Collection and Processing of Lymph Nodes from Large Animals for RNA Analysis: Preparing for Lymph Node Transcriptomic Studies of Large Animal Species
Summary
This protocol provides an overview of procedures for the isolation of RNA for the transcriptomic profiling of lymph node tissues from large animals, including steps in the identification and excision of lymph nodes from livestock and wildlife, sampling approaches to provide consistency across multiple animals, and considerations plus representative results for the post-collection preservation and processing for RNA analysis.
Abstract
Large animals (both livestock and wildlife) serve as important reservoirs of zoonotic pathogens, including Brucella, Mycobacterium bovis, Salmonella, and E. coli, and are useful for the study of pathogenesis and/or spread of the bacteria in natural hosts. With the key function of lymph nodes in the host immune response, lymph node tissues serve as a potential source of RNA for downstream transcriptomic analyses, in order to assess the temporal changes in gene expression in cells over the course of an infection. This article presents an overview of the process of lymph node collection, tissue sampling, and downstream RNA processing in livestock, using cattle (Bos taurus) as a model, with additional examples provided from the American bison (Bison bison). The protocol includes information about the location, identification, and removal of lymph nodes from multiple key sites in the body. Additionally, a biopsy sampling methodology is presented that allows for a consistency of sampling across multiple animals. Several considerations for sample preservation are discussed, including the generation of RNA suitable for downstream methodologies like RNA-sequencing and RT-PCR. Due to the long delays inherent in large animal vs. mouse time course studies, representative results from bison and bovine lymph node tissues are presented to describe the time course of the degradation in this tissue type, in the context of a review of previous methodological work on RNA degradation in other tissues. Overall, this protocol will be useful to both veterinary researchers beginning transcriptome projects on large animal samples and to molecular biologists interested in learning techniques for in vivo tissue sampling and in vitro processing.
Introduction
RNA-sequencing analysis of the transcriptome of lymph nodes provides the opportunity to characterize the immune response of animals to a variety of pathogens. While this methodology has been utilized extensively in mice, analyses have recently been expanding into larger mammals1,2. Livestock/large animal lymph nodes can be used to characterize host responses to an infection, not only for their use in vaccine or genetic susceptibility studies and for the identification of targets for drug development, but also as model systems for human studies on zoonotic diseases. For example, in the case of brucellosis (a zoonotic bacterial disease that impacts half a million people around the world each year), despite significantly increased costs, studies in sheep or goats are more relevant to the human infection and human vaccine development than laboratory animal models. Mouse infection models recapitulate the reticuloendothelial system infection but not the characteristic clinical signs3.
In large animal experiments as compared to laboratory animal studies, the process of tissue harvesting necessarily involves a longer delay between the euthanasia and the tissue collection, which presents a potential challenge for the preservation of high-quality RNA. Intact RNA is essential for the generation of biologically relevant transcriptomic data. The generation of high-quality RNA from tissue samples is particularly critical for large animal pathogen studies conducted in containment facilities. Such studies are inherently more difficult to perform as they not only require approved facilities and highly trained personnel but also carry significant financial costs, which, depending on the work, can range from tens to hundreds of thousands of dollars. These types of studies also involve a cross-disciplinary collaboration and cross-disciplinary knowledge for their completion, adding to their complexity. Therefore, training on, development of, and adherence to a streamlined system for the sample collection and preservation provides significant benefits for downstream molecular studies of tissues from infected animals.
The collection of larger lymph nodes presents additional challenges for the tissue collection compared to the similar sampling of murine lymph nodes. The preparation for the sample excision necessitates a basic understanding of the anatomy of the lymph node, including the relevant internal structures. The structure of a lymph node is comprised of lymphoid lobules surrounded by sinuses filled with lymph. These structures are enclosed within a tough, fibrous capsule.4 A lymphoid lobule is the "basic anatomical and functional unit of the lymph node" and is composed of follicles, a deep cortical unit, and medullary cords and sinuses4 (Figure 1A). B and T lymphocytes are home to the follicles and deep cortical units, respectively. These structures provide a 3D scaffold and facilitate the interaction between the lymphocytes and antigen or antigen presenting cells.
Grossly, follicles and deep cortical units can be identified on cut surface as they contain a denser reticular meshwork and appear darker than the sinuses, which are comprised of a more delicate reticular meshwork and appear lighter (Figure 1B). By convention, pathologists refer to the regions of the lymph nodes as the superficial cortex (follicles), the paracortex (deep cortical units) and the medulla (medullary cords and sinuses). A proper examination of all three regions has been deemed as best practice in routine pathological examination guidelines for lymph nodes5. Note that there is a considerable variation in the consistency, size, and color of lymph nodes, even within a single animal. As animals age, their lymph nodes will tend to decrease in size and become firmer than those of younger animals, typically due to an increase in their connective tissue and a reduction of the normal lymphoid structure6,7.
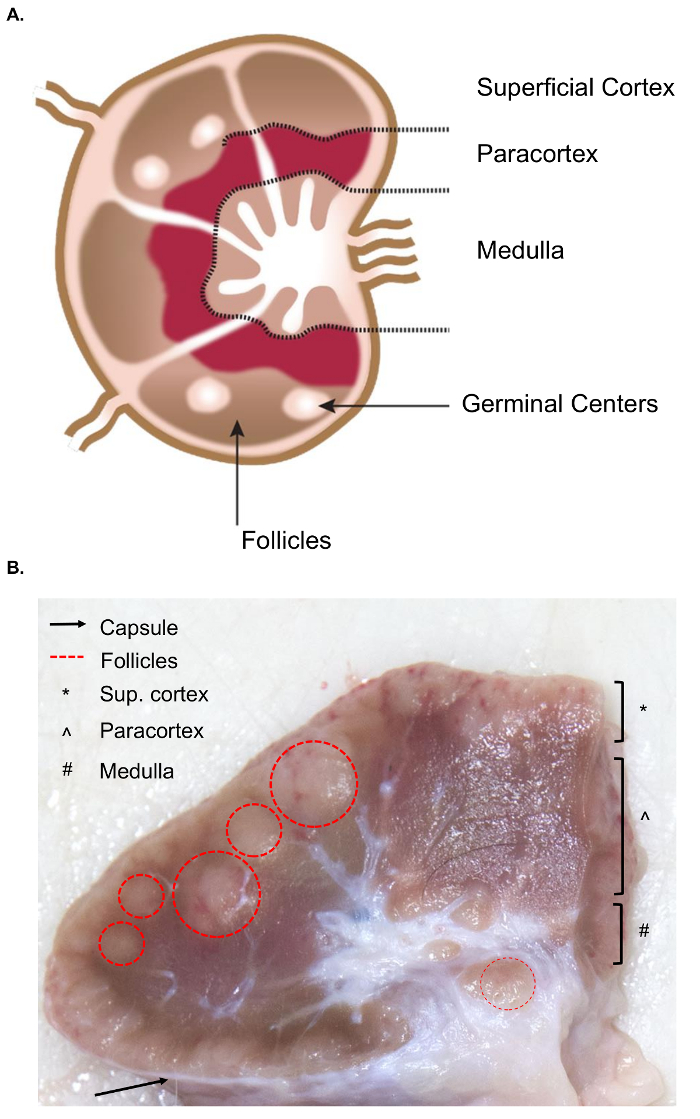
Figure 1. Anatomy of the lymph node. (A) This cartoon image shows the anatomy of the lymph node, depicting key structures. (B) This still image shows a bovine lymph node cut in cross-section. The relevant structures/layers that are visible to the naked eye are highlighted. Please click here to view a larger version of this figure.
Depending on the experimental question, different lymph nodes will be of interest for the collection and analysis. Peripheral lymph nodes are those located deep in the subcutaneous tissue. In cattle, peripheral or superficial lymph nodes often used in clinical and experimental practice include parotid, submandibular, retropharyngeal, prescapular, prefemoral (precrural) and superficial inguinal (supramammary in females, scrotal in males) (Figure 2). In Table 1, the properties of key superficial lymph nodes, as described in the cattle system8, are outlined. Below, some potential lymph node collection plans for infectious bacterial diseases of cattle are presented as a starting point for the investigation.
Brucella abortus/Brucella melitensis: Standard necropsies for B. abortus-infected cattle and B. melitensis-infected goats at the National Animal Disease Center recover supramammary, prescapular, and parotid lymph node tissue, both for the grinding for the bacterial enumeration and for the RNA preparation for the host RNA expression profiling. B. abortus can be regularly recovered in each of these lymph nodes in experimentally infected cattle9. The presence of bacteria in each of these lymph node types can be detected in B. melitensis-infected goats up to at least nine months post-infection using the RNA-based methodologies from our studies (Boggiatto et al., unpublished). Salmonella sp.: The prescapular, subiliac (prefemoral), and mesenteric lymph nodes have been useful during the profiling of cattle carcasses for a Salmonella prevalence10,11,12 and would be of potential interest for transcriptomic studies. E. coli O157:H7: Mesenteric lymph nodes (at the middle small intestine and distal small intestine locations) can be the sites of an occasional recovery of the bacteria in infected calves (but not in infected adult cattle)13. Leptospirosis (Leptospira sp.): A chronic persistence of the bacteria has been observed in the lymph nodes draining the mammary gland14. Mycobacterium bovis: In cattle, the bacteria have been recovered post-experimental infection from the mediastinal and tracheobronchial lymph nodes of calves15. Additionally, lymph node RNA has been utilized to examine large animal host responses to viruses, such as the porcine reproductive and respiratory syndrome virus2. Figure 2 depicts the location of a subset of these major lymph nodes in the cattle body.
