | 28025013 | |
| SYBR green supermix | BioRad | 1708880 | |
| specific primers | Sigma-Aldrich | specific primers were designed using Primer3Plus software (free resource) | |
| thermal-cycler | BioRad | MyiQ | |
| mouse monoclonal anti-CENPA | Abcam | ab13939 | |
| mouse monoclonal anti-Aurora B | Abcam | ab3609 | |
Referencias
- Dellenbach, P. Transvaginal, sonographically controlled ovarian follicle puncture for egg retrieval. Lancet. 1 (8392), 1467 (1984).
- Humaidan, P. Ovarian hyperstimulation syndrome: review and new classification criteria for reporting in clinical trials. Hum Reprod. , (2016).
- Pincus, G., Enzmann, E. V. The Comparative Behavior of Mammalian Eggs in Vivo and in Vitro : I. The Activation of Ovarian Eggs. J Exp Med. 62 (5), 665-675 (1935).
- Emery, B. R., Wilcox, A. L., Aoki, V. W., Peterson, C. M., Carrell, D. T. In vitro oocyte maturation and subsequent delayed fertilization is associated with increased embryo aneuploidy. Fertil Steril. 84 (4), 1027-1029 (2005).
- Nichols, S. M., Gierbolini, L., Gonzalez-Martinez, J. A., Bavister, B. D. Effects of in vitro maturation and age on oocyte quality in the rhesus macaque Macaca mulatta. Fertil Steril. 93 (5), 1591-1600 (2010).
- Requena, A. The impact of in-vitro maturation of oocytes on aneuploidy rate. Reprod Biomed Online. 18 (6), 777-783 (2009).
- Franciosi, F. In vitro maturation affects chromosome segregation, spindle morphology and acetylation of lysine 16 on histone H4 in horse oocytes. Reprod Fertil Dev. , (2015).
- Franciosi, F. Changes in histone H4 acetylation during in vivo versus in vitro maturation of equine oocytes. Mol Hum Reprod. 18 (5), 243-252 (2012).
- Choi, Y. H., Gibbons, J. R., Canesin, H. S., Hinrichs, K. Effect of medium variations (zinc supplementation during oocyte maturation, perifertilization pH, and embryo culture protein source) on equine embryo development after intracytoplasmic sperm injection. Theriogenology. , (2016).
- Hendriks, W. K. Maternal age and in vitro culture affect mitochondrial number and function in equine oocytes and embryos. Reprod Fertil Dev. 27 (6), 957-968 (2015).
- Carnevale, E. M. The mare model for follicular maturation and reproductive aging in the woman. Theriogenology. 69 (1), 23-30 (2008).
- Ginther, O. J. The mare: a 1000-pound guinea pig for study of the ovulatory follicular wave in women. Theriogenology. 77 (5), 818-828 (2012).
- Chiang, T., Duncan, F. E., Schindler, K., Schultz, R. M., Lampson, M. A. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol. 20 (17), 1522-1528 (2010).
- Duncan, F. E., Chiang, T., Schultz, R. M., Lampson, M. A. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod. 81 (4), 768-776 (2009).
- Larionov, A., Krause, A., Miller, W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 6 (62), (2005).
- Choi, Y. H., Hochi, S., Braun, J., Sato, K., Oguri, N. In vitro maturation of equine oocytes collected by follicle aspiration and by the slicing of ovaries. Theriogenology. 40 (5), 959-966 (1993).
- Hassold, T., Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2 (4), 280-291 (2001).
- Akiyama, T., Nagata, M., Aoki, F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A. 103 (19), 7339-7344 (2006).
- Homer, H. A. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 19 (2), 202-207 (2005).
- Rambags, B. P. Numerical chromosomal abnormalities in equine embryos produced in vivo and in vitro. Mol Reprod Dev. 72 (1), 77-87 (2005).
- Nabti, I., Marangos, P., Bormann, J., Kudo, N. R., Carroll, J. Dual-mode regulation of the APC/C by CDK1 and MAPK controls meiosis I progression and fidelity. J Cell Biol. 204 (6), 891-900 (2014).
- Shomper, M., Lappa, C., FitzHarris, G. Kinetochore microtubule establishment is defective in oocytes from aged mice. Cell Cycle. 13 (7), 1171-1179 (2014).
- Luzzo, K. M. High fat diet induced developmental defects in the mouse: oocyte meiotic aneuploidy and fetal growth retardation/brain defects. PLoS One. 7 (11), e49217 (2012).
- Ma, P., Schultz, R. M. Histone deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet. 9 (3), e1003377 (2013).
- Yang, F., Baumann, C., Viveiros, M. M., De La Fuente, R. Histone hyperacetylation during meiosis interferes with large-scale chromatin remodeling, axial chromatid condensation and sister chromatid separation in the mammalian oocyte. Int J Dev Biol. 56 (10-12), 889-899 (2012).
- Luciano, A. M. Oocytes isolated from dairy cows with reduced ovarian reserve have a high frequency of aneuploidy and alterations in the localization of progesterone receptor membrane component 1 and aurora kinase B. Biol Reprod. 88 (3), 58 (2013).
- Luciano, A. M., Lodde, V., Franciosi, F., Ceciliani, F., Peluso, J. J. Progesterone receptor membrane component 1 expression and putative function in bovine oocyte maturation, fertilization, and early embryonic development. Reproduction. 140 (5), 663-672 (2010).
- Terzaghi, L. PGRMC1 participates in late events of bovine granulosa cells mitosis and oocyte meiosis. Cell Cycle. , 1-14 (2016).
- Susor, A., Jansova, D., Anger, M., Kubelka, M. Translation in the mammalian oocyte in space and time. Cell Tissue Res. 363 (1), 69-84 (2016).
- Chen, J. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 25 (7), 755-766 (2011).
- Ma, J., Flemr, M., Strnad, H., Svoboda, P., Schultz, R. M. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod. 88 (1), 11 (2013).

