An In Vivo Mouse Model to Measure Naïve CD4 T Cell Activation, Proliferation and Th1 Differentiation Induced by Bone Marrow-derived Dendritic Cells
Summary
Here, we present a protocol for the in vivo determination of naïve CD4 T cell (T cell) activation, proliferation, and Th1 differentiation induced by GM-CSF bone marrow (BM)-derived dendritic cells (DCs). In addition, this protocol describes BM and T-cell isolation, DC generation, and DC and T-cell adoptive transfer.
Abstract
Quantification of naïve CD4 T cell activation, proliferation, and differentiation to T helper 1 (Th1) cells is a useful way to assess the role played by T cells in an immune response. This protocol describes the in vitro differentiation of bone marrow (BM) progenitors to obtain granulocyte macrophage colony-stimulating factor (GM-CSF) derived-dendritic cells (DCs). The protocol also describes the adoptive transfer of ovalbumin peptide (OVAp)-loaded GM-CSF-derived DCs and naïve CD4 T cells from OTII transgenic mice in order to analyze the in vivo activation, proliferation, and Th1 differentiation of the transferred CD4 T cells. This protocol circumvents the limitation of purely in vivo methods imposed by the inability to specifically manipulate or select the studied cell population. Moreover, this protocol allows studies in an in vivo environment, thus avoiding alterations to functional factors that may occur in vitro and including the influence of cell types and other factors only found in intact organs. The protocol is a useful tool for generating changes in DCs and T cells that modify adaptive immune responses, potentially providing important results to understand the origin or development of numerous immune associated diseases.
Introduction
CD4 T cells and antigen presenting cells (APCs) such as DCs are required mediators of immunity to microbial pathogens1,2. In peripheral lymphoid organs, CD4 T cells are activated upon recognition of specific antigens presented by APCs3,4,5. Activated CD4 T cells proliferate and differentiate into distinct specific effector Th cells that are necessary for the development of a correct adaptive immune response6,7. Control of these processes is critical for producing an adequate adaptive defense that kills the pathogen without producing harmful tissue damage8. Th cells are defined according to the expression or production of surface molecules, transcription factors, and effector cytokines and perform essential and precise functions in response to pathogens1. Cells of the Th1 cell subset express the transcription factor T-bet and the cytokine interferon γ (IFNγ) and participate in host defense against intracellular pathogens1. Quantification of naïve CD4 T cell activation, proliferation, and Th1 differentiation is a useful means of assessing the role T cells play in an immune response.
This protocol enables in vivo analysis of the capacity of in vitro-generated BM-derived DCs to modulate the activation, proliferation, and Th1 differentiation of naïve CD4 T cells. The protocol also serves to assess the capacity of naïve CD4 T cells to be activated, induced to proliferate, and Th1 differentiated (Figure 1). This versatile protocol circumvents the inability to specifically manipulate or select the studied cell population in purely in vivo protocols. The effects of diverse molecules and treatments on DCs can be studied by using BM from genetically modified mice5 or by treating or genetically manipulating isolated BM cells9. Similarly, T cell responses can be explored by obtaining T cells for adoptive transfer from different sources or after several manipulations3,8,10.
The main advantages of this protocol are twofold. T cell activation, proliferation, and Th1 differentiation are analyzed with a flow cytometry approach; and this is combined with in vivo studies, thus averting alterations that may occur in vitro and including cell types and other factors only found in intact organs11.
The use of vital dyes is a widely used technique to track cell proliferation while avoiding the use of radioactivity. The measurement of proliferation with these reagents is based on dye dilution after cell division. Moreover, these dyes can be detected at multiple wavelengths and are easily analyzed by flow cytometry in combination with multiple fluorescent antibodies or markers. We highlight the utility of this protocol by showing how T cell activation, proliferation, and Th1 differentiation can be analyzed by flow cytometry.
Protocol
Experimental procedures were approved by the Fundación Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) and the Comunidad Autónoma de Madrid in accordance with Spanish and European guidelines. Mice were bred in specific pathogen free (SPF) conditions and were euthanized by carbon dioxide (CO2) inhalation.
1. Isolation of Mouse Bone Marrow Cells from Tibias and Femurs
NOTE: The C57BL/6 congenic mouse strain carries the differential leukocyte marker Ptprca, generally recognized as CD45.1 or Ly5.1, whereas wild-type C57BL strains carry the Ptprcb allele, known as CD45.2 or Ly5.2. CD45.1 and CD45.2 variants can be distinguished by flow cytometry using antibodies. CD45.1, CD45.2, and CD45.1/CD45.2 mice can be used as cell sources or as recipients for adoptive transfer, permitting tracing of the distinct cell populations by flow cytometry. Preferentially use age-and sex-matched male or female mice below 12 weeks of age.
- Preparation of Femurs and Tibias
- Euthanize mice using the protocol approved by the institutional animal care committee.
- Disinfect the hind limbs by spraying the animal surface with 70% ethanol.
- Use sterile scissors, forceps and scalpels. With a scalpel, make a cut in the skin and remove the skin from the distal part of the mouse including the skin covering the posterior extremities. Peel the skin around the lower calf muscle and remove the skin from the legs entirely (Figure 2A, 2B).