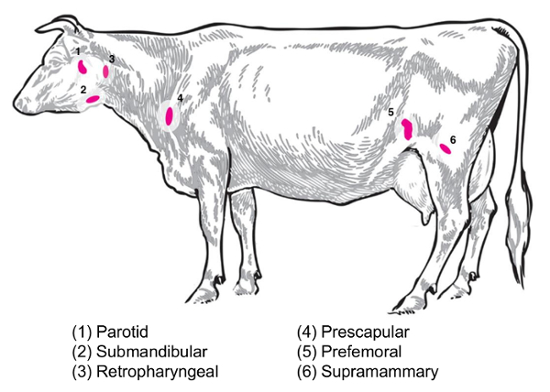
Figure 2: Cartoon depicting selected lymph node locations in Bos taurus. The numbered lymph nodes are annotated. Please click here to view a larger version of this figure.
In this paper and the associated video, we present a protocol for the isolation of large animal lymph nodes for RNA studies, designed to be informative for molecular biologists involved in transcriptomic studies of large animal infections. First, we provide an overview of the isolation procedure for the lymph nodes, using sampling from bovine and bison tissues as examples. Paired with this demonstration, as displayed in the video, is a workflow for a reproducible tissue sampling for RNA isolation. Next, we describe important considerations for the processing of an infected lymph node, with a focus on safety, consistency, and RNA quality.
The preparation of RNA from the tissue with an acidified phenol-guanidine isothiocyanate reagent is based on the original method of Chomczynski and Sacchi16,17, with a purification over silica-based spin columns in the presence of chaotropic agents based on the original work of Vogelstein and Gillespie18. We also examine the potential for the recovery of RNA for transcriptomics from cattle lymph nodes preserved by alternative methods. Finally, we explore the impact of the time variable on the RNA quality in large animal necropsies, including a representative experiment depicting the effect of an increase in time between the euthanasia and the sampling on the recovered RNA profile from bison and bovine lymph nodes. This article will be useful not only to molecular biologists but also to veterinary researchers commencing transcriptomic studies.
Protocol
The animal necropsy procedures depicted here are covered under approved IACUC protocols at the National Animal Disease Center, Ames, IA. All experiments were conducted in accordance with the approved guidelines for animal care and welfare.
1. Pre-planning Before Necropsy
- Before the necropsy, prepare the following materials for transport to the necropsy suite:
- 1.5 or 2 mL microcentrifuge tubes filled with ~ 1.0 mL of RNA preservation solution, in a rack or transport container. Be sure to follow the manufacturer's guidelines for the ratio of the volume of the preservation solution to the volume of the tissue.
- Disposable scalpels and clean, autoclaved forceps: one set for dissecting the skin and a different set for collecting the lymph nodes (one for each lymph node, per animal), thus preventing a cross-contamination of the skin and environmental flora with the lymph node tissues.
- 3 mm punch biopsy tools, necropsy knives, a sharps container for the used scalpels and biopsy tools, and cutting boards or disposable trays for use in the lymph node dissection.
- Use personal protective equipment, including PPE required for work at the appropriate BSL level for the system.
- Autoclave reusable metal tools, including the forceps, in metal pans prior to the necropsy, using the utensils/dry goods setting.
2. Identification and Sampling of Lymph Nodes in Cattle and Bison
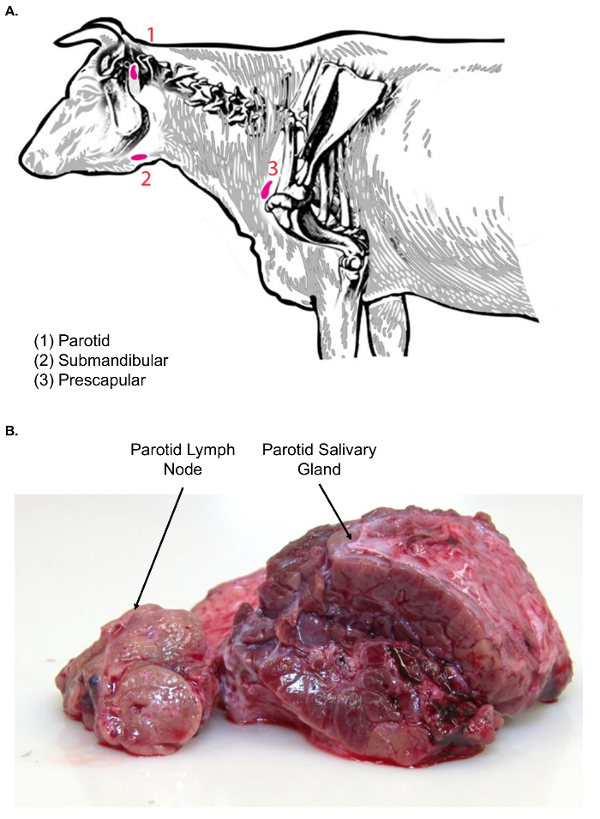
Figure 3. Lymph nodes of the bovine head and neck. (A) This cartoon image shows selected lymph nodes of the head and neck of Bos taurus. (B) This image shows the parotid lymph node in cross-section (left) as compared to the parotid salivary gland in cross-section (right). Note the difference in textures between the two tissue types. Please click here to view a larger version of this figure.
- Isolation of prescapular lymph nodes in cattle and bison
- Euthanize the animal (cattle or bison of any age, as appropriate for the experiment) based on the local IACUC protocol. Confirm the death by methods specifically described in the institutional IACUC protocol, including the lack of a heartbeat, the lack of respiration, and the absence of a corneal reflex. Move the animal to the necropsy floor.
Note: There are various methods of humane euthanasia used in the laboratory practice of large animals. However, intravenous administration of pentobarbital sodium and phenytoin sodium solution is the most commonly approved and utilized form. Consult the AVMA Guidelines for the euthanasia of animals.19
CAUTION: Follow all relevant biosafety protocols for a necropsy at the host institution, including the guidelines for the safe disposal of sharps in a puncture-proof container, and the decontamination of reusable metal knives and forceps. This protocol generates sharps waste from the use of disposable scalpels. Wear personal protective equipment as dictated by the institutional guidelines, including boots, coveralls, gloves, and eye protection. - Identify 3 participants to work on the animal tissues.
Note: Here, dissector 1 will be in charge of the large-scale sectioning of the desired animal tissues, dissector 2 will recover the lymph nodes from the different animal sections, and dissector 3 will work at a separate table to isolate sections from each lymph node and transfer them to tubes with a preservative solution. - Begin by placing the animal in lateral recumbency.
- Extend the shoulder by moving the front limb back at the level of the knee (carpus); this motion will bring the shoulder back and allow for an easier palpation of the lymph node. Feel for the presence of the lymph node "lump".
Note: During disease conditions, lymphadenopathy may be present, which makes the identification of lymph nodes easier, but this process can also affect the consistency, composition, and architecture of the lymph node. - Once located, and with the shoulder still extended, use a scalpel or necropsy knife to make a skin incision along the outline of the shoulder.
Note: Do not restrict the size of the incision, even if the lymph node has been located via palpation. A larger incision provides easier access to deeper tissues and makes the excision of the lymph node easier for the dissector. - Using a pair of forceps, retract the skin to reveal the point of the shoulder and neck musculature.
Note: The prescapular lymph node is often found in subcutaneous fat, especially in overconditioned animals. - Once again, palpate the location of the lymph node (Figure 3A).
Note: Unlike the subcutaneous fat, the lymph node will hold its shape when palpated. Additionally, lymph nodes are not contiguous with any surrounding tissue, as they are enclosed by a thin fibrous capsule. - After locating the prescapular lymph node, carefully dissect out the subcutaneous fat, using a scalpel and forceps, and isolate the lymph node.
- Look for the presence of a thin fibrous capsule, as depicted in Figure 1B, to distinguish between the lymph node and adipose tissue; lymph nodes are not contiguous with any surrounding tissue.
- Transfer the lymph node to the lymph node dissection table, using a plastic tray for the tissue transfer, for further sectioning.
Note: The appearance of these lymph nodes in bison is similar. The associated video depicts the appearance of the prescapular lymph node as dissected out of the right shoulder of an American bison.
- Euthanize the animal (cattle or bison of any age, as appropriate for the experiment) based on the local IACUC protocol. Confirm the death by methods specifically described in the institutional IACUC protocol, including the lack of a heartbeat, the lack of respiration, and the absence of a corneal reflex. Move the animal to the necropsy floor.
