CRISPR Guide RNA Cloning for Mammalian Systems
Summary
Here, a simple, efficient, and cost-effective method of sgRNA cloning is outlined.
Abstract
The outlined protocol describes streamlined methods for the efficient and cost-effective generation of Cas9-associated guide RNAs. Two alternative strategies for guide RNA (gRNA) cloning are outlined based on the usage of the Type IIS restriction enzyme BsmBI in combination with a set of compatible vectors. Outside of the access to Sanger sequencing services to validate the generated vectors, no special equipment or reagents are required aside from those that are standard to modern molecular biology laboratories. The outlined method is primarily intended for cloning one single gRNA or one paired gRNA-expressing vector at a time. This procedure does not scale well for the generation of libraries containing thousands of gRNAs. For those purposes, alternative sources of oligonucleotide synthesis such as oligo-chip synthesis are recommended. Finally, while this protocol focuses on a set of mammalian vectors, the general strategy is plastic and is applicable to any organism if the appropriate gRNA vector is available.
Introduction
CRISPR-associated protein 9 (Cas9) is an RNA-guided endonuclease which is directed towards a desired genomic target when complexed with an appropriately designed small guide RNA (gRNA)1,2. gRNAs comprise a 20-nucleotide sequence (the protospacer), which is complementary to the genomic target sequence. Next to the genomic target sequence is a 3' protospacer-associated motif (PAM), which is required for Cas9 binding. In the case of Streptococcus Pyogenes Cas9 (SpCas9), this has the sequence NGG. Upon binding the DNA target, Cas9 cleaves both strands of DNA, thereby stimulating repair mechanisms that can be exploited to modify the locus of interest. The activation of the error-prone non-homologous end-joining (NHEJ) pathway can cause the disruption of a target gene. Alternatively, repairs made via a homologous recombination can stimulate the targeted integration of a desired DNA sequence if a donor template is provided6. Nuclease-dead Cas9 (dCas9) variants can also be generated3 by mutating the residues within Cas9 that are essential for endonucleolytic activity. dCas9 remains competent for DNA binding but does not cut its bound target. By fusing various effector domains to dCas9 and directing it near a gene's transcriptional start site, dCas9 can be used for selective gene induction or partial gene silencing4,5.
While incredibly powerful, the ability to use Cas9 to target a genomic sequence of interest requires that a user first generates a gRNA unique to the target site(s). A variety of methods for building gRNA expression vectors have previously been described, many of varying efficiency and cost6,7,8. Stand-alone PCR amplicons may be used as gRNAs for rapid screening, going from oligonucleotide delivery to transfection within several hours; however, this requires Ultramer (over 100 bp) oligo synthesis, which is expensive9. It is also possible to purchase gBlocks that are more cost-effective than Ultramers. As Cas9 has rapidly become an integral component of the modern molecular biology toolkit, a simple, inexpensive, and highly efficient method for gRNA cloning would be a boon to the field. The method described here has been employed in our group and collaborating laboratories for the past several years to generate over 2,000 unique gRNAs4.
The outlined method focuses on techniques for the cloning of lentiviral compatible vectors containing a single gRNA or at most two gRNAs. For the generation of plasmids containing more than two gRNAs, or to clone a library of gRNAs, alternative approaches are recommended9,10,11,12.
Protocol
Note: The following protocol outlines how to perform gRNA design using online tools (steps 1.1 – 1.4), which are common to all methods of gRNA plasmid construction detailed in this manuscript. Once the desired gRNAs are identified, steps for ordering the necessary oligonucleotides are described along with several different methods for introducing the oligonucleotides into expression vectors (e.g., pSB700). Presented are 2 methods for cloning single guide-expressing vectors based on either ligation into a predigested expression vector (steps 2 – 7.3) or Golden Gate cloning (steps 8 – 8.4). Further outlined is a strategy for cloning paired guide-expressing vectors using polymerase chain reaction (PCR) followed by Golden Gate cloning (steps 11 – 16). Finally, common methods for performing an E. coli chemical transformation (steps 9 – 9.6) and an expression vector sequence validation (steps 10 to 10.2.3) are also outlined.