- Separate the quadriceps muscle from the femur using a scalpel. Disarticulate the hip joint without breaking the femur head. Remove the muscles from the tibia using a scalpel (Figure 2C, 2D). Separate the femur from the tibia without breaking the bone ends.
- Keep the bones in a Petri dish containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in ice-cold 1x Roswell Park Memorial Institute (RPMI) 1640 medium.
- Cell Isolation
NOTE: All subsequent steps must be performed under a culture hood and with sterile material to avoid contamination.- In a sterile Petri dish, carefully cut off the proximal and distal ends of each bone with a scalpel.
- Flush the bones repeatedly with a total volume of 10 mL of warm complete RPMI medium (RPMI + 10% FBS, 2 mM EDTA, 1% penicillin/streptomycin, 20 mM HEPES, 55 µM 2-mercaptoethanol, 1 mM sodium pyruvate, and 2 mM L-glutamine). Flush the bones from both ends using a 25 G needle attached to a 1 mL syringe.
- Transfer the effluate to a 50 mL conical tube fitted with a 70 µm nylon web filter. Carefully dislodge debris and cell conglomerates by gentle stirring and pipetting.
- Centrifuge the cell suspension at 250 x g for 10 min at room temperature (RT).
- Resuspend the cell pellet in 1 mL of cold red-blood-cell lysis buffer (0.15 M NH4Cl + 1 mM KHCO3 + 0.1 mM EDTA, adjusted pH to 7.2 with 1 N HCl, 0.4 µm sterile filtered, and stored at 4 °C). Maintain on ice for 5 min with the manual shaking.
- Add 10 mL of cold buffer (1x phosphate buffered saline (PBS) + 2 mM EDTA + 2% bovine serum albumin (BSA)) to inactivate and wash out the red-blood-cell lysis buffer.
- Centrifuge the cell suspension at 250 x g for 5 min at 4 °C.
- Wash cells with 25 mL of complete RPMI medium and centrifuge at 250 x g for 5 min at 4 °C.
- Resuspend the cells in 5 mL of complete RPMI medium. Mix 50 µL of the cell suspension 1:1 with trypan blue. Determine the cell number in a counting chamber under a microscope. Calculate the cell number using the formula: Cells/mL = [(non-stained cell count/4) x 2 x 10,000]12.
NOTE: This method yields 40 x 106 to 60 x 106 bone marrow cells per uninfected 8 to 12 week-old C57BL/6 mouse. - Centrifuge the cell suspension at 250 x g for 5 min at 4 °C.
2. DC Differentiation, Maturation and Antigen Loading
- BM Cell Harvesting
- Seed the complete culture on 6-well ultra-low-attachment surface plates at 106 cells per mL of medium, typically in 5 mL per well. Differentiated macrophages will attach to the culture-plates. After 12 h, transfer the supernatant, which mainly contains monocytes, to clean 6-well plates, discarding the old 6-well plates with the adhered macrophages.
- To promote cell differentiation, culture the non-adherent cells at 5 x 105 cells per mL in typically 5 mL per well of complete RPMI medium containing 20 ng/mL commercially obtained recombinant murine GM-CSF at 37 °C and 5% carbon dioxide (CO2) and observe daily.
- Every 3 days, spin down suspended cells and collect the adherent cells with 5 mM EDTA in PBS and spin down. Combine and resuspend at 5 x 105 cells/mL in fresh medium containing 20 ng/mL rm-GM-CSF.
- On the day 8, collect adherent and detached cells as above and resuspend at 5 x 105 cells/mL in medium containing 20 ng/mL GM-CSF and 20 ng/mL lipopolysaccharide (LPS) for 24 h in a non-tissue culture-treated sterile Petri dish, to induce high MHC-II, CD80 and CD86 expression.
- Check DC differentiation by flow cytometry as indicated in step 2.2.
- Flow Cytometry Differentiation and Maturation Analysis.
- Centrifuge the cell suspension at 250 x g for 5 min at 4 °C. Repeat this step twice.
- Block Fc receptors by incubating the resuspended cell pellet in mouse Fc receptor blocker (purified anti mouse CD16/CD32 IgG2b antibody) for 15 min at 4 °C according to the manufacturer's instructions.
- Centrifuge the cell suspension at 250 x g for 5 min at 4 °C.
- For each probe, resuspend 250,000 cells in 300 µL of flow cytometry buffer (1x PBS + 2 mM EDTA + 2% BSA) at 4 °C.
- Transfer 30 µL of cell solution per well to a 96-well-plate.
- Centrifuge the cell suspension at 250 x g for 5 min at 4 °C.
- Discard the supernatant and add 30 µL of antibody-containing buffer to each well following the manufacturer's indications. Incubate the cells with antibodies for 20 min at 4 °C.
NOTE: Usually employed markers for DCs, monocytes, and macrophages are CD11b, CD64, CD115, CD11c, MHCII, CD80, and CD86 (Figure 3). - Centrifuge the samples at 250 x g for 5 min at 4 °C and resuspend in 200 µL per well of cold flow cytometry buffer containing a marker to exclude dead cells (e.g., propidium iodide, 4′,6-diamidino-2-phenylindole (DAPI) or an amine-reactive dye) at the manufacturer's recommended concentration13.