- Isolation of parotid lymph nodes in cattle and bison
- Begin by placing the animal in lateral recumbency on the necropsy floor. Locate the temporomandibular joint (TMJ).
- If this is difficult to locate, move the mandible slowly and determine the location at which the mandible attaches to the skull; this is the TMJ.
- Using a scalpel or necropsy knife, carefully make an incision in the skin. Start the incision dorsal to the joint and extend it ventrally/vertically along the jawline.
- Dissect the skin away to reveal the subcutaneous tissue. The parotid salivary gland will be visible first, due to its large size, lobulated surface appearance, and red coloration.
- Using forceps and a scalpel, move the parotid salivary gland aside to find the parotid lymph node sitting just cranial and medial to the salivary gland (Figure 3A). Examine the parotid lymph node tissue for texture before further processing.
Note: Lymph nodes of the face (parotid and submandibular) are associated with salivary glands, which can confound the lymph node isolation. In comparison to lymph nodes, salivary glands are much larger and have a lobulated surface appearance. Moreover, on cut surface, salivary glands are homogenous in nature and do not exhibit the classic cortical and medullary architectural pattern of lymph nodes (Figure 3B). - Excise the gland with a scalpel. Transfer it to the lymph node dissection table, using a plastic tray for the tissue transfer, for further sectioning.
- Begin by placing the animal in lateral recumbency on the necropsy floor. Locate the temporomandibular joint (TMJ).
- Isolation of supramammary lymph nodes in female cattle and bison
- Place the animal in lateral recumbency.
- Elevate the hind leg to expose the inguinal region and locate the udder.
Note: In a healthy, non-lactating animal, palpation of these lymph nodes may not be possible. In a lactating animal, these lymph nodes may be palpable at the caudal border of the base of the udder. - Using a scalpel, make an incision just dorsal to the udder, running along the upper leg, to expose the supramammary glands.
- Dissect the subcutaneous tissue and find the supramammary lymph node embedded in a thin layer of subcutaneous fat. If not visually distinguishable, palpate the exposed tissue for the presence of a firm structure within the subcutaneous fat.
- Using forceps and a scalpel, carefully dissect out the lymph node from the fat. Transfer it to the lymph node dissection table, using a plastic tray for the tissue transfer, for further sectioning.
3. Sectioning and Storage of Lymph Nodes
- Pass the isolated lymph nodes from dissector 2 to dissector 3 at a dissection table adjacent to the main necropsy space. Place each lymph node on a fresh section of a cutting board. Pre-label the cutting boards in sections by lymph node, to facilitate the receipt of multiple lymph nodes in succession. For infectious samples, use disposable trays.
- Using forceps and a disposable scalpel or necropsy knife, remove a section of the lymph node. Using a sharp knife, make a sagittal cut to open up the lymph node.
- Examine the interior of the lymph node, especially in specimens from infected animals, looking for asymmetry, lesions, color differences, etc. Record the observations.
- (Option 1) Small tissue sections
- Use disposable scalpels to excise pieces of each lymph node that are less than 5 mm in thickness and transfer each piece to a tube with an RNA preservation solution with clean forceps.
- (Option 2) Lymph node biopsy or sectioning methodologies
Note: Biopsies or sectioning methodologies, when applied to parallel lymph node sections across different nodes and animals, provide consistency of sampled tissue for a bulk RNA analysis.- Wedge sectioning method
- After examining the sagittal section to capture cells across the profile of the lymph node, excise a pie-shaped wedge from the lymph node, from the center to the outer capsule. For a node 2 cm in width (standard size; see Table 1), a wedge cut to the center that is 4 mm in outer arc length and 5 mm in thickness will have a wet weight of ~ 100 mg.
- Using the scalpel, cut the wedge into small pieces, no thicker than 5 mm, and approximately 50 – 100 mg each in wet tissue weight.
Note: For consistency, process all wedge pieces from a single lymph node either in one batch or in separate tubes followed by RNA pooling to provide a representative RNA profile from the lymph node of interest. - Place the tissue pieces with forceps into tubes containing at least 10 volumes of RNA preservation solution, as compared to the volume of the tissue.
- Punch biopsy method
- Select a punch biopsy tool if the goal is to collect specific sections, such as specific follicles from the node. Select a tool consistent with the size of the sample desired for collection. Punch biopsy tools are available in a range of sizes, from 1 to 8 mm in diameter.
- Visually locate the regions of interest to sample in the lymph node.
- Use the punch biopsy tool to excise the sections of interest, pressing and turning the tool into the tissue to create the core. Transfer the tissue piece with forceps to a tube containing at least 10 volumes of RNA preservation solution. If thicker than 5 mm, use forceps and a scalpel to further divide the tissue into smaller pieces.
- Wedge sectioning method
- Once the samples have been transferred to preservation solution (e.g., RNALater), transport them back to the laboratory at room temperature.
- Store the samples at 4 °C overnight to allow for tissue penetration.
- The next day, pour off the excess RNA preservative solution and store the tissue samples at -80 °C. Remove as much preservative as possible before freezing the samples. Use a P1000 and/or P200 micropipette tip for an efficient RNA preservative solution removal.
CAUTION: Discard the decanted preservation solution appropriately, based on the infective properties of the pathogen utilized as well as the chemical and environmental hazards of the preservative solution as specified on the Safety Data Sheet.
Note: The guidelines provided here are provided for the RNA preservation solution described in the Table of Materials. Consult the manufacturer's guidelines if using alternative preservation solutions.
4. Processing from RNA Lymph Nodes
CAUTION: Wear a lab coat, gloves, and proper eye protection for the processing steps.
Note: The phenol-based reagent used here is described in the Table of Materials (and the protocol is based on the manufacturer's guidelines)20. The use of alternative phenol-based reagents may necessitate a modification of the procedure, based on the manufacturer's recommendations for the specific product purchased.
- Prior to accessing the samples, pre-aliquot the chilled (4 °C) phenol-based reagent to homogenization tubes (1.5 mL/tube), using a pipette.
CAUTION: Phenol is toxic, corrosive, and a health hazard. Chloroform is toxic and a health hazard. Complete processing steps 4.1 – 4.9 in a chemical fume hood, and wear gloves to prevent any contact with the chemicals. In the United States, chloroform is a hazardous waste; dispose of tubes containing hazardous waste according to local and institutional guidelines. Process tissue samples from animals infected with pathogens under the corresponding BSL-2, BSL-3, or BSL-4 guidelines. - Once a sample can be loosened from the microcentrifuge tubes with clean forceps, immediately transfer approximately 50 – 100 mg of the preserved tissue to the homogenization tube containing the phenol-based reagent.
- Limit the thawing prior to the addition to the phenol-based reagent. Use storage on dry ice when multiple samples are removed from the freezer for processing.
- Tightly cap the tube, place it on the homogenizer, and homogenize the sample with a rotor-stator tissue homogenizer in the phenol-based reagent. Complete the processing at room temperature. When complete, check for a cloudy appearance to ensure the sample was successfully homogenized; the tissue should be dispersed into a cloudy suspension.
Note: A 2 min RNA extraction setting is used (the pre-set RNA_02_1 setting for the homogenizer described in the Table of Materials; designed for frozen tissues). The rotor-stator specified in this protocol (see the Table of Materials) is large-toothed and adapted for the homogenization of a range of tissue types, including fibrous material. The RNA_02_01 setting is designed specifically for frozen (as opposed to fresh) tissues. As lymph nodes have fibrous connective tissue components, a similarly optimized rotor-stator homogenizer is recommended to achieve an effective dissociation. See the National Institute of Environmental Health Sciences21 for additional information about homogenization recommendations.- If large chunks of tissue remain after the homogenization, repeat the homogenization procedure. In order to efficiently recover the RNA, the tissue must be well dissociated.
- For more fibrous lymph node pieces, use forceps and scissors to pre-chop the lymph node piece into several smaller pieces prior to the transfer to the homogenization tube. As described in the introduction, lymph node tissues from older animals may be more fibrous.
Note: If a dual analysis of the host and pathogen RNA is desired, additional treatment (such as bead homogenization) may be necessary for the recovery of bacterial RNA, especially if Gram-positive pathogens are present in the tissues.
- Immediately remove the homogenized sample from the tubes and transfer it to RNase-free microcentrifuge tubes (2.0 mL in size) with a P1000 micropipette.
- Centrifuge the sample for 5 min at 12,000 x g at 4 °C to remove any sample debris. Using a P1000 micropipette tip, transfer the supernatant to a fresh microcentrifuge tube, leaving the pellet behind.
- Incubate the supernatant for 5 min at room temperature. Split each sample between two 1.5 microcentrifuge tubes.
- Add 0.2 mL of chloroform per 1 mL of the phenol-based reagent (i.e., 0.3 mL for a 1.5 mL sample); invert it by hand for 1 – 2 min to mix the phases, but do not vortex the sample. Incubate it for 2 min at room temperature.