- Design gRNA oligonucleotides using online tools as follows.
- Design gRNAs using online tools such as the sgRNA Scorer 2.0 online tool (https://crispr.med.harvard.edu/sgRNAScorerV2/)13 or alternatives such as the Broad sgRNA design tool (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design)14 and several others15.
- Use the gene name, symbol, or raw DNA target sequences to generate potential gRNA sequences.
Note: A spreadsheet (.csv/.xls) file with all the potential gRNAs against the gene or the provided sequence is created. The quality of the potential gRNAs will be reported as Score if the sgRNA Scorer 2.0 online tool is being used, or as % on-target efficiency if the Broad sgRNA design tool is being used. Both measures are derived at based on the on-target cutting activity. - Consider the Off-target activity in the gRNA selection when ranking the gRNAs.
Note: Optimal guides are those with the greatest on-target efficiency and the least off-target activity. For activation (CRISPRa), gRNAs are designed to target the region from 1 – 200 bp upstream of the transcriptional start site (TSS)4,15. For inhibition (CRISPRi), gRNAs are designed to target the region from 50 bp upstream of the TSS until 300 bp downstream of the TSS5. To disable a gene using Cas9 cutting, gRNAs are designed to target the first 10 – 50% of the coding sequence downstream of the initiating codon (as gRNAs targeting the 3’ end of the gene have been shown to be less effective)13. - Ensure no internal BsmBI sites are present if oligonucleotides are being used for the Golden Gate cloning, as this will result in the annealed oligonucleotides being digested13.
- Select the genomic target sequences and remove the 3’ NGG PAM (leaving only the protospacer sequence) for the single gRNA oligonucleotide sequence modification (e.g., starting sequence: CGCGTGCTCTCCCTCATCCATGG).
Note: If using a spreadsheet, the following formula will remove 3’ NGG: =LEFT(A2,LEN(A2)-3), where A2 is the cell containing the target sequence and the PAM.- Append CACCG to the 5’ end of the oligo.
Note: Upon ligation to the pSB700 backbone, this sequence will comprise the 3’ end of the U6 promoter driving the transcription of the gRNA. The “CACC” sequence ensures that the oligo is compatible with the overhangs of the BsmBI-digested pSB700 vector. The “G” is a requirement of Polymerase III promoters and ensures the efficient initiation of the transcription of the gRNA. Use the following formula in a spreadsheet to perform this task: =CONCATENATE(“CACCG”,(B2)), where B2 is the cell containing the protospacer sequence [e.g., adding the 5’ CACCG sequence to the protospacer: CACCGCGCGTGCTCTCCCTCATCCA (this is the final forward oligo that is ordered)]. - Create a reverse complement (rc) of the protospacer sequence.
Note: For example, the reverse complement of the aforementioned protospacer is TGGATGAGGGAGAGCACGCG. - Append AAAC to the 5’ end of the rc protospacer. Append an additional C to the 3’ end of the rc protospacer (reverse oligo).
Note: The “AAAC” sequence ensures that the oligo is compatible for cloning into the BsmBI-digested pSB700 vector. The additional “C” on the 3’ end is needed to anneal the sequence to the initiating “G” added to the forward oligo. Use the following formula in a spreadsheet to perform this task =CONCATENATE(“AAAC”,(C2),”C”), where C2 is the cell containing the reverse complement sequence [e.g., AAACTGGATGAGGGAGAGCACGCGC (this is the final reverse oligo that is ordered)].
- Append CACCG to the 5’ end of the oligo.
- Order the oligonucleotides, with no additional modifications on the oligonucleotides.
- Order the minimal amount of oligo required, as only very small amounts are needed for the cloning reactions.
- Dilute lyophilized primers to a final concentration of 100 μM in 1x TE buffer (10 mM Tris-Cl, pH 7.5 with 1 mM EDTA).
- Aliquot 1:1 forward and reverse oligonucleotides (e.g., 10 μL each) into PCR strip-cap tubes.
