A Method for Targeted 16S Sequencing of Human Milk Samples
Summary
A semi-automated workflow is presented for targeted sequencing of 16S rRNA from human milk and other low-biomass sample types.
Abstract
Studies of microbial communities have become widespread with the development of relatively inexpensive, rapid, and high throughput sequencing. However, as with all these technologies, reproducible results depend on a laboratory workflow that incorporates appropriate precautions and controls. This is particularly important with low-biomass samples where contaminating bacterial DNA can generate misleading results. This article details a semi-automated workflow to identify microbes from human breast milk samples using targeted sequencing of the 16S ribosomal RNA (rRNA) V4 region on a low- to mid-throughput scale. The protocol describes sample preparation from whole milk including: sample lysis, nucleic acid extraction, amplification of the V4 region of the 16S rRNA gene, and library preparation with quality control measures. Importantly, the protocol and discussion consider issues that are salient to the preparation and analysis of low-biomass samples including appropriate positive and negative controls, PCR inhibitor removal, sample contamination by environmental, reagent, or experimental sources, and experimental best practices designed to ensure reproducibility. While the protocol as described is specific to human milk samples, it is adaptable to numerous low- and high-biomass sample types, including samples collected on swabs, frozen neat, or stabilized in a preservation buffer.
Introduction
The microbial communities that colonize humans are believed to be critically important to human health and disease influencing metabolism, immune development, susceptibility to disease, and responses to vaccination and drug therapy1,2. Efforts to understand the influence of the microbiota on human health currently emphasize the identification of microbes associated with defined anatomic compartments (i.e., skin, gut, oral, etc.), as well as localized sites within these compartments3,4. Underpinning these investigative efforts is the rapid emergence and increased accessibility of next-generation sequencing (NGS) technologies that provide a massively parallel platform for analysis of the microbial genetic content (microbiome) of a sample. For many physiological samples, the associated microbiome is both complex and abundant (i.e., stool), but, for some samples, the microbiome is represented by low microbial biomass (i.e., human milk, lower respiratory tract) where sensitivity, experimental artefacts, and possible contamination become major issues. The common challenges of microbiome studies and appropriate experimental design have been the subject of multiple review articles5,6,7,8.
Presented herein is a robust NGS experimental pipeline based on targeted sequencing of the rRNA 16S V4 region9 to characterize the microbiome of human milk. Microbiome analysis of human milk is complicated not only by an inherently low microbial biomass10, but additionally by high levels of human DNA background11,12,13,14 and potential carryover of PCR inhibitors15,16 in extracted nucleic acid. This protocol relies on commercially available extraction kits and semi-automated platforms that can help minimize variability across sample preparation batches. It incorporates a well-defined bacterial mock community that is processed alongside samples as a quality control to validate each step in the protocol and provide an independent metric of pipeline robustness. Although the protocol as described is specific to the human milk samples, it is readily adaptable to other sample types including stool, rectal, vaginal, skin, areolar, and oral swabs10,17, and can serve as a starting point for researchers who wish to perform microbiome analyses.
Protocol
For all protocol steps, proper personal protective equipment (PPE) must be worn, and stringent contamination prevention approaches need to be taken. Observe flow of work from pre-amplification work areas to post-amplification work areas to minimize contamination of samples. All supplies used are sterile, free of RNase, DNase, DNA, and pyrogen. All pipette tips are filtered. A flowchart of the protocol steps is provided (Figure 1).
1. Sample Lysis
NOTE: Sample lysis and nucleic acid extraction are performed using a DNA/RNA extraction kit in a clean-room environment where both engineering and procedural controls are in place to minimize the introduction of environmental bacteria to the samples.
- Work area preparation
- Clean the biosafety cabinet (BSC) work area with appropriate surface cleaner to eliminate any nucleic acid contamination.
- Turn on the temperature-controlled vortexer, and set it to 37 °C.
- Prepare guanidinium thiocyanate lysis buffer (see Table of Materials)
- Check the lysis buffer for precipitates. Re-dissolve precipitates by warming at 37 °C.
- Prepare 600 µL of lysis buffer with 6 µL β-mercaptoethanol (β-ME) for each sample. Consider an extra 20% volume per sample.
- Sample preparation
- If whole milk is frozen, thaw it on ice. Aliquot 5 mL of whole milk into a 15-mL or a 5-mL sterile tube in BSC and keep it on ice.
- Spin the 5-mL milk aliquot for 10 min at 5,000 x g at 4 °C to pellet cells.
- Remove the fat layer, now the top layer in the tube, with a plastic spatula or large bore pipette tip.
- Without disturbing the pellet, remove all the supernatant except for 100 µL.
- Wash the pellet by resuspending in 1 mL of sterile phosphate buffered saline (PBS).
- Prepare 1 negative control by adding 1 mL of sterile PBS to a 5-mL tube.
- Transfer the suspension to a clean 1.5-mL centrifuge tube, and spin in a microcentrifuge for 1 min (5,000 x g at room temperature (RT)).
- Use a 1,000 µL sterile filtered pipette tip to discard the entire supernatant/fatty layer.