- Centrifuge it for 15 min at 12,000 x g at 4 °C.
- Carefully remove the upper, clear aqueous phase with a micropipette tip (P1000 or P200) which will form the upper layer, without disrupting the interphase or the red phenol-chloroform layer. Transfer it to a new microcentrifuge tube.
- Mix it with an equal volume of 100% ethanol; mix it thoroughly by inverting the tube 4 – 5 times. The tube will often appear cloudy.
- With a micropipette tip, transfer the liquid to a silica-based commercial spin column, to further purify and elute the RNA, following the manufacturer's instructions22. Elute the RNA generated from ~ 50 mg of tissue into 50 – 100 µl of RNase-free water.
Note: While the phenol-based reagent extraction removes DNA in the organic phase, for the downstream usage of RNA in qRT-PCR and/or RNA-sequencing applications, an additional treatment of RNA samples with DNase is recommended. - Aliquot the eluted RNA to separate tubes for use in a quantification and quality assessment, to reduce any freeze-thawing of the main RNA sample. Transfer the tubes to -80 °C for storage.
- In the case of lymph node samples derived from animals infected with zoonotic pathogens, especially at the BSL-3 containment level, validate the resulting RNA sample for the absence of pathogens if the sample will be moved to a lower biosafety level containment area for quality analysis, RT-PCR, and/or RNA-sequencing library preparation.
Note: It is critical to consider local regulations, and in the case of the CDC (Centers for Disease Control and Prevention) inactivation guidelines for work with select agents, it is necessary to validate all inactivation procedures at the researcher's local site.- Remove 1/10th volume of each eluted RNA sample from step 4.12 (i.e., 5 µl from 50 µl of the total RNA). Using a sterile technique, mix the sample with 100 µl of bacterial growth broth in a sterile 1.5 mL microcentrifuge tube. Using a sterile plastic spreader, spread the RNA-broth mixture across the surface of an agar plate.
Note: Select a broth type and agar plate consistent with the growth of the pathogen with which the animals were challenged. - Incubate the agar plate under conditions specific to the growth of the pathogen with which the animals were challenged. Check for the absence of recovered bacteria before removing the samples from the BSL-3 containment.
Note: The resulting eluted RNA, after quantification and tests of integrity, is suitable for downstream applications, including an RNA-sequencing and RT-PCR analysis.
- Remove 1/10th volume of each eluted RNA sample from step 4.12 (i.e., 5 µl from 50 µl of the total RNA). Using a sterile technique, mix the sample with 100 µl of bacterial growth broth in a sterile 1.5 mL microcentrifuge tube. Using a sterile plastic spreader, spread the RNA-broth mixture across the surface of an agar plate.
- Assess the RNA recovery and purity using a spectrophotometer. To assess the RNA quality, load 1 – 2 µl of the eluted RNA sample to the spectrophotometer for an analysis. Record a spectrum of the sample in the ultraviolet (UV) range.
- From the UV spectrum, record the A260/A280 ratio, the A260/A230 ratio, and the A260 value for the sample (A = Absorbance).
- As single-stranded RNA at a concentration of 40 µg/mL has an absorbance of 1.0 at 260 nm, calculate the RNA concentration (µg/mL) by multiplying the A260 x 40 for the purified samples.
- As a quick method of assessment of RNA integrity, to exclude any degraded samples from further analysis, mix 2 µl of each RNA sample with 4 µl of 1.5x formamide loading dye [95% deionized formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA (pH 8.0), 0.025% (w/v) SDS] in 1.5 mL microcentrifuge tubes23,24.
- Heat the tubes at 70 °C for 1 min, and then transfer the tubes to ice for 1 min.
- Load the samples to a 1% agarose gel prepared in 1x TBE (for a full description of a native agarose gel electrophoresis for RNA, see Rio, Ares Jr., Hannon, and Nilsen)24. Separate the samples by standard gel electrophoresis methods and confirm the presence of 28S and 18S ribosomal RNA bands.
- Follow up the gel electrophoresis with quantitative analysis methods for the RNA integrity determination [e.g., a Bioanalyzer and RIN (RNA Integrity Number) analysis] as described in the Representative Results and in Mueller, O. et al.25 and Schroeder, A. et al.26.
5. Alternative Extraction Method from Formalin-fixed, Paraffin-embedded (FFPE) Tissues
Note: Although FFPE tissue preservation does not represent the most robust method of nucleic acid preservation, the protocol presented below can be a way to study some transcriptional changes when other preserved tissues are unavailable.
- Prepare formalin reagents for the tissue preservation by dispensing formalin into plastic containers. Tissues should be preserved in formalin with a ratio of 20:1 formalin:tissue volume/weight, respectively.
CAUTION: Formalin is a known human carcinogen and although not acutely toxic, chronic exposure (typically through inhaled fumes) can represent a significant health hazard. Proper ventilation and PPE (i.e., the use of nitrile gloves, to be changed within 15 min after the initial exposure) are required to minimize the health risks. Refer to the OSHA Formaldehyde Standard (29 CFR 1910.1048) for additional information on exposure monitoring and risk assessment.
Note: Using too little formalin can impact the ability of the tissue to become fully preserved when submerged in preservative. - After the isolation of the tissue of interest, carefully trim the tissue to less than 5 mm in thickness.
- Using forceps, carefully submerge the trimmed tissue into formalin to minimize any splashing.
Note: The tissues should be completely submerged in formalin to allow for a complete fixation. It is critical that all surfaces of the tissue are completely covered with formalin.
- Using forceps, carefully submerge the trimmed tissue into formalin to minimize any splashing.
- Once the tissues have been submerged in formalin, allow for the tissue fixation to occur for at least 24 h at room temperature.
Note: Formalin penetrates tissues slowly, at a rate of 1 mm per hour from all surfaces27,28. Therefore, keeping the thickness of the tissue sections to less than 5 mm results in a rapid fixation of the entire tissue. - After the fixation, transfer the tissues to an appropriate reagent for the paraffin embedding.
Note: For example, the tissues examined in this manuscript were transferred to 70% ethanol prior to the embedding. - Embed the tissues into paraffin.29
- Section the tissue to 10 – 80 µm in thickness (use 10-20 µm if miRNA is desired to be extracted)29.
- Using scalpel and forceps, remove a 40 mg tissue piece from each sample to be processed and place it into a sterile, nuclease-free microfuge tube.
- Utilize a commercial kit designed for deparaffinization and nucleic acid recovery of FFPE tissue samples to extract RNA from the preserved samples.
Note: The kit referenced in the Table of Materials utilizes xylene and ethanol washes to remove paraffin, a protease treatment step, and a glass-fiber filter to purify RNA.
Representative Results
The use of the considerations presented in this article (steps 1 – 4 of the protocol) will aid in the recovery of RNA from large animal samples that is suitable for a downstream analysis in host gene expression studies. The RNA quality for downstream applications is assessed by multiple standard measures. For spectrophotometry, the A260/A280 ratio provides a measure of the protein contamination, and the A260/A230 ratio provides another means of purity assessment that will detect chemical contaminants such as phenol. In each case, ratios of ~ 2.0 are evidence of high-quality RNA, and decreased ratios are evidence of contamination. Importantly, these ratios do not provide any information about RNA degradation, which can be assessed qualitatively (steps 4.15 – 4.17) and quantitatively (step 4.18, such as through an RNA integrity number or RIN score).
It is important to note that the expected quality level of RNA recovered from lymph node tissue sections is, on average, lower than for RNA recovered from bovine white blood cell samples. In a comprehensive series of RNA extractions from bovine tissues and cell lines, Fleige and Pfaffl30 find that the average RIN (RNA integrity numbers as measured on a Bioanalyzer) vary between < 5 for jejunum and rumen tissue and > 9 for white blood cells and corpus luteum, with lymph node RIN scores falling in the middle at an average of 6.9. In general, Fleige and Pfaffl30 note that solid tissues produce RIN scores ranging from 6 – 8, and that tissues with higher concentrations of connective tissue exhibit a higher mean degradation. The density of connective tissue in the lymph node, plus the intrinsic level of RNases in this tissue type, may be responsible for the inability to consistently recover RNA with RIN scores > 9 from lymph nodes.
However, an example of a Bioanalyzer profile for the RNA recovered from a bison supramammary lymph node using the methodology presented here is displayed in Figure 4A, demonstrating the recovery of RNA with a RIN score of 8.6 (primarily intact and suitable for essentially all downstream applications). RNA generated from this procedure has been used to successfully generate libraries for use in RNA-sequencing; this protocol allows for the regular isolation of RNA samples from bison and bovine lymph nodes with RIN values > 8 (and often > 9). For a sample set of eight extracted RNA samples, the average absorbance ratio at 260 nm/280 nm was 2.1 and at 260 nm/230 nm was 2.1, indicating a low protein contamination and a low contamination with chemicals like phenol, respectively. The average nucleic acid recovery from each extraction was 97 ± 10 µg per ~ 100 mg or a yield of ~ 1 µg RNA/mg of lymph node tissue.