Note: This will allow the rapid cloning of many gRNAs at a time using a multichannel pipette.- Vortex and spin down the oligo mixtures (100 x g for 15 s). Incubate the reaction at room temperature for 5 min before setting up the ligation.
Note: It is not necessary to heat and then cool the oligo mixtures to facilitate their annealing. It is worth noting that 3 – 4 pairs of gRNA oligonucleotides may be pooled and annealed in a single reaction (e.g., mix 3 μL of forward and 3 μL of reverse oligonucleotides together from up to 4 gRNA oligo pairs, giving a total volume of 24 μL). The volume of the oligonucleotides used for the annealing may be adjusted to accommodate a user’s needs.
Note: Steps 6 – 7.3 describe the predigestion of pSB700. For an alternative method, see step 8, which describes a single-step BsmBI restriction ligation.
- Vortex and spin down the oligo mixtures (100 x g for 15 s). Incubate the reaction at room temperature for 5 min before setting up the ligation.
- Digest 1 – 5 μg of the selected pSB700 guide expression vector with BsmBI (0.5 μL per 1 μg) for 1 h at 55 °C15. Conduct the digestion of the pSB700 gRNA expression vector in a total volume of 40 μL (Table 1).
- Run the digestion products on 1.5% (wt/vol) low-melting agarose gel.
- Cut out the digested vector backbone band which corresponds to a fragment of ~9 kb in size and place the gel slice in a 1.5 mL microcentrifuge tube (Figure 1).
- Use a commercial gel purification kit to extract the DNA from the agarose gel according to the manufacturer’s instructions.
- Elute the DNA into 10 – 15 μL of TE buffer (10 mM Tris, pH 7.5; 10 mM Tris, pH 8.0) to obtain a concentrated eluate.
- Ligate the annealed gRNA oligonucleotides from step 4 into the BsmBI-digested pSB700 guide expression vector in a 20 μL reaction (Table 2) for the cloning of the gRNA oligonucleotides into the pre-digested pSB700 expression vector.
- First, add water, buffer, and DNA, then vortex the mixture and add enzyme, vortex 1 final time after adding the enzyme and spin down the solution (100 x g for 15 s). Incubate the reactions at room temperature overnight.
- Include a no-insert negative control reaction that has a BsmBI-digested vector alone, without an annealed gRNA oligo insert. 1 μL of water may be used in place of the 1 μL of gRNA oligonucleotides for the no-insert negative control.
- Proceed to the transformation (step 9).
Note: Step 8 is an alternative method to the predigestion of pSB700 as described in steps 6 – 7.3.
- Introduce gRNA oligonucleotides (outlined in steps 1 – 5.1) into the pSB700 vector through the use of Golden Gate cloning for the single-step BsmBI restriction ligation.
- Assemble the master mix (1x)—if multiple gRNAs are being cloned, prepare the master mix without the insert oligonucleotides added (Table 3). Aliquot the master mix into PCR tubes and use a multichannel pipette to add the gRNA oligonucleotides from step 4.2. Include a no-insert control using 1 μL of water.
- By including the BsmBI enzyme and T4 DNA ligase within the same reaction, simultaneous restriction digestion and ligation are achieved (i.e., Golden Gate cloning). Use the following Simple Gate protocol to digest the vector and ligate the inserts in 1 reaction: keep the reaction at 37 °C for 2 h, at 50 °C for 10 min, at 65 °C for 15 min, at 80 °C for 15 min, and finally, hold it at 4 °C.
- Of note, verify that the oligonucleotides do not contain BsmBI sites before using this method. If the gRNAs that are being cloned contain BsmBI restriction sites, they will be digested upon the addition of BsmBI. This will prevent the user from obtaining any transformants.
- After the initial Simple Gate reaction, add an additional 0.5 μL of BsmBI enzyme to each reaction and place them back at 55 °C for 1 h. After the digestion, hold the reaction at 4 °C or freeze it until ready to transform. This second round of restriction enzyme incubation removes any undigested or religated wild-type pSB700 vector backbone from the reaction mixture so that only vectors that have received the oligo inserts remain intact and will yield transformants.
- Proceed to the transformation (step 9).