- If not extracting the same day, snap freeze the cell pellet by putting it in an ethanol/dry ice slurry, and immediately transfer it to the -80 °C freezer.
- Sample disruption and homogenization (bead-beating)
- Add 600 µL of lysis buffer containing β-ME to the pellet, and transfer the suspension to a bead tube.
- Each extraction batch of 12 contains 10 samples, 1 negative control (prepared in step 3.7 above), and 1 positive control (prepared in the next step).
- Prepare 1 positive control with lysis buffer and 20 µL of the bacterial mock sample (the mock community used has a concentration, once extracted, of approximately 0.2 ng/µL of DNA).
- Vortex all bead tubes vigorously for 15 s.
- Heat samples on the temperature-controlled vortexer at 37 °C for 10 min, with shaking at 700 - 800 rpm.
- Load tubes into the automated sample disruptor adapter set.
- Bead beat for 1 min at 30 Hz.
- Disassemble the adaptor set and manually switch the adaptor set with the sample plates from the left robotic arm to the right robotic arm of the instrument.
- Bead beat for another 1 min at 30 Hz.
- Centrifuge tubes at 17,200 x g, for 3 min at 25 °C.
- Remove all of the supernatant with a 200 µL pipette, being careful not to get any of the fatty layer at the top of the sample or the bead residual at the bottom of the sample, and apply to a homogenizer column.
- Centrifuge homogenizer columns at maximum speed, 17,200 x g, for 3 min at 25 °C.
- Transfer 350 µL of eluate to a 2-mL manufacturer's microcentrifuge tube for use with the automated instrument for purification of DNA and RNA (do not use any other tube). Be careful not to transfer any residual fatty layer.
- If the flow-through volume is less than 350 µL, add lysis buffer with β-ME to a final volume of 350 µL.
- Leave the samples at RT for up to 2 h before proceeding to nucleic acid isolation using the automated DNA purification instrument, or freeze at -80 °C.
- Cleaning work area
- Clean all surfaces in use with a non-enzymatic decontamination solution, leave for 10 min, then spray with 70% ethanol and wipe down the surface.
2. Isolate DNA/RNA
- Work area preparation
- Turn on the heat block and set the temperature to 70 °C.
- Turn on the temperature-controlled vortexer and set the temperature to 37 °C.
- Warm up the elution buffer (EB) containing 10 mM Tris-Cl, pH 8.5, in a 50-mL tube to 70 °C.
- Warm up 350 µL of frozen sample lysates in 2 mL tubes at 37 °C until completely thawed without any precipitate (approximately 10 min).
- DNA purification
- Vortex and centrifuge the 2-mL tubes with sample briefly (3000 x g for 10 s).
- Insert 2 mL tubes into the shaker following the automated DNA/RNA purification instrument's loading chart, per the manufacturer's instructions.
- Get the rotor adaptors and set them up on the tray based on the number of samples.
- Label each rotor adaptor based on the sample's identification (ID).
- Cut off the lids and smooth the edges of individual spin columns for DNA and RNA.
- Insert the DNA spin column without the collection tube into the rotor adaptor. Discard the collection tube.
- Label 1.5 mL collection tubes, and insert into rotor adaptors.
- Set rotor adaptors into the centrifuge following the automated DNA/RNA purification instrument's loading chart.
- Insert manufacturer's 1,000 µL filter-tips into tip racks.
- Add RNase-free water to a 2-mL manufacturer's microcentrifuge tube based on the number of samples (per specific machine protocol instructions).
- Insert the tube into tube slot "A" of microcentrifuge tube slots.
- Discard any reagent that is left over in reagent bottles and fill with a minimum volume of 10 mL.
- Insert reagent bottles into the reagent bottle rack (except EB bottle).
- Add warm EB from a 50-mL tube to the reagent bottle position 6.
- Check 1.5 mL tubes are placed tightly in the rotor adaptors.
- Close the instrument's lid and select: "RNA"→ "extraction kit" → "Animal, tissues and cells" → "kit's name 350 µL Part A Custom DNA" → Edit Elution Volume to 100 µL (default) or 50 µL for low biomass samples → "Start."
- When complete, remove rotor adaptors out of the centrifuge and place them on the tray.
- Discard the DNA spin column from rotor adaptor position 3.
- Do NOT discard rotor adaptors, RNA is in position 2.
- Remove 1.5 mL collection tubes containing eluted DNA at position 3, and store in −20 °C.
- Collect sample-containing tubes from the shaker and store in −20 °C, if any sample is left over, otherwise discard.
- Continue with the protocol "kit's name 350 µL part B RNA" for further purification of RNA.
- RNA purification will be done from the approximately 350 µL flow-through that is in the middle position of the rotor adaptors.
- RNA purification
- Insert the RNA spin column without its collection tube and lid into the rotor adaptor.
- Label new 1.5 mL collection tubes and insert them into rotor adaptor as indicated in the manual.
- Set the rotor adaptors into the centrifuge following the automated DNA/RNA purification instrument's loading chart.
- Close the instrument's lid and select: "RNA" → "Manufacturer's Kit" → "Animal, tissues and cells" → "Standard Part B RNA" → "Start."