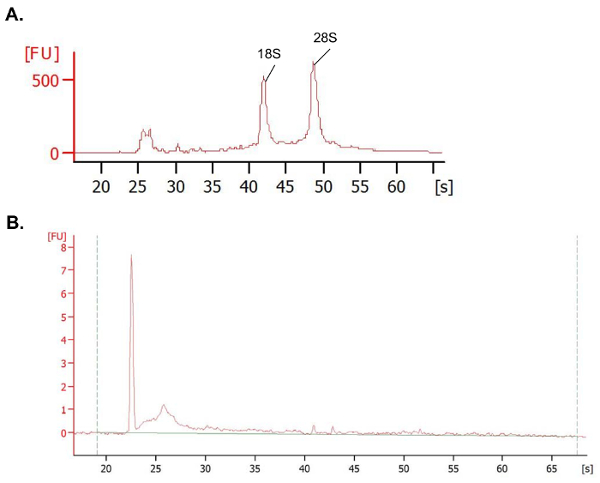
Figure 4. Representative quality traces depicting RNA quality from extraction methodologies. (A) This panel shows a total RNA trace of high-quality RNA generated from a bison lymph node, using the procedure presented in this manuscript. (B) This panel shows a trace of a bovine FFPE-sample selected from Table 4, depicting very degraded RNA. Please click here to view a larger version of this figure.
Due to the challenges associated with the rapid isolation of tissues from large carcasses, especially in the case of sampling from wildlife species, the impact of the time delay between the euthanasia and the placement of the tissue pieces in an RNA preservation solution is of potential concern in the analysis. The information in the literature about the stability of RNA in lymph node tissues, in particular, is limited. Therefore, to provide information about acceptable parameters for tissue collection, the stability of RNA in bison lymph node post-euthanasia was characterized.
For this experiment, time 0 was considered as the time the animal was pronounced dead by the lack of a heartbeat and an absence of a corneal reflex. Following the transport of the carcass and the beginning of necropsy, ~ 15 min had elapsed before the retrieval of the first lymph node sample (15 – 20 min was standard in our observations for the recovery of field animals; time courses will vary depending on the location). The supramammary lymph node was identified, and a small section of the tissue was removed and maintained at room temperature. Sections were subsequently taken at approximately 15 min intervals and transferred to the RNA preservative; in the middle of the time course, a second sample of lymph node was retrieved from the animal for the second set of 4 time points in the series (Figure 5A; closed circles, Figure 5C). RNA was extracted in parallel from the lymph node pieces using the method described above. This 1 h experiment reveals that the lymph node RNA retained the majority of its integrity across the time course, with only slight reductions of the RIN score by the end of the time course (Figure 5B and 5C). Similarly, RNA from bovine prescapular and parotid lymph nodes retained its integrity as measured by the RIN score over ~ 1 h time courses post-mortem (Figure 5C; the animal information is provided in Table 2). Additionally, RNA was extracted from supramammary lymph node sections that were collected 8 h after death, holding whole sections in cell culture media at either room temperature or at 37 °C to simulate the holding time in an animal carcass. Ribosomal RNA was observable even after 8 h holding times (Figure 5D).
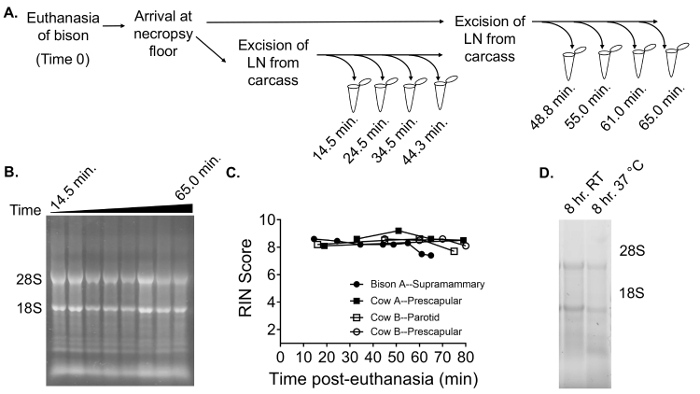
Figure 5. RNA quality in bison lymph node samples collected across time courses post-death. (A) This panel shows an experimental overview of a 1 h time course procedure. (B, C) These panels show an examination of the RNA quality for samples taken across a 1 h time course from a supramammary lymph node isolated from an American bison. Gel reflects RNA samples denatured in formamide and separated on a 1% non-denaturing gel. Panel (C) depicts the changes in RIN scores for each sample. The data from additional experiments on bovine prescapular and parotid lymph nodes is overlaid as indicated. (D) This panel shows an example of RNA gel, as described for the gel used in panel (B), for supramammary lymph node samples processed after 8 h of incubation at either room temperature (Lane 1) or at 37 °C (Lane 2) in cell culture media. Please click here to view a larger version of this figure.
These representative findings are consistent with RNA stability results for other tissue types and sources. Table 3 provides a summary of the results of a sample of published papers that have investigated the RNA stability over pre-preservation time courses. Note that only trends from RIN scores and/or rRNA gel observation (total RNA) are included in the table, although some papers provide an additional analysis of mRNA integrity, such as through the determination of ratios of 5' vs. 3' segments of selected RNAs (e.g., Fajardy et al.)31. However, it is important to remember that longer times between the death and sample preservation can result in changes in the RNA expression profiles, as demonstrated, for example, for the placental tissue31. Compared to stable transcripts, more unstable RNAs can be depleted. Therefore, it is important to utilize a rapid, streamlined workflow for the tissue recovery in order to reduce any delay once the animal is available for necropsy, in order to achieve the most biologically valid RNA profiles. It would be a mistake to assume that the presence of intact ribosomal RNA indicates the absence of any changes in the transcriptome profile. Still, the representative results suggest that even with the increased processing times necessary for large animal sampling, it is possible to prepare RNA that is suitable for a gene expression analysis from lymph node samples.
Additionally, the impact of the post-thawing time variable was examined; during this time, RNA is susceptible to degradation from the nucleases present in the tissue. The quality of the RNA samples processed using this protocol with either 0 or 64 min of hold time at room temperature post-thawing at step 4.1 was measured by an RIN score. This experiment demonstrates that the protocol is quite insensitive to the thaw time, making it robust for the processing of multiple samples (Figure 6A).
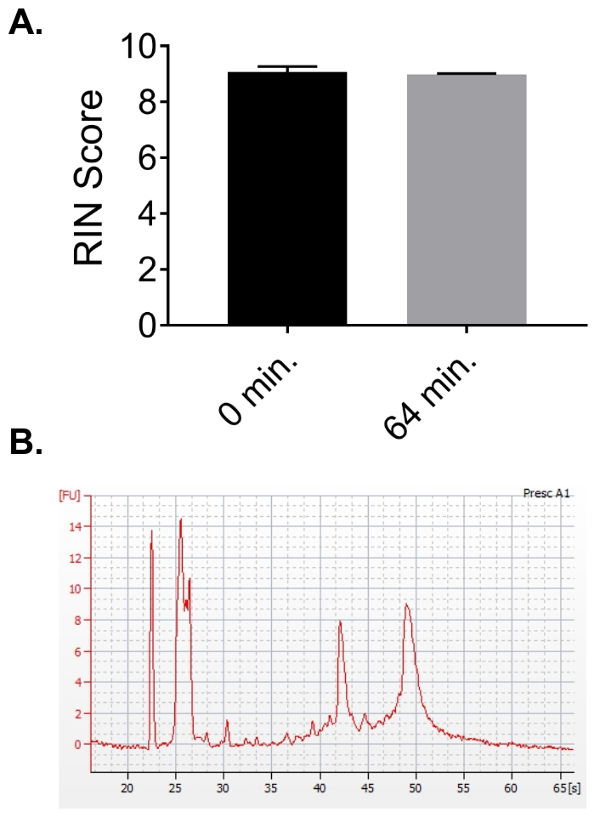
Figure 6. Additional analysis of RNA quality parameters. (A) This panel shows the RNA quality results for RNA purified either immediately after thawing (0 min) or after holding for 64 min at room temperature prior to the processing. The bars represent the average of 3 tissue pieces for each condition, ± the standard deviation. All tissues were stored in an RNA preservative solution prior to thawing, as described in the protocol. (B) This panel shows a Bioanalyzer trace of a sample from a poorly homogenized lymph node piece containing large sections of connective tissue. The small size of the 28S and 18S peaks, as compared to the peaks in the 5S range, is evident. This can also occur due to the degradation of the RNA, although the relatively flat baseline between the 5S range and the 18S peak is alternatively suggestive of poor extraction; see Avraham, R. et al.48 for additional details. Please click here to view a larger version of this figure.