- Monitor cell differentiation by transferring the samples to flow cytometry tubes and performing the flow cytometry analysis excluding dead cells on days 0, 3, 6, and 9.
- DC Antigen loading.
- On day 8, centrifuge the samples at 250 x g for 5 min at 4 °C and resuspend the pellets in 200 µL medium (RPMI + 10% FBS without antibiotics). Repeat this step twice.
- Incubate the DCs with 10 µg/mL OVAp (323-339; ISQAVHAAHAEINEAGR) per 1 x 107 DCs in 1 mL of medium (RMPI + 1% FBS) in a plastic tube for at least 30 min at 37 °C. Use non OVAp-loaded DCs as a negative control condition.
- Centrifuge the samples at 250 x g for 5 min at 4 °C and resuspend the pellets in 200 µL complete RPMI medium. Repeat this step twice.
- Centrifuge the samples at 250 x g for 5 min at 4 °C and resuspend the pellets in 200 µL medium (RPMI + 10% FBS) at 2 x 106 cells/mL.
3. Isolation of Naïve CD4 T Cells from OTII Mice
NOTE: This method yields approximately 100 x 106 spleenocytes per uninfected 8 to 12 week-old C57BL/6 mouse. Both males and females can be used. CD4 T cells can be isolated from spleen or lymph nodes; CD4 T cells account for approximately 25% and 50% of cells in these organs, respectively. Perform the following steps in sterile conditions.
- Isolate inguinal, axillary, brachial, cervical, and mesenteric lymph nodes and the spleen from OTII transgenic mice (Figure 4) and transfer them to a plastic Petri dish containing 10 mL of complete RPMI medium.
- Rinse the cell strainer with 2 mL of complete RPMI medium and remove it from the 50 mL tube. Transfer lymph nodes and spleen to a 70 µm cell strainer placed in a 50 mL tube. Homogenize using a syringe plunger (Figure 5) and add medium up to 15 mL.
- Centrifuge the samples at 250 x g for 5 min at 4 °C and resuspend the pellet in 15 mL of complete RPMI medium. Repeat this step twice.
- For spleen cell suspension, resuspend the cell pellet and lyse the red blood cells as indicated in step 1.2.5.
- Centrifuge the cell suspension at 250 x g for 5 min at 4 °C.
- Wash cells with 25 mL of complete RPMI medium and centrifuge at 250 x g for 5 min at RT.
- Resuspend the cells in 5 mL of medium. Mix 50 µL of the cell suspension 1:1 with Trypan blue. Determine the cell number using a counting chamber under a microscope. Calculate the cell number using the formula: cells/mL = [(non-stained cell count/4) x 2 x 10,000]12.
- Repeat step 2.2.2.
- Following the manufacturer's instructions, incubate the cells with 1:800 dilutions of biotinylated antibodies to CD8α, IgM, B220, CD19, MHCII (I-Ab), CD11b, CD11c, CD44, CD25, and DX5 for 30 min on ice for later negative selection of CD4 T cells.
- Following the manufacturer's instructions and based on the number of cells and the capacity of the beads, calculate the required amount of streptavidin-coated magnetic microbeads and isolate CD4 T cells using a magnetic cell separator and the appropriate separation columns.
- Resuspend the cells in 5 mL of complete RPMI medium and keep them on ice while counting live cells as described in step 3.7.
- Extract 2 x 105 cells and check the purity of the collected cells using anti CD4, CD3, B220 and CD8 fluorescent antibodies in a flow cytometer using manufacturer-recommending antibody dilutions.
- To stain isolated naïve CD4/OTII T cells, resuspend 106 cells in 500 µL buffer (1x PBS + 2 mM EDTA + 2% BSA). Prepare 500 µL buffer (1x PBS + 2 mM EDTA + 2% BSA) containing double the final manufacturer-recommended concentration of vital cell tracer dye. Mix both solutions by slowly adding the cell tracer solution to the cell suspension. Incubate the cells for 5 min at 37 °C.
- Wash the cells with 5 mL of complete RPMI medium and centrifuge at 250 x g for 5 min at 4 °C. Repeat this step twice. Resuspend the cells in 1 mL of complete RPMI medium at 1 x 107cells/mL.
4. In Vivo Activation, Proliferation and Th1 Differentiation Assay
NOTE: The vital cell tracer dye carboxyfluorescein diacetate succinimidyl ester (CFSE) and other succinimidyl ester-based dyes, which are excited and emit fluorescence at different wavelengths, are widely used to assess lymphocyte proliferation by flow cytometry due to their high fluorescence intensity, long-life, low variability, and low toxicity14.
- Adoptively transfer intravenously (i.v.) 1 x 106 live vital cell tracer dye-stained naïve CD4/OTII T cells in 100 µL of PBS per mouse. Labeled T cells can be adoptively transferred by tail vein or retro-orbital injection15; in both cases, the cells will home to lymphoid organs (Figure 6A).
- Challenge the recipient mice one day later by adoptively transferring 100,000 LPS-matured OVAp-loaded DCs by subcutaneous injection (s.i.) in the footpads (Figure 6B).
- Harvest popliteal lymph nodes from recipient mice and process them until the obtainment of single cell suspensions following the same steps as previously described in 3.1 and 3.2. Do this on day 2 post-immunization to measure CD4/OTII T cell activation, and on day 5 to measure proliferation plus Th1 differentiation.