- When completed, remove rotor adaptors out of the centrifuge and place them on the tray.
- Discard RNA spin column from position 3.
- Remove 1.5 mL collection tubes containing 30 µL eluted RNA at position 3, and store at −80 °C.
- Remove reagent bottles.
- Dispose of rotor adaptor contents through appropriate hazardous waste channels.
- Clean work station
- Spray all the automated DNA/RNA purification instrument's accessories, such as reagent racks, tray, and any other surface in use with a non-enzymatic decontamination solution, leave for 10 min and rinse with deionized (DI) water, then let them dry.
- Spray the automated instrument with only manufacturer approved non-enzymatic decontamination solution, wipe the inside of the centrifuge along with all surfaces in use, leave for 10 min, and then wipe with 70% ethanol. Do not use other types of decontamination solutions as they can damage the instrument.
3. Targeted 16S PCR Set-up
NOTE: The set-up for the 16S PCR is carried out in a designated pre-amplification workspace located within the clean-room. The reagents and samples are prepared and then loaded onto a liquid handler to perform the PCR for each sample in triplicate (30 samples, which include true samples and extraction positive and negative controls, plus 2 PCR water controls in triplicate, for a total of 96 combined samples and controls). Once the PCR reactions are assembled and sealed, the sample plate is transferred to a thermal cycler located in a post-amplification area for cycling.
- Work area preparation
- Clean the PCR workstation. Spray all surfaces in use with an RNase, DNase, DNA decontaminant, followed by DI water two times, and finally 70% ethanol.
- Prepare the 16S PCR Worksheet (see Targeted 16S PCR Worksheet) with an accurate sample list, and assign different barcoded primers to each sample9. Print out the worksheet and the plate maps (see Plate 1).
- PCR master mix preparation
NOTE: 16S primer selection is based on the region of interest for the particular study, and may change by study or sample type. Therefore, before ordering primers, it is important to confirm what area of the 16S rRNA best amplifies the microbes of interest for the particular study. This protocol as written is for amplification of the 16S V4 region. If different primers/region are selected, then the annealing temperature in the thermal cycling may require adjustment.- Work in the PCR workstation to prepare everything.
- Take out 50 – 100 µL of DNA samples from -20 °C, and all the reagents needed, and thaw them on ice. Vortex and briefly spin down.
- The primers are pre-diluted to the working concentration of 5 µM in a minimum volume of 20 µL.
- Prepare the PCR master mix in the specific 5 mL tube with only the forward primer according to the calculation on the worksheet.
- Preparing the robotic liquid handler for automated PCR setup
- For samples, take out a 32-well instrument's sample adaptor, and load 50 – 100 µL of DNA samples according to the 96-well plate map according to the manufacturer's instructions.
- For each PCR plate, set up 2 negative controls by placing 30 µL of PCR water in a clean sample tube.
- Place all the samples on the 32-well instrument's sample adaptor with caps locked in open position.
- For reverse primers9
- Remove the cap of each reverse primer with specific barcode #'s one at a time (change gloves in between to avoid cross-contamination).
- Place a maximum of 32 primers on the instrument's reagent adaptor.
- Take out a 96-well PCR plate and label it with study name, sample number, date, and the technician's initials.
- Loading the robotic liquid handler
- Place the reagent adaptor in position B1. In order to avoid an edge effect, carefully place one edge of the adaptor against the grip side, and slowly bring the other edge down. Make sure to push on all the corners of the adaptors.
- Place the sample adaptor in Position C1. Make sure to push on all the corners of the adaptors.
- Vortex the 5-mL master mix, open the cap, and place it in position A on the instrument's master mix and reagent block.
- Place the PCR plate on the 96-well instrument's adaptor that is intended to hold half skirted PCR plates.
- Start the run and save as a new file.
- Follow the prompts and check mark each prompt: one, tips are available, two, waste box is available, and three, start.
- After the run is complete, verify there were no errors by checking the machine for error messages.
- Seal the plate with the 12 or 8 strip caps, vortex vigorously, and spin down for 1 min using a 96-well plate spinner, and place it on ice or in the refrigerator.
- Remove tubes containing reverse primers and samples.
- Take the 96-well plate to the post-amplification room and load the plate onto the thermal cycler and cycle according to the table of thermal-cycling conditions (see Table 1).
- For samples, take out a 32-well instrument's sample adaptor, and load 50 – 100 µL of DNA samples according to the 96-well plate map according to the manufacturer's instructions.
4. Targeted 16S Post-PCR Quality Control Using Tape-based Platform for Gel Electrophoresis
NOTE: Post-PCR quality control (QC) and all subsequent steps are carried out in a designated post-amplification area of the lab. The DNA is analyzed in an automated DNA/RNA fragment analyzer.
- Work area preparation
- Clean the workstation by spraying all surfaces in use with an RNase, DNase, DNA decontaminant, followed by DI water two times, and finally 70% ethanol.
- Gather all supplies and equipment needed.
- Prepare reagents
- Place sample buffer and ladder in the temperature-controlled vortexer at 25 °C for a minimum of 30 min.