- Transform 0.5 μL of the reaction mixture into 8 μL of competent E. coli [e.g., NEB DH5a (1 – 3 x 109 cfu/µg of pUC19 DNA) or NEB Stable 1 – 3 x 109 cfu/µg of pUC19 DNA). For lentiviral plasmids, NEB Stable cells will provide more consistent plasmid yields.
- Remove E. coli from storage at -80 °C and thaw it on ice for 5 – 10 min.
- Add 0.5 μL of the reaction mix to 8 μL of competent E. coli and keep the mixture on ice for 30 min.
- Heat shock the mixture at 42 °C for 45 s.
- Rest it on ice for 2 min.
- Recover the culture on a rotary shaker in 250 μL of SOC media for 1 h at 37 °C for NEB DH5a and at 30 °C for NEB Stables.
- Plate 80 μL of the culture on an appropriate antibiotic resistance Lysogeny Broth (LB) plate and incubate it overnight at 37 °C for NEB DH5a and at 30 °C for NEB Stables [e.g., pSB700 is plated on LB with ampicillin (100 μg/mL) plates] (Figure 3).
- Validate the sequence of gRNA expression plasmid: verify the sequence of each colony by Sanger sequencing using the primer 5’-GAGGGCCTATTTCCCATGATTCC-3’ which primes at the U6 promoter upstream of the gRNA oligo insert.
- Pick 2 – 3 colonies per reaction for sequencing (Table 4).
Note: Several sequencing providers now offer services that can perform Sanger sequencing directly from bacterial colonies, which greatly reduces the time and costs by eliminating the need to purify the plasmids prior to sequencing. Alternatively, a plasmid isolation kit may be used to recover the plasmid DNA from the individual bacterial clones. Purified plasmids may then be submitted for sequencing. If a pooled ligation reaction is performed, use Table 4 as a guide for the number of colonies that should be selected and submitted for sequencing. - Following the sequence confirmation, isolate the gRNA expression plasmids.
- Use a sterile pipette tip to inoculate the desired colony into a 5 mL culture of LB medium containing 100 μg/mL of ampicillin for pSB700 expression vectors.
- Grow the cells in a rotary incubator at 100 rpm at 37 °C overnight (30 °C if NEB Stables are being used).
- Isolate the plasmid DNA from the cultures using a plasmid prep kit, following the manufacturer’s protocol.
Note: While the above methods enable users to create single gRNA-containing vectors, this protocol will allow users to create vectors that contain 2 gRNAs, each driven by their own RNA polymerase III promoter.
- Pick 2 – 3 colonies per reaction for sequencing (Table 4).
- For a paired gRNA design, use the output file from the sgRNA design tools outlined above (steps 1.1 – 1.4) and select 2 guides of interest.
- For an activation or repression, select pairs of guides that are at least 30 nt apart.
- For gRNAs used for cutting, select guides that target 2 different exons within the gene.
- For the instructions below, designate the first gRNA in the array gRNA-A and the second gRNA in the array gRNA-B, to create a paired gRNA oligonucleotide sequence modification.
Note: The oligo used to create gRNA-A will be called fA, and the reverse oligo used to introduce gRNA-B will be referred to as rB. The cloning method below automatically appends an initiating G at the 5’ end of each of the gRNAs.- Create the appropriate fA oligo as follows.
Note: The 20 nucleotides comprising the gRNA targeting sequence for gRNA-A will constitute the initial fA sequence upon which the following steps will build (e.g., protospacer – aggggctacaccactcattg).- Append TTTTCGTCTCTCACCG to the 5’ end of fA (e.g., of the sequence TTTTCGTCTCTCACCGaggggctacaccactcattg).
- Append GTTTTAGAGCTATGCTGAAAAGCA to the 3’ end of fA (e.g., of the sequence TTTTCGTCTCTCACCGaggggctacaccactcattgGTTTTAGAGCTATGCTGAAAAGCA). This is the final version of the fA oligo that will be ordered.
- Create the necessary rB oligo as follows.