- To analyze T cell activation on day 2, stain cells with fluorescent antibodies against CD4, CD69, and CD25 (optionally CD45.1 and CD45.2) and determine the percentage of C69+CD25+ T cells in the CD4 population. Analyze by flow cytometry (Figure 7).
- To check T cell proliferation on day 5, stain cells using appropriate fluorescent antibodies against CD4 and optionally CD45.1 and CD45.2. Analyze the decay of the vital cell tracer dye signal in the CD4 population by flow cytometry (Figure 8). Several parameters can be determined in these studies, such as the number of cell divisions undergone by cells labelled with the vital cell tracer dye, the percentage of dividing cells in each fluorescence peak, and the percentage of dividing cells in the total cell population.
- To determine Th1 differentiation at day 5, plate 400,000 cells per well of a 96-well plate in 200 µL of complete RPMI medium containing 1 µM ionomycin and 10 ng/mL phorbol 12 myristate 13 acetate (PMA) for 6 h at 37 °C in the incubator. For the last 4 h, add 3–10 µg/mL brefeldin A to block cytokine secretion to the medium.
- Centrifuge the plates for 5 min at 250 x g at 4 °C. Discard the supernatant by rapid inversion of the 96-well plate.
- Repeat step 2.2.2.
- Stain cell membranes by incubating for 15 min at 4 °C in 30 µL of buffer (1x PBS + 2 mM EDTA + 2% BSA) containing antibodies to CD4 and optionally to CD45.1 and CD45.2 at manufacturer-recommended dilutions. Wash the cells by adding 150 µL of buffer (1x PBS + 2 mM EDTA + 2% BSA) and centrifuge the plates for 5 min at 250 x g at 4 °C and discard supernatant.
- Fix the cells by adding 100 µL of 2% paraformaldehyde/1% sucrose for 20 min at RT. Centrifuge the plates for 5 min at 1,000 x g and 4 °C. Discard the supernatant.
- Permeabilize the cells by adding 50 µL of intracellular permeabilization buffer (1x PBS + 0.1% BSA, 0.01 M HEPES, 0.3% saponin) and incubate for 30 min at 4 °C.
- Add 150 µL of PBS and centrifuge for 5 min at 1,000 x g at RT. Discard the supernatant.
- Stain intracellular cytokines by adding 30 µL of fluorophore-conjugated anti IFNγ antibody in buffer (PBS + 0.1% saponin) and incubate for 30 min at RT.
- Wash the cells by adding 150 µL of buffer (PBS + 0.1% saponin). Centrifuge the plates for 5 min at 1,000 x g at RT and discard the supernatant. Repeat this step twice.
- Analyze cell populations by flow cytometry (Figure 8).
Representative Results
Figure 1 illustrates the steps described in this protocol. Figure 2 illustrates the isolation and culture of mouse BM cells. The addition of GM-CSF and LPS to these cultures allows the in vitro generation and maturation of DCs. Figure 3 illustrates the flow cytometry analysis of the differentiation and maturation of the obtained DCs. OVAp-loaded GM-CSF BM-derived DCs and isolated vital cell trace dye-stained CD4/OTII T cells are adoptively transferred to mice. Figure 4 shows the location of the lymph nodes used for the isolation of CD4/OTII T cells. Figure 5 shows a 50 mL tube fitted with the 70 µm nylon filter and a syringe plunger used to homogenize the lymph nodes and spleen. Figure 6 shows i.v. injection of the CD4/OTII T cells into the retro-orbital plexus and the s.c. injection of the OVAp-loaded GM-CSF BM-derived DCs into the footpad. CD4/OTII T cells distribute to the different lymphoid organs, whereas GM-CSF DCs migrate to the popliteal lymph nodes where GM-CSF DCs activate naïve CD4/OTII T cells by the presentation of OVAp. Naïve CD4/OTII T cells then proliferate and differentiate towards the Th1 phenotype. Figure 7 illustrates the quantification of naïve CD4/OTII T cell activation from the analysis of membrane CD69 and CD25 expression. Figure 8 shows the quantification of CD4/OTII T cell proliferation. Figure 9 illustrates the quantification of CD4/OTII T cell differentiation towards the Th1 phenotype.