- While the reagents are warming up, prepare PCR samples by pooling the 3 PCR replicates for each sample into a single tube
- Load samples on instrument.
- Briefly vortex and spin down tubes with pooled PCR product.
- Place the instrument's 96-well plate on a rack at RT and label it.
- Briefly vortex and spin down the sample buffer and ladder.
- Add 3 µL sample buffer to each well of the 96 well plate (need at least 53 µL/screen tape).
- Run an even number of samples; if there is odd number of samples, add 4 µL of sample buffer to an empty well.
- Add 1 µL of ladder to a ladder well.
- Following the map, add 1 µL of pooled PCR product to its designated well.
- Cover plate with foil seal
- Vortex the plate using the instrument's vortexer and adaptor at 2,000 rpm for 1 min.
- Spin down to position the sample at the bottom of the tube, if there are bubbles spin down again.
- Launch the software.
- Load the screen tape and loading tips into the instrument.
- Load samples into the fragment analyzer with a 96-well adaptor.
- Set up a run with the controller software.
- Make sure to select even numbered wells.
- Write sample IDs.
- Check that all the wells of the screen tape are highlighted in gray.
- Make sure the analyzer recognizes all the pipette tips.
- Select start and specify a filename to save the results (study name, sample numbers, and date).
- Refer to the manufacturer's instruction for more details.
- After the run is complete, discard tip, screen tape, and tubes.
- Analyze data for 16S peak nM (between 315 – 450 bp for V4). This peak will vary depending on the primers selected.
5. Library Calculation, Pooling, Clean-up, and QC
- After determining amplicon size and molarity of all samples, pool the libraries to achieve the final desired volume and nM for the pooled library (see Sample Calculation).
- Clean-up and concentrate the pooled library using a silica-membrane-based purification kit for PCR products, according to the manufacturer's protocol (see Table of Materials).
- Elute the DNA with a final volume of 50 µL.
- Measure the concentration of the cleaned library immediately by using a double stranded DNA high sensitivity kit for a fluorometer, according to the manufacturer's protocol.
- Dilute 2 µL of the pooled and cleaned library with 198 µL of the dilution buffer plus dye (1:100 dilution).
- Record the measured value from the fluorometric device and convert it according to the dilution factor.
- Using the concentration reading (ng/µL) for the pooled library, calculate the library molarity in nM. Use 1 µL undiluted library to measure the OD260/280 on a microvolume spectrophotometer.
NOTE: A value of 1.8 – 2.0 indicates adequate purity of the library. - Store the final library at -20 °C until ready for next generation sequencing.
Representative Results
The protocol presented here includes important quality control (QC) steps to ensure that the data generated meet benchmarks for protocol sensitivity, specificity, and contamination control. The protocol's first QC step follows PCR amplification of the 16S V4 region (Figure 2). One µL of PCR product from each sample was analyzed by electrophoresis to confirm that it was within the expected size range of 315 – 450 bp (Figure 2, red arrow). Some human milk samples generated lower amounts of specific product (Figure 2A, compare lanes 3 and 9 – 11 with lanes 4 – 8), suggesting either low levels of extracted microbial DNA in those samples, or carry-over of PCR inhibitors during extraction. For samples that produce less than 2.0 nM of product in the 315 – 450 bp range (Figure 2A, lane 7), PCR inhibitor cleanup is carried-out using a single step kit and the sample is re-amplified. Success rates for recovery of sample amplification after cleanup is approximately 40%. Quantitation of specific product for each sample (Figure 2B) is essential for determining its required volume for equal molar pooling of samples for sequencing. A pooled library for targeted sequencing is usually dominated by a specific PCR product (Figure 3). If there is a significant amount of non-specific product in the library, a gel-purification step should be added to the workflow.
In the example presented in Figure 2A, faint bands are observed for buffer controls (BC; lanes 2 and 12) and the PCR water negative control (PC; lane 1), indicating possible environmental or reagent contamination. Such bands are not uncommon and typically represent low amounts of PCR product (i.e., <1 nM) and produce few read counts during sequencing (<1,000). Representative sequencing results (Figure 4) confirm that these samples do indeed have very low sequencing read counts (Figure 4A, lanes 1 and 11; Figure 4B, Buffer and PCR Water lanes) and, importantly, the taxa composition for the control samples is distinct from the human milk samples (Figure 4A; compare lanes 1 and 11 with lanes 2 – 10). High read counts in the negative controls, together with significant overlap in taxa composition between controls and samples, suggests cross-contamination and the need for improved contamination control.
Sequencing results (Figure 4) demonstrate high diversity in the taxa associated with the human milk microbiome and variability in the number of sequencing read counts for each sample (Figure 4A, lanes 2 – 10). In contrast, the sequencing results for the bacterial mock that was processed along with the human milk samples demonstrated taxa composition and read counts that were comparable to results obtained for the mock in previous workflow runs (compare Figure 4A, lane 12 with Figure 4B, mock lanes). The consistent results for the mock lanes suggest that the observed variability for the human milk samples is an authentic experimental result, and not a function of intrinsic workflow variability.