Note: The 20 nucleotides comprising the gRNA targeting sequence for gRNA-B will constitute the initial rB sequence upon which the following steps will build (e.g., of the starting sequence gtcccctccaccccacagtg).- Take the reverse complement of rB =(revcom)rB (e.g., cactgtggggtggaggggac).
- Append TTTTCGTCTCTAAAC to the 5’ end of (revcom)rB (e.g., TTTTCGTCTCTAAACcactgtggggtggaggggac).
- Append CGGTGACCCAGGCGGCGCACAAG to the 3’ end of (revcom)rB (e.g., TTTTCGTCTCTAAACcactgtggggtggaggggacCGGTGACCCAGGCGGCGCACAAG). This is the final version of the rB oligo that will be ordered.
Note: The following formulas may be used with common spreadsheet software to automatically edit the oligo sequences: =CONCATENATE("TTTTCGTCTCTCACCG",(A2)," GTTTTAGAGCTATGCTGAAAAGCA"), where A2 is the cell containing the fA sequence, and =CONCATENATE("TTTTCGTCTCTAAAC",(B2)," CGGTGACCCAGGCGGCGCACAAG"), where B2 is the cell containing the (revcom)rB sequence.
- Create the appropriate fA oligo as follows.
- Order the oligonucleotides from a synthesis vendor of choice. No additional modifications are required.
- Use the prepared plasmid DNA to perform PCR with both fA and rB primers. This will attach both the forward and the reverse gRNAs to the pSN007 PCR fragment generated from this plasmid and will allow each to be expressed from its own promoter (U6 and 7SK promoters—different promoters are used in this design to prevent viral recombination).
- Use the Phusion GC Special PCR Protocol to create the needed fragment: keep the mixture at 98 °C for 1 min (1 cycle), at 98 °C for 15 s, at 53 °C for 15 s, at 65 °C for 2 min, and at 72 °C for 4 min (30 cycles), and then hold it at 4 °C (Table 5).
- Run the PCR product on a 1% agarose gel and verify that 1 band is seen at ~490 bp (Figure 2).
- Cut and extract this PCR product using a gel extraction kit of choice. Elute the DNA into 15 μL of TE buffer (10 mM Tris, pH 7.5; 10 mM Tris, pH 8.0) or distilled water.
- Measure the concentration of the extracted product along with that of the pSB700 vector with a spectrophotometer for the DNA molarity calculation. Normalize both the PCR product and the vector to a concentration of 40 femtomoles/μL.
Note: Several websites are available to assist with the calculation of the molarity of a given DNA solution based on the length of the DNA and its concentration in solution (e.g., Promega: http://www.promega.com/a/apps/biomath/index.html?calc=ugpmols). The calculator uses the formula:
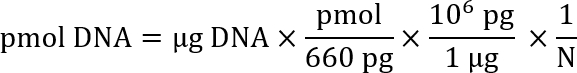
Here, N is the length of the nucleotide sequence.
- Set up a simultaneous restriction digestion and ligation reaction using the protocol outlined in steps 8 – 8.4 to clone the pSN007 PCR fragment into a pSB700 vector.
- Instead of adding 1 μL of primer solution to the reaction, add 1 μL of 40 fmole/μL of the PCR product. In addition, use 1 μL of the pSB700 vector at a concentration of 40 fmole/μL.
Note: Equal molar ratios lead to consistent outcomes, although even in cases where exact ratios are not used, colonies can be obtained.
- Instead of adding 1 μL of primer solution to the reaction, add 1 μL of 40 fmole/μL of the PCR product. In addition, use 1 μL of the pSB700 vector at a concentration of 40 fmole/μL.
- Upon completing the Golden Gate reaction process (steps 8 – 8.4), proceed to the transformation and sequence verification (steps 9 – 10.2.3).
Representative Results
The methods outlined in this protocol are for the creation of either single or paired gRNA expression vectors. Single gRNA expressing vectors can be created by either predigesting the vector backbone (Figure 1) followed by ligating it in a series of short oligonucleotides or using Golden Gate cloning to simultaneously digest the vector backbone and ligate it in a series of short oligonucleotides in a single reaction. Also provided is a method of producing vectors that contain 2 gRNAs, each driven by its own independent promoter, by cloning a custom pSN007 PCR fragment (Figure 2).