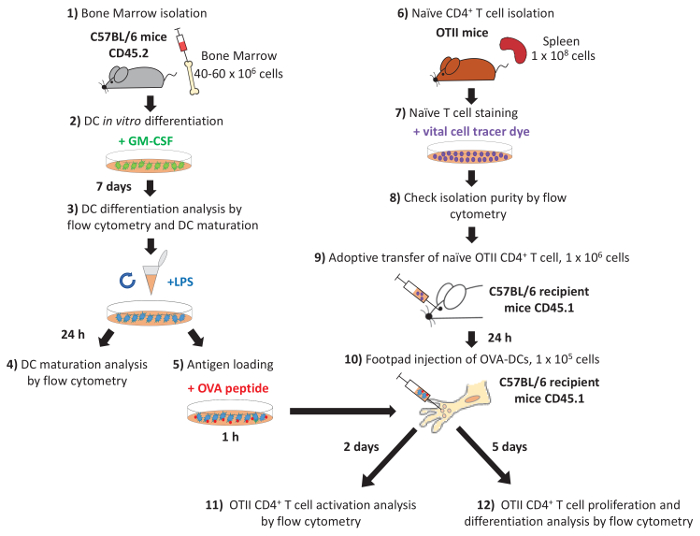
Figure 1: Protocol scheme. 1) Isolate BM cells from mouse femurs and tibias. 2) Culture BM cells with GM-CSF to generate GM-CSF BM derived-DCs. 3) Check DC differentiation and mature GM-CSF BM derived-DCs with LPS. 4) Check DC maturation. 5) Load DCs with OVAp. 6) Isolate CD4/OTII T cells form mouse spleen. 7) Stain CD4/OTII T cells with vital cell tracer dye. 8) Check isolation purity and vital cell tracer dye staining of isolated CD4/OTII T cells. 9) Adoptively transfer isolated CD4/OTII T cells i.v. to recipient mice. 10). Adoptively transfer OVAp-loaded and matured DCs s.c. to recipient mice. 11) Check CD4/OTII T cell activation. 12) Check CD4/OTII T cell proliferation and differentiation. Please click here to view a larger version of this figure.
Figure 2: Bone marrow isolation process from mouse femurs and tibias. (A) Skin incision with scissors. (B) Removal of skin from the limb. (C) Muscle incision with scissors. (D) Removal of muscle from the limb with a scalpel. Please click here to view a larger version of this figure.
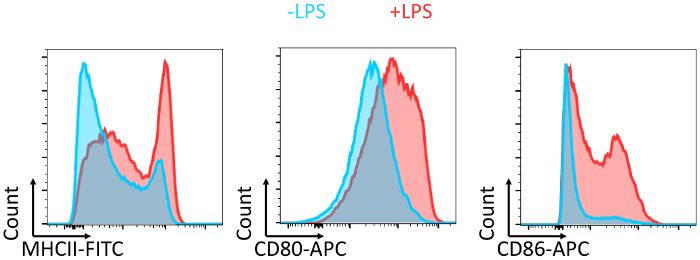
Figure 3: Flow cytometry analysis of the maturation of GM-CSF BM-derived DCs. Surface expression of MHCII, CD80, and CD86 in LPS activated (+LPS) and non-activated (-LPS) GM-CSF BM-derived DCs. Numbers show the mean fluorescence intensity in arbitrary units (a.u.). Please click here to view a larger version of this figure.
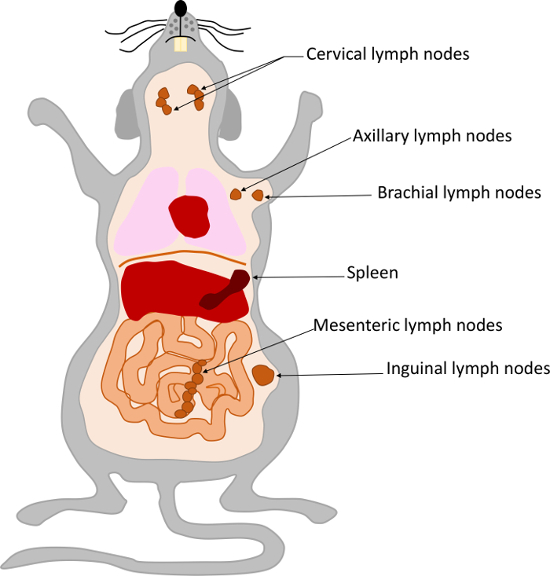
Figure 4: Location of the inguinal, axillary, brachial, cervical, and mesenteric lymph nodes and the spleen in mice. Please click here to view a larger version of this figure.
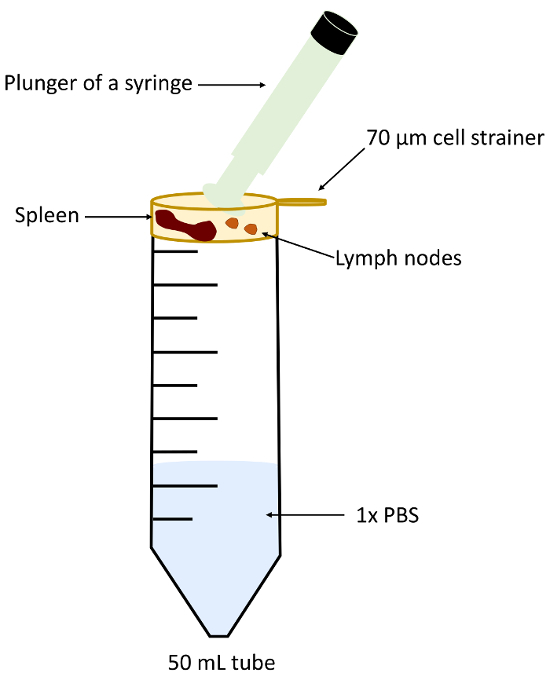
Figure 5: Experimental set up for lymph node and spleen homogenization. Please click here to view a larger version of this figure.