Figure 1: Flow chart of the Targeted 16S Sequencing Pipeline. Please click here to view a larger version of this figure.
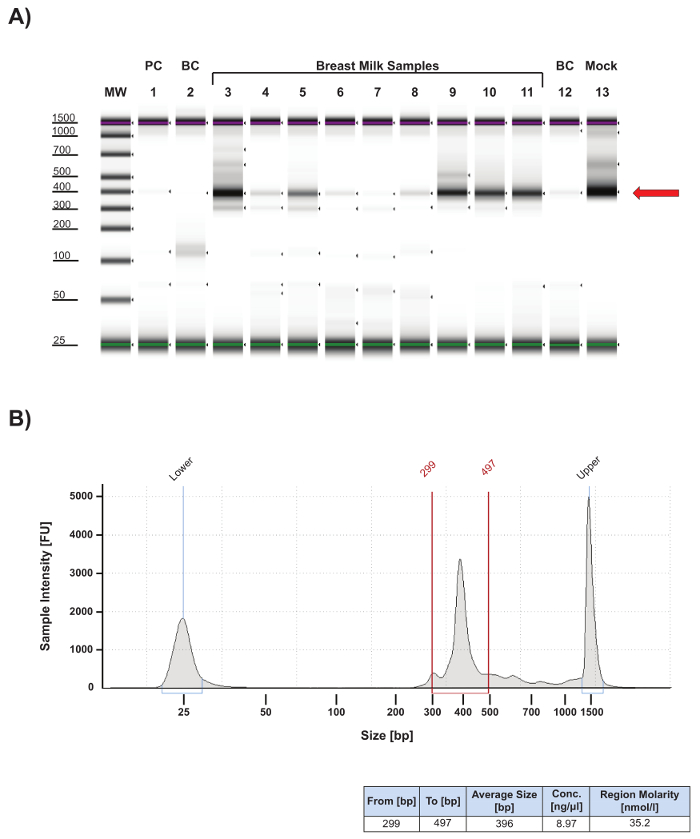
Figure 2: Quality control analysis of 16S V4 amplicons. (A) Gel image of 16S V4 amplicons resolved by electrophoresis using an automated DNA/RNA fragment analyzer. 16S V4 amplicons were generated according to Caporaso et al.9, and one µL of each PCR product was analyzed using high sensitivity DNA reagents according to the manufacturer's guidelines. Most human milk samples (lanes 3 – 6 and 8 – 11) and the bacterial mock (lane 13) produced a primary PCR product at the expected size of approximately 400 bp (red arrow). The human milk sample in lane 7 failed to produce a significant amount of specific product and was subject to cleanup and re-amplification. Minimal product was detected for the PCR negative control (PC, lane 1), and lysis buffer negative controls (BC, lanes 2 and 12) indicated minimal contamination present in the analyzed samples. MW, molecular weight markers: upper red and lower green bars identify the 1,500 bp and 25 bp size markers, respectively, in each lane. (B) Top Electropherogram of lane 3 from gel in (A). The primary PCR product falls within the peak region defined by the red vertical bars and comprises fragments ranging in size from 299 – 497 bp resulting in an average PCR product size of 396 bp. Gating is done on a slightly wider range than the anticipated amplicon size (in this case 315 – 450 bp) to be sure to include the entire sample peak. The upper and lower peaks correlate with fragment sizes of 25 bp and 1,500 bp, respectively. Bottom: chart summarizing the size parameters for the peak region, the concentration in ng/µL of the PCR product within the peak region, and the molarity in nM for the specific PCR product. This information is then used to calculate how much of each sample will be pooled in an equal molar library for sequencing (see Sample Calculation). Please click here to view a larger version of this figure.
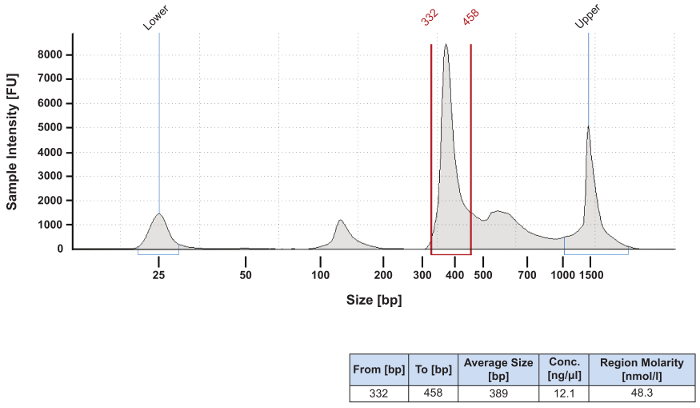
Figure 3: Electropherogram of a pooled and concentrated sequencing library. Equal molar amounts of individual samples to be sequenced were combined into a pooled library. The library was then cleaned and concentrated to a total volume of 50 µL using a silica-membrane-based PCR clean up kit. Final preparation of the library for sequencing on the next generation sequencer was conducted according to the manufacturer's protocol. This library was successfully sequenced despite the presence of additional bands. If there is a concern about PCR products outside the expected size range, the manufacturer's protocol suggests the addition of a gel size selection step. This QC step is not usually performed. Please click here to view a larger version of this figure.