A successful cloning for any of the outlined protocols will result in the appearance of significantly more colonies for transformations with the appropriate insert DNA than on the no-insert control plate (Figure 3).
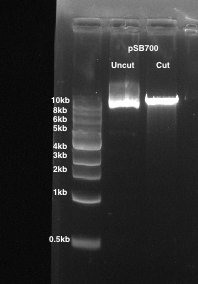
Figure 1: The digestion of a pSB700 gRNA expression vector. 1% agarose gel is used. Lane 1: uncut pSB700. Lane 2: pSB700 cut with BsmBI.
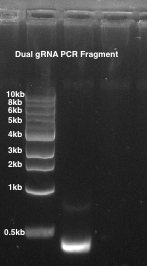
Figure 2: Paired gRNA PCR. 1% agarose gel is used. Lane 1: a paired guide pSN007 PCR fragment of ~490 bp.
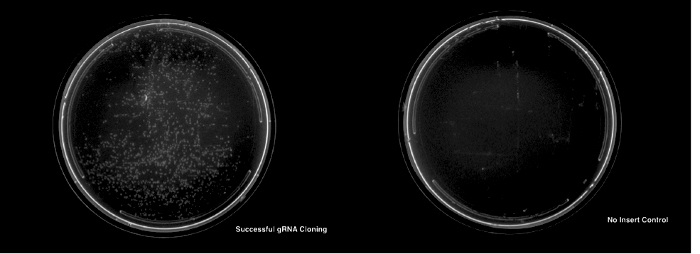
Figure 3: The successful cloning and transformation of a gRNA plasmid. The left panel shows an LB and ampicillin plate with successfully transformed colonies. The right panel shows a no-insert control plate showing no colonies. Please click here to view a larger version of this figure.
Supplemental File. Please click here to download this file.
| pSB700 guide expression vector | 1-5 μg |
| NEB Buffer 3.1 | 4 μL |
| BsmBI | 1 μL |
| Distilled water | up to 40 μL |
| Total volume | 40 μL |
Table 1: The restriction digestion of a pSB700 gRNA expression vector with BsmBI.
| Annealed gRNA oligos (either single or pooled reaction) | 1 μL |
| BsmBI-digested pSB700 guide expression vector | 1 μL (~100-250 ng) |
| 10x T4 DNA Ligase Reaction Buffer | 2 μL (1x final concentration) |
| T4 DNA Ligase | 1 μL (120 units) |
| Distilled water | 15 μL |
| Total volume | 20 μl |
Table 2: A ligation reaction of gRNA oligonucleotides into a predigested pSB700 gRNA expression vector.
| Reagent | Volume (1x) |
| H2O | 6 μL |
| T4 Ligase | 0.5 μL |
| ATP | 1 μL |
| BsmBI | 0.5 μL |
| NEB Buffer 3.1 | 1 μL |
| Vector (e.g., pSB700) (40 fmol) | 1 μlL |
| Insert – forward and reverse oligos or pSN007 PCR fragment for dual guide (40fmol) | 1 μL (0.5 μL forward and 0.5 μL reverse) |
Table 3: A single-step BsmBI restriction and gRNA ligation reaction mix.
| For 1 guide | sequence 3 colonies |
| For 2 guides | sequence 5-10 colonies |
| For 3 guides | sequence 10-15 colonies |
| For 4 guides | sequence 15-20 colonies |
Table 4: The recommended number of colonies to sequence for pooled gRNA cloning reactions.
| PCR Component | 50 µL reaction |
| 10 µM Forward Primer | 2.5 µL |
| 10 µM Reverse Primer | 2.5 µL |
| 5x Phusion HF or GC Buffer | 10 µL |
| 10 mM dNTPs | 1 µL |
| Template DNA | 100 ng |
| Phusion DNA Polymerase | 0.5 µL |
| Nuclease-free water | Up to 50 µL |
Table 5: A paired gRNA PCR mixture.