As a point of comparison to the isolation of RNA from lymph nodes, a trial recovery of RNA from FFPE mesenteric lymph node pieces from cattle was conducted. As described above, the samples were fixed for 1 week in formalin prior to the paraffin embedding, followed by 4 months of storage in paraffin at room temperature. The tissues were sectioned into five 10 µm sections, and using the FFPE extraction procedure, approximately 62 ng/µL of RNA per sample (± 14 ng/µL) was recovered. The absorbance ratios at 260 nm/280 nm averaged 1.9, indicating a product with low levels of protein contamination, although the A260/A230 ratios were unfavorable (Table 4). Note that the RIN scores observed for these samples were very low (< 2), indicating a degradation of the RNA products (Table 4; Figure 4B), consistent with the description of Srinivasan et al.45. In assays utilizing qPCR, typically small sections of RNA, or more specifically cDNA (< 200 bp), are amplified. Therefore, the amplification of small products can still occur from degraded samples, and qPCR analysis can be performed. For example, it was possible to amplify a segment of the ribosomal protein S9 (RPS9) transcript from these FFPE-recovered samples by qPCR. The CT values were consistent across the samples and averaged 19.2 ± 1.8. Keep in mind the limitations that are present when utilizing the FFPE-prepared samples, however, as larger products are not necessarily present, and the degradation may not have occurred at a uniform rate across all samples. Therefore, this material can be used for studies with caveats, but significant limitations are present for any downstream applications like RNA-sequencing.
| Lymph Node Type | Location | Length | Ancho | Regions Drained by Nodes |
| Prescapular | Just medial to the shoulder joint; embedded in subcutaneous fat and under a layer of muscle | 1-10 cm | 1.5-2 cm | Skin of neck; shoulder; and skin, muscles, and joints of the lower thoracic limb (ref. 8) |
| Prefemoral (also called precrural) | Just dorsal and medial to the stifle, approximately 12-15 cm above the patella | 8-10 cm | 2.5 cm | Skin of the pelvic limb, abdomen, and caudal portions of the thorax (ref. 8) |
| Retropharyngeal | Found in the midline dorsal to the pharynx | 4-5 cm | 2-3.5 cm | Tongue, mucous membranes of the oral cavity, gums, lips, hard palate, salivary glands, most of the muscles of the neck |
| Parotid | Located subcutaneously ventral to the temporomandibular joint, caudal to masseter muscle (ref. 8) | 6-9 cm | 2-3 cm | Skin, subcutis, most of the muscles of the head, muscles of eye and ear, eyelids, lacrimal gland, rostral portion of nasal cavity |
| Submandibular (may be 1-3 present) | Located subcutaneously on the medial aspect of the angle of the mandible, associated with salivary glands | 3-4 cm | 2-3 cm | Muzzle, lips, cheeks, hard palate, rostral part of nasal cavity, tip of tongue, skin and muscle of the head (ref. 8) |
| Supramammary (female superficial inguinal, typically 2 present) | Found caudally and dorsally relative to the mammary glands | 6-10 cm | Udder, vulva, vestibule of the vagina, skin on the medial and caudal aspects of the thigh, medial surface of the leg | |
| Scrotal (male version of superficial inguinal) | Found below the prepublic tendon within a layer of fat around the neck of the scrotum | 3-6 cm | Scrotum, prepuce, and penis |
Table 1. Overview of bovine superficial lymph nodes.
| Animal Description | Location in Manuscript | Age | Sex | Health Status |
| Videoed Bison bison | Bison in video–Lymph node recovery | 5-6 yo | Female | Healthy |
| Videoed Bos taurus | Bovine in video–Lymph node recovery | 7-8 yo | Castrated male | Healthy |
| Bison bison for RNA stability study | Fig. 5A-C | 5-6 yo | Female | Healthy |
| Bos taurus A for RNA stability study | Fig. 5C, Fig. 6A | 7-8 yo | Castrated male | Healthy |
| Bos taurus B for RNA stability study | Fig. 5C | 7-8 yo | Castrated male | Healthy |
| Bos taurus for FFPE quality study | Table 4, Fig. 4B | 0.5 yo | Castrated male | Healthy (control from O157:H7 infection study) |
| Note that the methodology has been used on multiple additional cattle, bison, and goat lymph node samples, beyond those that are highlighted here. | ||||
| Additionally, we used the same methodology on lymph node collected from a 1.5 mo.-old female calf. | ||||
Table 2. Characteristics of animals used in pilot studies.
| Tissue Type | Preservation Method | Holding Conditions | Degradation Conclusion | Reference |
| Bovine adipose, skeletal muscle, liver | Snap freezing (liquid N2) | 4 °C, vacuum packaged tissue | Muscle RNA more stable (even out to 8 days of storage); adipose and liver mainly stable to 24 h. as assessed by appearance of rRNA bands | Bahar et al. (2007)32 |
| Human colon, colorectal cancer | RNA preservation solution | Room temp. | RIN preserved for 2 h., but subset of genes show changes in expression from 0.5-2 h. | Yamagishi et al. (2014)33 |
| Mouse liver, spleen | RNA preservation solution, or snap freezing (dry ice slurry) | Inside the mouse carcass (37 °C) | Spleen samples were stable out to 1.5 h; liver samples exhibited earlier degradation (24% decrease in RIN at 105 min.) | Choi et al. (2016)34 |
| Mouse liver | None (Homogenized fresh in guanidinium thiocyanate-phenol-chloroform) | 25 °C, 37 °C | Limited degradation out to 4 h. at 25 °C, but extensive degradation at 37 °C for 4 h. (as observed using RIN scores). | Almeida et al. (2004)35 |
| Human ileum | RNA preservation solution, or snap freezing | 4 °C, 37 °C | Generally stable at 1.5 h. at 37 °C; Stable out to 6 h. at 4 °C (as observed using RIN scores). Note that the authors also provide a summary of selected RNA stability resources in Table S1 that can serve as an additional reference for those interested in further review of the tissue RNA integrity literature. | Lee et al. (2015)36 |
| Human liver | Snap freezing (liquid N2 or isopentane) | Room temp. | Degradation observed at 1 h. (up to 0.85 ΔRIN); more degradation in smaller samples of liver than in larger samples. | Kap et al. (2015)37 |
| Human colorectal cancer | Snap freezing (liquid N2/isopentane) | 4 °C | Degradation observed by 1 h.; RIN score of only 4.2 after 90 min. of delay in freezing time | Hong et al. (2010)38 |
| Human liver | RNA preservation solution, Also flash freezing (liquid N2) | Room temp, 4 °C | Samples were stable even out to 1 d. on ice; degradation observed after 1 d. at room temp. (but not after 3 h.; as observed using RIN scores) | Lee et al. (2013)39 |
| Horse adipose, skeletal muscle | Snap freezing (liquid N2) | 13 °C | Authors recommend that best integrity for muscle was within 2 h.; adipose preservation within 0.5 h. recommended by authors (as observed by rRNA gel appearance) | Morrison et al. (2014)40 |
| Salmon brain, kidney, liver, muscle | RNA preservation solution | Room temp. | Brain RNA stable to 4-8 h., kidney and muscle exhbiit some degradation by 8 h. (gel, RIN score both used) | Seear & Sweeney (2008)41 |
| Rabbit connective tissue (ligament, tendon, cartilage) | Snap freezing (liquid N2) | 4 °C, Room temp. | No "overt" degradation out to 96 h. postmortem, as observed by rRNA gel appearance | Marchuk et al. (1998)42 |
| Rat brain, lung, heart, liver | Appears to be extracted fresh | Inside the rat carcass held in 20 °C incubator | Highest RNA stability observed in brain (could observe rRNA bands out to 7 days postmortem, although degradation was present), moderate stability in lung and heart, low stability in liver; as observed by rRNA gel appearance | Inoue et al. (2002)43 |
| Human tonsil, colon | With and without RNA preservation solution, then snap-frozen | Room temp. (22 °C), 4 °C RNA preservation solution, cold saline, or ice (0 °C) | Stable to 16 h. on ice or in RNA preservation solution; slight degradation at 6 or 16 h. in cold saline or at RT (measured by RIN scores) | Micke et al. (2006)44 |
Table 3. Sample of previous findings on RNA stability in post-mortem tissue samples. The entries represent selected manuscripts from the literature on RNA stability, with a range of tissue types represented, including murine, human biopsy, and veterinary examples. The methods for preservation, the mode of storage pre-extraction, and a brief summary of the results are provided for each example. Unless indicated otherwise, the samples were stored at conditions ranging from ice to 37 °C, followed by snap freezing or an infusion with RNA preservation solution, post-time course (in other words, stability was not being measured in the presence of an RNA preservation solution during the incubation, unless indicated).
| Sample ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A260/A280 | 1.96 | 1.98 | 1.98 | 1.91 | 1.98 | 1.89 | 1.89 | 1.91 | 1.9 | 1.84 | 1.84 | 1.97 |
| A260/A230 | 0.67 | 0.14 | 0.19 | 0.26 | 0.61 | 0.69 | 1.33 | 0.11 | 0.21 | 0.39 | 0.1 | 0.44 |
| RIN | 1.6 | 1.1 | 1.3 | 1.2 | 1 | 1.3 | 2 | 1.1 | 1.2 | 1 | 1 | 1 |
Table 4. Representative results from RNA isolated from bovine FFPE mesenteric lymph nodes. The table depicts the results from spectrophotometric and quality analyses from a series of FFPE bovine lymph node samples processed using the method described in this manuscript. In each case, the results reflect highly degraded RNA.