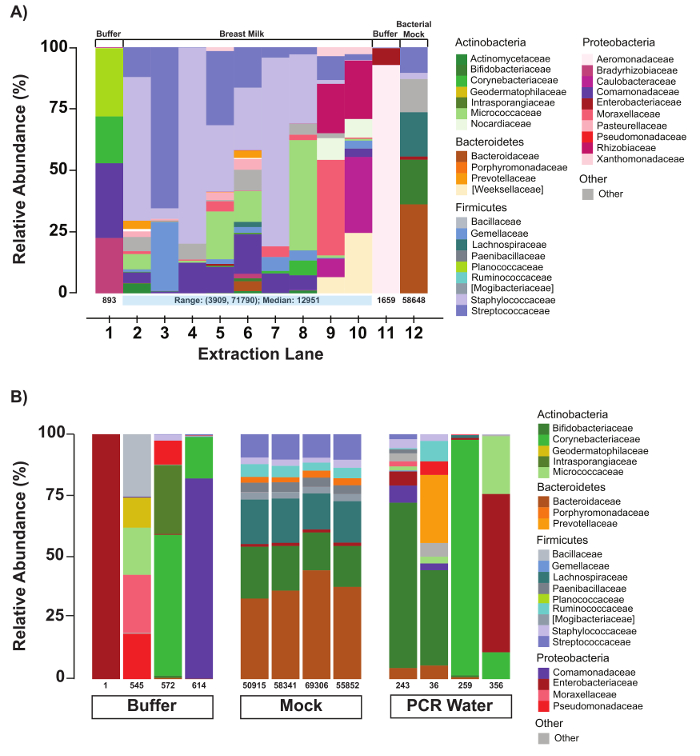
Figure 4: Evaluation of negative and positive controls. (A) Relative abundances of bacterial taxa of an extraction batch with controls and human milk samples. As a QC measure, compositions of each extraction batch as loaded on the automated DNA/RNA purification instrument are generated immediately following a sequencing run. Numbers under each sample bar indicate the number of filtered reads for the respective sample. The compositions of the buffer controls are distinct from that of the human milk samples. (B) Relative abundances of bacterial taxa in buffer, mock, and PCR controls. Number of reads and composition are evaluated for all negative (buffer and PCR water) and positive (bacterial mock) controls. The compositions of the buffer and water vary, but the mock community remains quite stable. Please click here to view a larger version of this figure.
Discussion
Targeted next-generation sequencing of 16S rRNA is a widely used, rapid technique for microbiome characterization18. However, many factors, including batch effects, environmental contamination, sample cross-contamination, sensitivity, and reproducibility can adversely affect experimental results and confound their interpretation7,19,20. To best facilitate robust 16S analyses, microbiome workflows must incorporate good experimental design, the use of appropriate controls, spatial segregation of workflow steps, and application of best practices. The protocol described here incorporates each of these parameters and provides important experimental tools to address the challenges above and implement a 16S workflow for diverse samples.
Good experimental design is critical for 16S microbiome analyses. This includes proper collection and storage of samples, as well as selection of 16S primers appropriate for the region of interest. For example, the V4 region (515F/806R) is selected for human milk because it has good amplification of Bifidobacterium, which plays an important role in development of the neonatal gut microbiome21. Other primer sets (e.g., 27F/338R, 515F/926R) may be more appropriate for studies of other microbial communities. An important note is that the annealing temperature for the targeted 16S PCR and the expected amplicon size may vary based on primer selection.
Other places the protocol may be modified are based on results of the QC steps incorporated in the work flow. A few options exist for troubleshooting when either no or little DNA is detected following the targeted 16S PCR amplification step. 1) The sample can be put through a PCR inhibitor removal step. Amplification following PCR inhibitor removal using a single step kit performed per the manufacturer's protocol is successful approximately forty percent of the time, 2) more extracted DNA can be added to the targeted 16S PCR, or 3) a new and potentially larger aliquot of milk can be processed if available. If there is a concern about PCR products outside the expected size range following the silica-membrane-based purification of the library, a gel purification step can be added. Finally, the QC steps are critical to determine if there is evidence of contamination, which is discussed in detail below. If significant contamination is detected, then depending on where the contamination is introduced, either the PCR can be repeated or if necessary, the sample can be re-extracted. Fortunately, with good laboratory practices, these are rare events. Finally, while this protocol is written to highlight caveats in the amplification of low biomass samples and specifically human milk, the protocol can easily be modified for the amplification of oral, rectal, vaginal, and skin swabs or sponges as well as stool. If other sample types are chosen, then consideration is given to which extraction kits are optimal for the specific sample type.
Batch effects due to kits, reagents, or sequencing runs are important sources of variability in microbiome studies. DNA extraction kits, along with other reagents, possess low levels of bacterial DNA, which may vary substantially by lot20,22,23,24. For a large project, using a single lot of kits, reagents, selecting kits designed to minimize kit contamination may simplify analysis7. Samples of both subjects and controls are processed side by side. It is best if all the samples for a single study, both subject and control, can be incorporated into a single sequencing run. If a large number of samples are to be processed in batches, samples that are representative of both subjects and controls are included in each batch and processed together. It is also important to organize batch processing to minimize contamination of low-biomass samples (i.e., human milk) by samples that are high biomass (i.e., stool). In such cases, process low biomass sample types first, and then high biomass samples for the same study.