Discussion
A number of time- and cost-saving modifications have been made to the gRNA expression vector construction. A single-step restriction and ligation protocol is outlined as well as a method of paired gRNA expression vector construction. The paired gRNA expression is achieved through a PCR amplification using oligonucleotides containing both a forward gRNA target sequence (n20) and the reverse complement of another target (n20). This then introduces a secondary RNA Pol III promoter (7SK) as well as a sgRNA tail into conventionally expressing single gRNA expression plasmids.
A careful oligonucleotide design will ensure a successful PCR for the paired gRNA construction as well as a sticky-end ligation. Following the single-step restriction and ligation reaction, the addition of further BsmBI will ensure all unmodified expression vectors in the reaction are cut. This will significantly reduce the background upon the transformation. Using NEB-Stable competent E. coli and a growth at 30 °C will increase the yield of a successful transformation.
The advantages this technique has over common practices include a simplification of the process of oligonucleotide annealing, a single-step restriction of the gRNA expression vector and the ligation of oligonucleotides, and the ability to utilize conventional single gRNA expression vectors for the expression of paired guides. While the protocol described here focuses on a set of mammalian vectors, the general strategy is plastic and is applicable to any organism, provided the appropriate gRNA vector is available. Overall, the protocol described saves time and costs.
However, it should be noted that the outlined procedure of purchasing individual oligonucleotides does not scale well for the generation of libraries containing thousands of gRNAs. For those purposes, alternative sources of oligonucleotide synthesis such as oligo chip synthesis are recommended.
It is beneficial to include a no-insert control when cloning gRNAs. This reaction can be easily set up in parallel with the reactions of interest and can be used to determine whether an incomplete digestion of the starting vector has contributed to the colonies observed at the end of the procedure. Furthermore, if one of the above cloning protocols has failed due to an incompletely digested vector backbone or poor ligation efficiency, we suggest trying the alternative cloning method we have outlined.
When using Golden Gate cloning to simultaneously digest the vector and ligate in the small oligonucleotides within a single reaction, it is important to check that the gRNA that are being cloned into the vector does not have a BsmBI site inside of it, as this will lead to the gRNA being cut and an absence of colonies upon transformation.
The gRNA cloning strategy we have outlined enables a rapid and cost-effective generation of gRNAs at ~10 U.S. dollars per guide, with the bulk of the costs coming from the oligonucleotide synthesis and the sequence verification. While the outlined method is designed to allow users to generate gRNAs for use with SpCas9, the protocol can easily be adapted for use with Cas9 orthologues or other RNA-guided endonucleases such as Cpf1 or C2C2, with slight modifications to the vector backbone and the oligonucleotide overhang sequences16,17.
The protocol outlined above will provide sequence-verified gRNA expression plasmids in 3 d (starting with appropriately designed oligonucleotides), which is significantly faster than current methods. This includes the gRNA design of 1 h (steps 1 – 2.3), the dilution and aliquot of the oligonucleotides of 10 min (steps 3 – 5.1), the digestion and purification of the pSB700 gRNA expression vector of 2 h (steps 6 – 6.4), the cloning of the gRNA oligonucleotides into a predigested pSB700 gRNA expression vector overnight at room temperature (steps 7 – 7.3), the single-step BsmBI restriction ligation of 3 h and 40 min (steps 8 – 8.4), the transformation of 16 h (steps 9 – 9.6), and the validation of the sequence of the gRNA expression plasmid of 24 h (with the duration depending on the sanger sequencing availability and the speed following the submission of the bacterial colony; step 10). The paired gRNA design and the sequence modification takes 1 h (steps 11 – 13). The paired gRNA PCR takes 3.5 h (steps 14 – 14.4). The cloning of the pSN007 PCR fragment into a pSB700 vector takes 3h and 40 min (step 15 – 16).