Discussion
The majority of transcriptomic studies and the associated protocols focus on mouse, rat, or post-mortem human samples. However, investigations in livestock and wildlife provide a wide range of opportunities for the characterization of the immune response to disease, both as applicable to veterinary medicine and, in regard to zoonotic diseases, to human public health. This protocol provided an outline of key considerations for high-integrity RNA extraction from tissues from large animals, such as cattle, bison, goats, and sheep. The size of the animals, as well as the conditions for the sample collection, make the recovery a more extensive process, with a longer post-mortem processing time than for mouse lymph node recovery. Similarly, the large size of these lymph nodes necessitates protocols that recover parallel samples from different animals as well as samples that are representative of the immunological/pathological changes within the lymph node. The suggestions for sample collection in this protocol provide for the recovery of intact RNA as well as for representative profiles based on the sectioning of the lymph node.
As a demonstration of the importance of a standardized sampling strategy and protocol, as presented here, we surveyed 25 papers from the PubMed Central database that characterized the transcriptomes of livestock lymph nodes. One paper indicated the processing of a large lymph node section at one time, one discussed specific laser capture microdissection methods, one mentioned that a "representative sample" was collected from the lymph node, and only one paper46 indicated that the piece of lymph node collected (10 mm3 in size) included both cortex and medulla regions. While other papers noted that small samples (generally 30 – 100 mg) were processed for RNA analysis, 84% of the papers did not mention a sampling strategy for the collection of these tissue pieces. As RNA-sequencing is still relatively expensive, the analysis of multiple sections per lymph node is unlikely to be financially viable in most cases, especially since biological replicate samples are typically prioritized for statistical analysis. By collecting a sample across a wedge of a sagittally sectioned lymph node, RNA from lymph node regions rich in different types of immune cells is captured in all samples. This protocol can, therefore, serve as a reference for a documented and reproducible sampling strategy for lymph node transcriptomics.
In the preparation of RNA from tissues, there are multiple key steps for procedural decision-making. First, the methodological literature is mixed in terms of the use of preservation solutions vs. the use of flash-frozen tissue samples (and subsequent cold-temperature grinding). Especially in the case of large animal necropsies, there are multiple potential procedural benefits to the use of a preservation solution, as incorporated in this protocol. First, tissue samples can be stored during the necropsy in preservation solution without the need to immediately transfer the tubes to liquid nitrogen. Second, the use of a preservation solution provides additional protection prior to sample processing for RNA extraction, particularly if the sample partially or briefly thaws before homogenization. The results in Figure 6A indicate that this protective effect extends to lymph node tissue. In contrast, flash-frozen samples must be maintained in the frozen state until the moment of homogenization in a phenol-containing buffer. Finally, for field necropsies of large animals, where liquid nitrogen is not readily available, samples can be stored in preservation solution until they can be returned to the lab.
Additional advantages of the molecular components of the procedure presented here include the presence of phenol during the initial tissue processing, which serves as a disinfectant during the aerosol-generating process of homogenization for samples from infected animals, and the use of the spin column procedure to remove residual phenol and guanidine isothiocyanate from the sample. The representative results demonstrate that the procedure generates high-quality RNA as assessed by A260/A280 and A260/A230 ratios, gel electrophoresis, and RIN scores, and is robust in the face of time course challenges of livestock and wildlife sampling. The pairing of tissue preservation (e.g., in RNALater), homogenization in phenol-based reagents (e.g., TRIzol), and silica column purification has been successfully utilized in the literature to profile gene expression patterns in ovine abomasal lymph nodes infected with gastrointestinal nematodes46 and in cattle lymph nodes infected with a herpesvirus47, with RIN scores greater than 8 and greater than 7 across all samples, respectively.
In the case of a resulting RNA with unacceptable RIN scores for downstream analysis, the following variables, which are standard for RNA processing, should be assessed: the total processing time post-mortem, the condition of the tissues, the processing time/delay post-thaw, the technique during the RNA extraction, and the RNA handling post-extraction. The total processing time post-mortem should be assessed especially in the case of tissues recovered from animals found dead, as opposed to euthanized animals. As described above, RNA is relatively stable in lymph nodes, but long delays in processing could lead to degradation, especially for unstable transcripts. A confounding time variable could be present if samples are compared between carcasses that have remained in the field for disparate amounts of time. The condition of the tissues should be assessed, because the degradation of tissue integrity, due to pathologic changes associated with infection, may reduce the RNA stability. For example, lower RNA quality has been observed in the placentome of Brucella-infected animals post-abortion (Boggiatto et al., submitted). The processing time/delay post-thaw is mitigated by the use of preservation solutions, as described in this protocol. Be certain that the thickness of the tissue pieces is low enough to allow a proper and rapid penetration of the preservative solution into the tissue (< 5 mm thickness using the preservative mentioned in the Table of Materials). During the RNA extraction, the buffers should be RNase-free and contact with gloves and other contaminated items (RNases are present on the skin) must be avoided. For the grinding of the lymph node tissue, the largest source of potential RNases will be in the tissue itself, but it is beneficial to reduce the introduction of contaminants at all stages of the procedure. To handle the RNA post-extraction, as described in the protocol, aliquot the RNA samples into tubes before the freezing and storage at -80° C, to eliminate the need to freeze-thaw the samples for a quality and quantity analysis.
Low yields in the procedure, in addition to resulting from the degradation of RNA samples, can be due to an inefficient homogenization of lymph node tissue at step 4.3 in the protocol. Note that the incubation in an RNA preservation solution can change the texture of the tissue (increase firmness), although most lymph node tissues dissociated effectively in pilot runs with the large toothed rotor-stator dissociator described here. As noted in step 4.3.1, the homogenization in phenol-based reagent should be repeated if the dissociation is incomplete. Mortar-and-pestle grinding, followed by a resuspension of the ground sample in a phenol-based reagent, is another alternative for laboratories without access to a homogenizer with these specifications. However, a homogenization in disposable tubes presents multiple potential advantages over the grinding of frozen tissue in a mortar and pestle, including a more feasible and shorter workflow for processing multiple samples, an absence of sample loss when grinding, no cleanup, and no potential contact with powered samples, in the case of tissues from infected animals.
Avraham et al.48 describe that a large 5S RNA peak in the final RNA product, paired with a low recovery of 28S and 18S ribosomal RNAs, can be a sign of an incomplete lysis of the cells (which in turn, for lymph node processing, can be due to an incomplete dissociation of the tissue). Figure 6B provides an example of an RNA profile generated from a piece of bovine parotid lymph node that was very high in connective tissue and did not homogenize effectively. RNA yields should be calculated and monitored across samples in a set for analysis to confirm that similar recoveries are obtained per mg of tissue, and samples for direct comparison by RNA-sequencing should be processed in batch, using the same homogenization strategy for all samples. Note that the wedge sampling approach also aids in avoiding a collection of a sample that is exclusively a region of extensive connective tissue.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank James Fosse for his excellent work on all videography and video processing; Michael Marti for his excellent work in the generation of digitized cattle images; Lilia Walther for her help with RNA extraction and Bioanalyzer runs; Mitch Palmer and Carly Kanipe for their helpful review and feedback on lymph node images; and the animal care and veterinary staff at the National Animal Disease Center for all of their hard work and assistance with animal husbandry and the preparation for necropsies.
Materials
| RNA preservation solution (we used RNALater for all experiments) | ThermoFisher | AM7020 | |
| 1.5 ml or 2 ml polypropylene microcentrifuge tubes | Fisher Scientific | 05-408-129 | |
| Disposable scalpels | Daigger Scientific | EF7281 | |
| Tissue forceps, rat tooth | Fisher Scientific | 12-460-117 | Other tissue forceps available including curved tip, tapered edge, etc. , depends on user preference |
| 3 mm punch biopsy needles | Fisher Scientific | NC9949469 | |
| Sharps container (small and transportable for necropsy) | Stericycle | 8900SA | 1 qt. size shown here |
| Cutting boards or disposable trays | Fisher Scientific | 09-002-24A | Available in a variety of sizes, depends on user preference |
| Personal protective equipment | Varies with pathogen (gloves, respirator masks, goggles, etc.) | ||
| Phenol-based RNA extraction reagent (we used TRIzol Reagent for all experiments) | ThermoFisher | 15596026 | |
| Silica column-based RNA extraction kit (we used the PureLink RNA Mini kit for all experiments) | ThermoFisher | 12183018A | Designed for up to 100 mg tissue |
| 100% Ethanol (200 proof for molecular biology) | Sigma-Aldrich | E7023 | |
| Tissue homogenizer with enclosed homogenization tubes (we used the gentleMACS dissociator for all experiments) | Miltenyi Biotec | 130-093-235 | |
| Agarose (General, for gel electrophoresis) | Sigma-Aldrich | A9539 | |
| 1X TBE | Fisher Scientific | BP24301 | Can also make from scratch in the laboratory |
| Deionized formamide | EMD Millipore | S4117 | |
| Sodium dodecyl sulfate | Sigma-Aldrich | L3771 | |
| Bromophenol blue | Sigma-Aldrich | 114391 | |
| Xylene cyanol | Sigma-Aldrich | X4126 | |
| EDTA (Ethylenediaminetetraacetic acid) | Sigma-Aldrich | EDS | |
| UV-Vis Spectrophotometer (we used the NanoDrop Spectrophotometer) | ThermoFisher | ND-2000 | |
| Device for quantitative RNA assessment (we used the Bioanalyzer, with associated components and protocols) | Agilent | G2939BA | |
| FFPE RNA extraction kit (we used the RecoverAll Total Nucleic Acid Isolation Kit for Formalin Fixed, Paraffin Embedded Tissue) | ThermoFisher | AM1975 | |
| Plastic spreader (L-shaped spreader) | Fisher Scientific | 14-665-231 | Only needed for sterility testing for samples from infected animals |
| Necropsy knives | Livestock Concepts | WI-0009209 |
Referencias
- Tizioto, P. C., et al. Immunological response to single pathogen challenge with agents of the bovine respiratory disease complex: an RNA-sequence analysis of the bronchial lymph node transcriptome. PLoS One. 10 (6), e0131459 (2015).