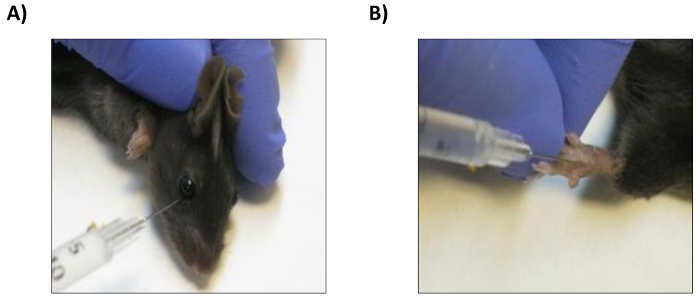
Figure 6: Adoptive transfer of CD4/OTII T cells and OVAp-loaded GM-CSF BM-derived-DCs. (A) Intravenous injection of CD4/OTII T cells into the retro-orbital plexus. (B) Subcutaneous injection of OVAp-loaded GM-CSF BM-derived DCs into the footpad. Please click here to view a larger version of this figure.
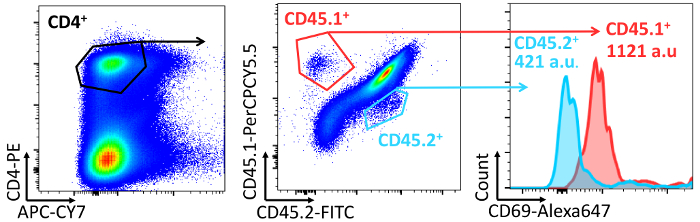
Figure 7: Quantification of T cell activation in vivo. Flow cytometry plots and graph showing the analysis and quantification of the membrane expression of CD69 in CD4 T cells. CD45.1/CD45.2 mice were adoptively transferred i.v. with mouse CD45.1/CD4/OTII and CD45.1/CD4/OTII cells 24 h before the s.c. adoptive transfer of OVAp-loaded GM-CSF BM-derived-DCs. T cell activation was measured at 2 days post-DC transfer by flow cytometry after staining with fluorescent-tagged antibodies to the indicated antigens. Please click here to view a larger version of this figure.
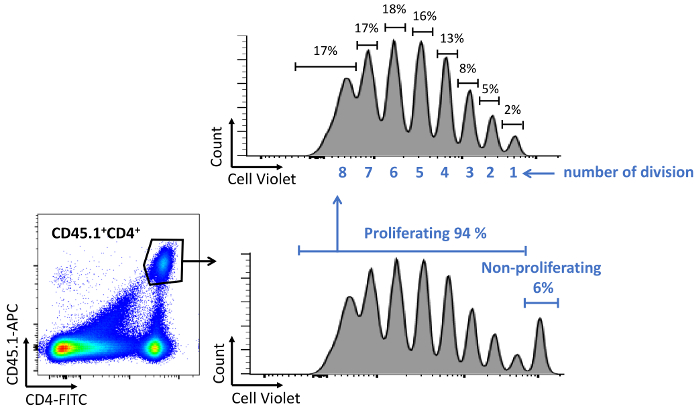
Figure 8: Quantification of T cell proliferation in vivo. Flow cytometry plots and graphs showing the analysis and quantification of the decline vital cell tracer dye staining in proliferating CD4 T cells. CD45.2 mice were adoptively transferred i.v. with mouse CD45.1/CD4/OTII cells 24 h before the s.c. adoptive transfer of OVAp-loaded GM-CSF BM-derived-DCs. T cell proliferation was measured at 5 days post-DC transfer by flow cytometry after staining with the indicated fluorescent antibodies. T cell proliferation can be determined by measuring the percentage of proliferating cells, the percentage of cells in each division peak, or by counting the number of division peaks as shown in the graphs. Please click here to view a larger version of this figure.
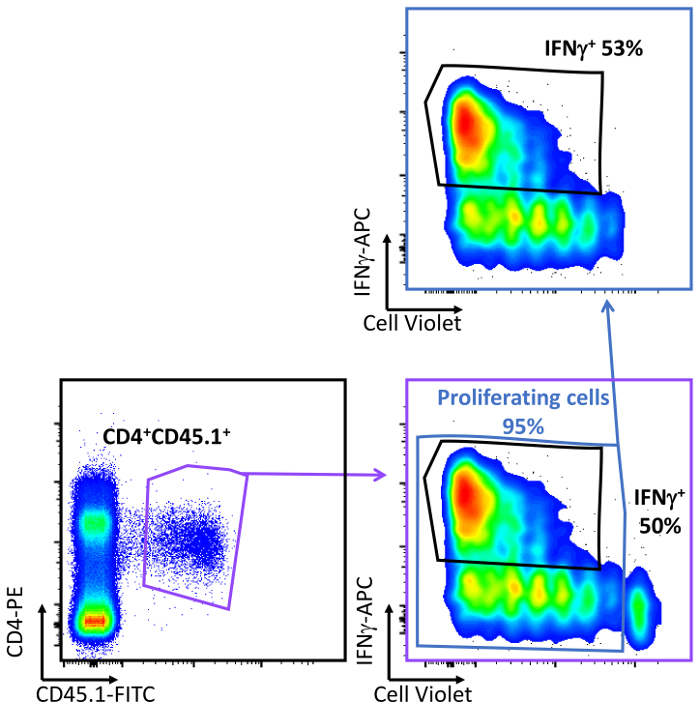
Figure 9: Quantification of T cell differentiation towards Th1 phenotype in vivo. Flow cytometry plots and graphs showing the analysis and quantification of the percentage of IFNγ-expressing CD4 T cells. CD45.2 mice were adoptively transferred i.v. with mouse CD45.1/CD4/OTII cells 24 h before the s.c. adoptive transfer of OVAp-loaded GM-CSF BM-derived-DCs. T cell differentiation was measured at days post-DC transfer by flow cytometry after staining with the indicated fluorescent antibodies. T cell differentiation towards the Th1 phenotype was determined as IFNγ-expressing cells as a percentage of total CD4 T cells and from only the proliferating CD4 T cells. Please click here to view a larger version of this figure.