The most likely issues that may be faced with this protocol relate to inefficiency in the Golden Gate assembly and transformation. Include a no-insert control during the Golden Gate assembly to visualize the efficiency of the assembly. During the transformation, the puc19 positive control will provide a transformation efficiency control. NEB-Stable E. coli should be used for lentiviral compatible plasmids and frequently require up to > 16 h at 30 °C to yield visible colonies.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
Sathiji Nageshwaran is supported by the Friedreich's Ataxia Research Alliance (07340305-01) and National Ataxia Foundation Fellowships (7355538-01). Alejandro Chavez was funded by the National Cancer Institute grant 5T32CA009216-34 and the Burroughs Wellcome Fund Career Award for Medical Scientists. George M. Church is supported by the U.S. National Institutes of Health (NIH) National Human Genome Research Institute grant RM1 HG008525 and the Wyss Institute for Biologically Inspired Engineering. James J. Collins acknowledges support from the Defense Threat Reduction Agency grant HDTRA1-14-1-0006 and the Paul G. Allen Frontiers Group. Alejandro Chavez developed the dual-guide cloning method. Sathiji Nageshwaran, Alejandro Chavez, and Nan Cher Yeo wrote the manuscript with input from all authors.
Materials
| BsmBI | New England Biolabs | R0580L | |
| T4 DNA ligase | Enzymatic/New England Biolabs | L6030-LC-L/M0202S | |
| Buffer 3.1 | New England Biolabs | B7203S | |
| Adenosine 5'-Triphosphate (ATP) | New England Biolabs | P0756S | |
| QIAprep spin miniprep kit | Qiagen | 27104 | |
| Chemically competent E. coli | New England Biolabs | C3019l, C2987l, or C3040H | |
| Standard microcentrifuge tubes, 1.5 mL | Eppendorf | 0030 125.150 | |
| Axygen 8-Strip PCR tubes | Fischer Scientific | 14-222-250 | |
| Thermocycler with programmable temperature-stepping control | BioRad, | 1851148 | |
| UV spectrophotometer (NanoDrop 2000c) | Thermo Scientific | ||
| pSB700 plasmid | Addgene | #64046 | |
| NEB Stable Competent E. coli (High Efficiency) | New England Biolabs | c3040 | |
| Lysogeny broth (LB) | |||
| Ampicillin | |||
| TE buffer (1 mM EDTA, 10 mM Tris-Cl, pH 7.5) |
Referencias
- Gasiunas, G., Barrangou, R., Horvath, P., Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 109 (39), E2579-E2586 (2012).
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J., Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 337 (6096), 816-821 (2012).
- Shalem, O., Sanjana, N., Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nature Reviews Genetics. 16 (5), 299-311 (2015).
- Chavez, A., et al. Highly efficient Cas9-mediated transcriptional programming. Nature Methods. 12 (4), 326-328 (2015).
- Gilbert, L., et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 154 (2), 442-451 (2013).
- Yang, L., Yang, J., Byrne, S., Pan, J., Church, G. CRISPR/Cas9-Directed Genome Editing of Cultured Cells. Current Protocols in Molecular Biology. , 31.1.1-31.1.17 (2014).
- Yang, L., Mali, P., Kim-Kiselak, C., Church, G. CRISPR-Cas-Mediated Targeted Genome Editing in Human Cells. Methods in Molecular Biology. 1114, 245-267 (2014).
- Vidigal, J., Ventura, A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nature Communications. 6, 8083 (2015).
- Joung, J., et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nature Protocols. 12 (4), 828-863 (2017).
- Kabadi, A., Ousterout, D., Hilton, I., Gersbach, C. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Research. 42 (19), e147 (2014).
- Sakuma, T., Nishikawa, A., Kume, S., Chayama, K., Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Scientific Reports. 4 (1), (2014).
- Chari, R., Yeo, N., Chavez, A., Church, G. sgRNA Scorer 2.0: A Species-Independent Model To Predict CRISPR/Cas9 Activity. ACS Synthetic Biology. 6 (5), 902-904 (2017).
- Doench, J., et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature Biotechnology. 32 (12), 1262-1267 (2014).
- Konermann, S., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517 (7536), 583-588 (2014).
- Engler, C., Marillonnet, S., Valla, S., Lale, R. Golden Gate Cloning. DNA Cloning and Assembly Methods. , 119-131 (2013).
- Zetsche, B., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 163 (3), 759-771 (2015).
- Abudayyeh, O., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 353 (6299), (2016).