Low biomass samples pose unique challenges to microbiome studies, as contamination from the environment, reagents, instruments, and the researcher can make it difficult to distinguish between authentic community members present in low abundance7,25,26 and those that are artificially introduced to the sample through the experimental process19. The workflow described here incorporates important experimental negative controls at both the sample preparation step (buffer-only lysis control), and the PCR step (PCR water control) (see Figure 4). These controls help identify contamination sources and facilitate effective corrective measures at the bench or in silico27,28,29,30. Negative controls are carefully evaluated and reported with the study results7.
To minimize contamination, spatial segregation of experimental activities into a clean pre-PCR amplification area and post-amplification area is important. Optimally, the clean room has both an area for PCR master mix preparation and a sample preparation/addition of master mix area, and may incorporate a separate dead air box or a biological safety cabinet housing dedicated consumables and small equipment needed for the master mix preparation. The clean room design incorporates a positive airflow system with high efficiency particulate air (HEPA) filtration. Use of personal protective equipment is essential to maintaining a controlled, low-microbial environment, and includes hairnets, lab coats, gloves, and shoe covers. Kits/reagents and samples are ideally stored in separate dedicated refrigerators/freezers. PCR setup is also carried out in the clean room in designated workstations; clear separation of primer stocks and reagents from extracted DNA is maintained until samples are loaded on the automated pipetting platform. Once a PCR setup is complete, the plate is transferred to the post-amplification area and loaded onto a thermal cycler.
It is important to restrict the flow of work activity from clean areas to post-amplification areas; there is no retrograde movement of reagents, instruments, or supplies from post-amplification areas to the clean area. Personnel that have entered any of the post-amplification areas are barred from entry to the clean area for 24 hours (until the next day). In addition to the above workflow considerations, cleaning protocols must be implemented in both clean and post-amplification areas to minimize nucleic acid contamination of work surfaces and instrumentation. If physical barriers or separate rooms are not possible, all efforts must be taken to set up the work in areas as far apart as possible.
In addition to contamination, microbiome studies are challenged by sensitivity, variability, and reproducibility31. This protocol addresses these issues by incorporating a defined bacterial mock community that is extracted, amplified, and sequenced along with each batch of samples (see Figure 4b). This control provides a constant internal reference that evaluates the reproducibility of the experimental results generated, and can be used to troubleshoot problems that arise. For example, the quality of the extracted mock DNA can provide a metric for effective sample lysis and DNA extraction, which genomic DNA controls miss. Quality control of PCR amplicons for the mock sample can also indicate PCR efficiency and specificity. Furthermore, because the mock comprises multiple bacteria types, the relative sensitivity for a processed batch of samples can be inferred by the representation of taxa in the sequencing results for the mock sample. An ideal mock community will evaluate the ability to detect key bacterial species in the compartment being analyzed, and therefore the composition of the mock community may need to vary by study. As shown in Figure 4a, there is considerable variability among sample sequencing results, but the sequence results for the bacterial mock community is highly reproducible (see Figure 4b).
While the mock community in Figure 4 is a unique mixture of 33 strains from a combination of commercially available and local clinical isolates, a commercially available mock community has recently been developed32.
Although the workflow described here is limited in its ability to broadly address reproducibility across different microbiome studies, it does provide an important experimental approach that allows researchers to incorporate appropriate experimental controls and monitor reproducibility within their own results.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
We would like to thank Helty Adisetiyo, PhD and Shangxin Yang, PhD for the development of the protocol.Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Materials
| AllPrep RNA/DNA Mini Kit | Qiagen | 80204 | DNA/RNA extraction kit |
| Eliminase | Fisher Scientific | 435532 | RNase, DNase, DNA decontaminant |
| Thermo Mixer | Fisher Scientific | temperature-controlled vortexer | |
| Buffer RLT plus | Qiagen | 1053393 | guanidinium thiocyanate lysis buffer/ Part of Allprep kit |
| ß-Mercaptoethanol | Sigma Aldrich | 63689-25ML-F | ß-ME is a reducing agent that will irreversibly denature RNases by reducing disulfide bonds |
| LME Beads | MP Biomedicals | 116914050 | bead tube |
| QIAgen TissueLyzer | Qiagen | 85300 | automated sample disruptor adapter set |
| QIAshredder column | Qiagen | 79654 | |
| QIAgen RB tube | manufacturer's microcentrifuge tube in kit | ||
| QIAcube and related plasticware | Qiagen | 9001292 | automated DNA/RNA purification instrument |
| DNA exitus plus | Applichem | A7089 | non-enzymatic decontamination solution |
| EB Buffer | Qiagen | 19086 | elution buffer |
| QIAgility and related plasticware | Qiagen | 9001532 | robotic liquid handler |
| PCR water | MO BIO | 17000- | |
| 5PRIME HotMasterMix | Quantabio | 2200400 | |
| Barcoded reverse primers | Eurofin | No Catalog #'s | designed and ordered |
| 96 well PCR plate | USA scientific | 1402-9708 | |
| Tapestation 2200 and related plasticware | Agilent | G2964AA | automated DNA/RNA fragment analyzer |
| D1000 reagents for Tapestation | Agilent | 5067-5585 | Sample buffer and ladder are part of this kit |
| OneStep PCR Inhibitor Removal Kit | Zymo Research | 50444470 | PCR inhibitor removal is done per the manufacturer's instructions. |
| QIAquick PCR Purification Kit | Qiagen | 28104 | DNA clean up kit: silica-membrane-based purification of PCR products |
| Qubit dsDNA HS Assay Kit | Thermo Fisher | Q32854 | dimethylsulfoxide-based dilution buffer and dye are part of this kit. |
| Qubit Fluorometer | Thermo Fisher | Q33216 | |
| NanoDrop | Thermo Fisher | microvolume spectrophotometer | |
| MiSeq 300 V2 kit | Illumina | 15033624/15033626 | |
| MiSeq | Illumina | No Catalog #'s | next generation sequencer |
Referencias
- Ahern, P. P., Faith, J. J., Gordon, J. I. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 40 (6), 815-823 (2014).