Discussion
This protocol allows for the characterization of the capacity of BM-derived DCs to modulate the activation, proliferation, and differentiation of naïve CD4 T cells. Moreover, it can also be used to assess the susceptibility of CD4 T cells to modulation by BM-derived DCs. With this protocol, changes in these events can be measured in vivo.
Depending on the hypothesis under investigation, several combinations of T cells and DCs can be used. For example, one can analyze the consequences of knocking in or knocking out specific genes or the efficiency of a specific stimulus of CD4 T cell activation, proliferation, or Th1 differentiation. These procedures can be performed on DCs or CD4 T cells or on both cell types. Thus, CD4 T cells from wild-type or genetically modified mice can be combined with DCs derived from wild-type mice and vice versa.
This protocol can be adapted to distinguish the origin of the different cell populations by using flow cytometry and antibodies against CD45.1 and CD45.2. Indeed, CD45.1/CD45.1 and CD45.2/CD45.2 mice can be used as the source of any of the cell types used or as recipients for adoptive transfer in any combination. For example, CD4 T cells can be obtained from CD45.1/CD45.1/OTII mice while CD45.2/CD45.2 mice can be the origin of BM for DC generation. In this experiment, both cell types can be adoptively transferred to CD45.1/CD45.2 mice. Furthermore, CD4/OTII T cells from CD45.1/CD45.1 type one mice and CD45.2/CD45.2 type two mice can be combined in the same or different CD45.1/CD45.2 type three recipients to compare the behavior of type one and type two CD4 T cell populations in competing and non-competing conditions, respectively.
This method describes the generation of GM-CSF BM-derived DCs. However, alternative protocols are available for DC maturation and differentiation; for example, papain can be used instead of LPS or fms-related tyrosine kinase 3 ligand (Flt3-L) instead of GM-CSF, allowing study of the capacity of phenotypically distinct DCs to activate CD4 T cell adaptive immunity. With the same intention, DCs for antigen loading and presentation can be directly isolated from mice. Combination of this protocol with others9 allows the manipulation of naïve CD4/OTII T cells by viral infection16 before their adoptive transfer to recipient mice. BM can be also transduced with retroviruses9 to study the effect of controlled cell modifications on DCs before their use in the protocol.
Additionally, this protocol can be adapted for intravital microscopy studies17 by using fluorescent protein-expressing cells. For example, CD4/OTII mice can be crossed with GFP- or cherry-expressing mice to obtain GFP or Cherry/OTII CD4 mice. These mice can then be used as a source of CD4 T cells and can be combined with BM-derived DCs from cherry- or GFP-expressing mice, as required, in vivo experiments.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Simon Bartlett for English editing. This study was supported by grants from Instituto de Salud Carlos III (ISCIII) (PI14/00526; PI17/01395; CP11/00145; CPII16/00022), the Miguel Servet Program and Fundación Ramón Areces; with co-funding from the Fondo Europeo de Desarrollo Regional (FEDER). The CNIC is supported by the Ministry of Economy, Industry and Competitiveness (MEIC), and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505). RTF is founded by Fundación Ramón Areces and CNIC, VZG by ISCIII, BHF by Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12) and JMG-G by the ISCIII Miguel Servet Program and imas12.