- Miller, L. C., et al. Analysis of the swine tracheobronchial lymph node transcriptomic response to infection with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. BMC Veterinary Research. 8, 208 (2012).
- Silva, T. M. A., Costa, E. A., Paixao, T. A., Tsolis, R. M., Santos, R. L. Laboratory animal models for brucellosis research. Journal of Biomedicine and Biotechnology. 2011, 518323 (2011).
- Willard-Mack, C. L. Normal structure, function, and histology of the lymph nodes. Toxicologic Pathology. 34, 409-424 (2006).
- Elmore, S. A. Histopathology of the lymph nodes. Toxicologic Pathology. 34 (5), 425-454 (2006).
- Luscieti, P., Hubschmid, T. h., Cottier, H., Hess, M. W., Sobin, L. H. Human lymph node morphology as a function of age and site. Journal of Clinical Pathology. 33 (5), 454-461 (1980).
- Hadamitsky, C., et al. Age-dependent histoarchitectural changes in human lymph nodes: an underestimated process with clinical relevance?. Journal of Anatomy. 216 (5), 556-562 (2010).
- Sisson, S., Grossman, J. D., Getty, R. . Sisson and Grossman’s The Anatomy of Domestic Animals, Volume 1. , (1975).
- Olsen, S. C., Johnson, C. Comparison of abortion and infection after experimental challenge of pregnant bison and cattle with Brucella abortus strain 2308. Clinical and Vaccine Immunology. 18 (12), 2075-2078 (2011).
- Brichta-Harhay, D. M., et al. Microbiological analysis of bovine lymph nodes for the detection of Salmonella enterica. Journal of Food Protection. 75 (5), 854-858 (2012).
- Samuel, J. L., O’Boyle, D. A., Mathers, W. J., Frost, A. J. Isolation of Salmonella from mesenteric lymph nodes of healthy cattle at slaughter. Research in Veterinary Science. 28 (2), 238-241 (1980).
- Arthur, T. M., et al. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. Journal of Food Protection. 71 (8), 1685-1688 (2008).
- Cray, W. C., Moon, H. W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Applied and Environmental Microbiology. 61 (4), 1586-1590 (1995).
- Thiermann, A. B. Experimental leptospiral infections in pregnant cattle with organisms of the Hebdomadis serogroup. American Journal of Veterinary Research. 43 (5), 780-784 (1982).
- Palmer, M. V., Waters, W. R., Whipple, D. L. Investigation of the transmission of Mycobacterium bovis from deer to cattle through indirect contact. American Journal of Veterinary Research. 65 (11), 1483-1489 (2004).
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA, and proteins from cell and tissue samples. BioTechniques. 15 (3), 532-537 (1993).
- Chomczynski, P., Sacchi, N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 162 (1), 156-159 (1987).
- Vogelstein, B., Gillespie, D. Preparative and analytical purification of DNA from agarose. Proceedings of the National Academy of Sciences of the United States of America. 76 (2), 615-619 (1979).
- . . AVMA Guidelines for the Euthanasia of Animals. , (2013).
- . . TRIzol Reagent Manual. , (2016).
- . . Disruption and Homogenization of Tissue for the Extraction of RNA. , (2014).
- . . PureLink RNA Mini Kit Protocol . , (2012).
- . . RNA Gel-loading Buffer. , (2006).
- Rio, D. C., Ares, M., Hannon, G. J., Nilsen, T. W. Nondenaturing Agarose Gel Electrophoresis of RNA. Cold Spring Harbor Protocols. , (2010).
- Mueller, O., et al. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis. 21 (1), 128-134 (2000).
- Schroeder, A., et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 7 (3), (2006).
- Suvarna, K. S., Layton, C., Bancroft, J. D. . Bancroft’s Theory and Practice of Histological Techniques. , (2012).
- Medawar, P. B. The rate of penetration of fixatives. Journal of Microscopy. 61 (1-2), 46-57 (1941).
- Canene-Adams, K. Preparation of formalin-fixed paraffin-embedded tissue for immunohistochemistry. Methods in Enzymology. 533, 225-233 (2013).
- Fleige, S., Pfaffl, M. W. RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine. 27, 126-139 (2006).
- Fajardy, I., et al. Time course analysis of RNA stability in human placenta. BMC Molecular Biology. 10 (21), (2009).
- Bahar, B., et al. Long-term stability of RNA in post-mortem bovine skeletal muscle, liver and subcutaneous adipose tissues. BMC Molecular Biology. 8, 108 (2007).
- Yamagishi, A., et al. Gene profiling and bioinformatics analyses reveal time course differential gene expression in surgically resected colorectal tissues. Oncology Reports. 31 (4), 1532-1538 (2014).
- Choi, S., Ray, H. E., Lai, S. H., Alwood, J. S., Globus, R. K. Preservation of multiple mammalian tissues to maximize science return from ground based and spaceflight experiments. PLoS One. 11 (12), e0167391 (2016).
- Almeida, A., Thiery, J. P., Magdelenat, H., Radvanyi, F. Gene expression analysis by real-time reverse transcription polymerase chain reaction: influence of tissue handling. Analytical Biochemistry. 328 (2), 101-108 (2004).
- Lee, S. M. L., Schelcher, C., Thasler, R., Schiergens, T. S., Thasler, W. E. Pre-analytical determination of the effect of extended warm or cold ischaemia on RNA stability in the human ileum mucosa. PLoS One. 10 (9), e0138214 (2015).
- Kap, M., et al. The influence of tissue procurement procedures on RNA integrity, gene expression, and morphology in porcine and human liver tissue. Biopreservation and Biobanking. 13 (3), 200-206 (2015).
- Hong, S. H., et al. Effects of delay in the snap freezing of colorectal cancer tissues on the quality of DNA and RNA. Journal of the Korean Society of Coloproctology. 26 (5), 316-325 (2010).
- Lee, S. M., et al. RNA stability in human liver: comparison of different processing times, temperatures and methods. Molecular Biotechnology. 53 (1), 1-8 (2013).
- Morrison, P. K., et al. Post-mortem stability of RNA in skeletal muscle and adipose tissue and the tissue-specific expression of myostatin, perilipin and associated factors in the horse. PLoS One. 9 (6), e100810 (2014).
- Seear, P. J., Sweeney, G. E. Stability of RNA isolated from post-mortem tissues of Atlantic salmon (Salmo salar L.). Fish Physiology and Biochemistry. 34 (1), 19-24 (2008).
- Marchuk, L., Sciore, P., Reno, C., Frank, C. B., Hart, D. A. Postmortem stability of total RNA isolated from rabbit ligament, tendon, and cartilage. Biochimica et Biophysica Acta. 1379 (2), 171-177 (1998).
- Inoue, H., Kimura, A., Tuji, T. Degradation profile of mRNA in a dead rat body: basic semi-quantification study. Forensic Science International. 130 (2-3), 127-132 (2002).
- Micke, P., et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Laboratory Investigation. 86 (2), 202-211 (2006).
- Srinivasan, M., Sedmak, D., Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. The American Journal of Pathology. 161 (6), 1961-1971 (2002).
- Ahmed, A. M., et al. Variation in the ovine abomasal lymph node transcriptome between breeds known to differ in resistance to the gastrointestinal nematode. PLoS One. 10 (5), e0124823 (2015).
- Russell, G. C., et al. Host gene expression changes in cattle infected with Alcelaphine herpesvirus 1. Virus Research. 169 (1), 246-254 (2012).
- Avraham, R., et al. A highly multiplexed and sensitive RNA-seq protocol for simultaneous analysis of host and pathogen transcriptomes. Nature Protocols. 11, 1477-1491 (2016).