- Postler, T. S., Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. , (2017).
- Hall, M. W., et al. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes. 3, 2 (2017).
- Perez Perez, G. I., et al. Body Site Is a More Determinant Factor than Human Population Diversity in the Healthy Skin Microbiome. PLoS One. 11 (4), e0151990 (2016).
- Hamady, M., Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 19 (7), 1141-1152 (2009).
- Human Microbiome Project, C. A framework for human microbiome research. Nature. 486 (7402), 215-221 (2012).
- Kim, D., et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 5 (1), 52 (2017).
- Hugerth, L. W., Andersson, A. F. Analysing Microbial Community Composition through Amplicon Sequencing: From Sampling to Hypothesis Testing. Front Microbiol. 8, 1561 (2017).
- Caporaso, J. G., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 (8), 1621-1624 (2012).
- Pannaraj, P. S., et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. , (2017).
- Ho, F. C., Wong, R. L., Lawton, J. W. Human colostral and breast milk cells. A light and electron microscopic study. Acta Paediatr Scand. 68 (3), 389-396 (1979).
- Parmely, M. J., Beer, A. E., Billingham, R. E. In vitro studies on the T-lymphocyte population of human milk. J Exp Med. 144 (2), 358-370 (1976).
- Sabbaj, S., et al. Human immunodeficiency virus-specific CD8(+) T cells in human breast milk. J Virol. 76 (15), 7365-7373 (2002).
- Jimenez, E., et al. Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women. J Hum Lact. 31 (3), 406-415 (2015).
- Ghosh, M. K., et al. Quantitation of human immunodeficiency virus type 1 in breast milk. J Clin Microbiol. 41 (6), 2465-2470 (2003).
- Lim, N. Y., Roco, C. A., Frostegard, A. Transparent DNA/RNA Co-extraction Workflow Protocol Suitable for Inhibitor-Rich Environmental Samples That Focuses on Complete DNA Removal for Transcriptomic Analyses. Front Microbiol. 7, 1588 (2016).
- Bender, J. M., et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med. 8 (349), 349ra100 (2016).
- Cox, M. J., Cookson, W. O., Moffatt, M. F. Sequencing the human microbiome in health and disease. Hum Mol Genet. 22 (R1), R88-R94 (2013).
- Lauder, A. P., et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 4 (1), 29 (2016).
- Salter, S. J., et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
- Walker, A. W., et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 3, 26 (2015).
- Glassing, A., Dowd, S. E., Galandiuk, S., Davis, B., Chiodini, R. J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 8, 24 (2016).
- Kennedy, K., Hall, M. W., Lynch, M. D., Moreno-Hagelsieb, G., Neufeld, J. D. Evaluating bias of illumina-based bacterial 16S rRNA gene profiles. Appl Environ Microbiol. 80 (18), 5717-5722 (2014).
- Weiss, S., et al. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 15 (12), 564 (2014).
- Aho, V. T., et al. The microbiome of the human lower airways: a next generation sequencing perspective. World Allergy Organ J. 8 (1), 23 (2015).
- Charlson, E. S., et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 184 (8), 957-963 (2011).
- Bittinger, K., et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 15 (10), 487 (2014).
- Jervis-Bardy, J., et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 3, 19 (2015).
- Knights, D., et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 8 (9), 761-763 (2011).
- Lazarevic, V., Gaia, N., Girard, M., Schrenzel, J. Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. 16, 73 (2016).
- Kurilshikov, A., Wijmenga, C., Fu, J., Zhernakova, A. Host Genetics and Gut Microbiome: Challenges and Perspectives. Trends Immunol. 38 (9), 633-647 (2017).
- . Mock microbial communities Available from: https://www.atcc.org/en/Products/Microbiome_Standards.aspx?utm_id=t170601524l1 (2017)