Materials
| Ethanol | VWR Chemicals | 20,824,365 | 5 L |
| Scissors | Fine Science Tools (F.S.T.) | 14001-12 | |
| Forceps | Fine Science Tools (F.S.T.) | 11000-13 | |
| Fine Forceps | Fine Science Tools (F.S.T.) | 11253-20 | |
| Scalpel | Fine Science Tools (F.S.T.) | 10020-00 | Box of 100 blades |
| Fetal Bovine Serum | SIGMA | F7524 | |
| Penicillin/streptomycin | LONZA | DE17-602E | |
| Roswell Park Memorial Institute medium (RPMI) | GIBCO | 21875-034 | |
| Sterile Petri dishes | Falcon | 353003 | |
| 25-gauge needle | BD Microlance 3 | 300600 | |
| 1 ml syringe | Novico | N15663 | |
| 15 ml conical tubes | Falcon | 352096 | |
| 50 ml conical tubes | Falcon | 352098 | |
| 70 μm nylon web filter | BD Falcon | 352350 | |
| Red blood lysis buffer | SIGMA | R7757 | 100 Ml |
| EDTA | SIGMA | ED2SS-250 | |
| Bovine Serum Albumin | SIGMA | A7906 | 100 g |
| Trypan blue | SIGMA | 302643-25G | |
| Culture-plates | Falcon | 353003 | |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Cambrex | BE17-737E | |
| Beta-mercaptoethanol | Merck | 8-05740-0250 | |
| Sodium pyruvate | LONZA | BE13-115E | |
| L-glutamine | LONZA | BE17-605E | |
| Recombinant murine Granulocyte Macrophage colony-stimulating factor (GM-CSF) | Prepotech | 315-03 | |
| lipopolysaccharide (LPS) | SIGMA-ALDRICH | L2018 | |
| 96-well-plate | Costar | 3799 | |
| v450 anti-mouse CD11b antibody | BD Biosciences | 560455 | |
| PE anti-mouse CD64 antibody | BioLegend | 139303 | |
| PE anti-mouse CD115 antibody | BioLegend | 135505 | |
| FITC anti-mouse CD11c antibody | BioLegend | 117305 | |
| FITC anti-mouse MHCII antibody | BioLegend | 125507 | |
| APC anti-mouse CD86 antibody | BioLegend | 105011 | |
| APC anti-mouse CD80 antibody | BioLegend | 104713 | |
| Flow Cytometry tubes | Zelian | 300800-1 | PS 12X75 5mL |
| OTII Ovoalbumine peptide | InvivoGen | 323-339 | |
| anti-mouse biotinylated CD8α antibody | Tonbo Biosciences | 30-0081-U500 | |
| anti-mouse biotinylated IgM antibody | BioLegend | 406503 | |
| anti-mouse biotinylated B220 antibody | Tonbo Biosciences | 30-0452-U500 | |
| anti-mouse biotinylated CD19 antibody | Tonbo Biosciences | 30-0193-U500 | |
| anti-mouse biotinylated MHCII (I-Ab) antibody | BioLegend | 115302 | |
| anti-mouse biotinylated CD11b antibody | Tonbo Biosciences | 30-0112-U500 | |
| anti-mouse biotinylated CD11c antibody | BioLegend | 117303 | |
| anti-mouse biotinylated CD44 antibody | BioLegend | 103003 | |
| anti-mouse biotinylated CD25 antibody | Tonbo Biosciences | 30-0251-U100 | |
| anti-mouse biotinylated DX5 antibody | BioLegend | 108903 | |
| streptavidin-coated magnetic microbeads | MACS Miltenyi Biotec | 130-048-101 | |
| Magnetic cell separator | MACS Miltenyi Biotec | 130-090-976 | QuadroMACS Separator |
| Separation columns | MACS Miltenyi Biotec | 130-042-401 | |
| PE anti-mouse CD4 antibody | BioLegend | 100408 | |
| APC anti-mouse CD3 antibody | BioLegend | 100235 | |
| FITC anti-mouse CD8 antibody | Tonbo Biosciences | 35-0081-U025 | |
| Cell Violet Tracer | Thermofisher | C34557 | |
| APC anti-mouse CD25 antibody | Tonbo Biosciences | 20-0251-U100 | |
| Alexa647 anti-mouse CD69 antibody | BioLegend | 104518 | |
| PerCPCY5.5 anti-mouse CD45.1 antibody | Tonbo Biosciences | 65-0453 | |
| APC anti-mouse CD45.1 antibody | Tonbo Biosciences | 20-0453 | |
| PECY7 anti-mouse CD45.1 antibody | Tonbo Biosciences | 60-0453 | |
| FITC anti-mouse CD45.2 antibody | Tonbo Biosciences | 35-0454 | |
| Ionomycin | SIGMA-ALDRICH | I0634 | |
| Phorbol 12 Myristate 13 Acetate (PMA) | SIGMA | P8139 | |
| Brefeldin A (BD GolgiPlug) | BD | 555029 | |
| Paraformaldehyde | Millipore | 8-18715-02100 | |
| Intracellular permeabilization buffer | eBioscience | 00-8333 | |
| APC anti-mouse IFNg antibody | Tonbo Biosciences | 20-7311-U100 | |
| Fc-block (anti-mouse CD16/CD32) | Tonbo Biosciences | 70-0161-U100 | |
| B6.SJL CD45.1 mice | The Jackson Laboratory | 002014 | |
| BD LSRFortessa™ Cell Analyzer | BD Biosciences | 649225 | |
| DAPI Solution | Thermofisher | 62248 |
Referencias
- Zhu, J., Yamane, H., Paul, W. E. Differentiation of effector CD4 T cell populations (*). Annual Review of Immunology. 28, 445-489 (2010).
- Bustos-Moran, E., Blas-Rus, N., Martin-Cofreces, N. B., Sanchez-Madrid, F. Orchestrating Lymphocyte Polarity in Cognate Immune Cell-Cell Interactions. International Review of Cell and Molecular Biology. 327, 195-261 (2016).
- Gonzalez-Granado, J. M., et al. Nuclear envelope lamin-A couples actin dynamics with immunological synapse architecture and T cell activation. Science Signaling. 7 (322), ra37 (2014).
- Rocha-Perugini, V., Gonzalez-Granado, J. M. Nuclear envelope lamin-A as a coordinator of T cell activation. Nucleus. 5 (5), 396-401 (2014).
- Rocha-Perugini, V., et al. CD9 Regulates Major Histocompatibility Complex Class II Trafficking in Monocyte-Derived Dendritic Cells. Molecular and Cellular Biology. 37 (15), (2017).
- Kaech, S. M., Wherry, E. J., Ahmed, R. Effector and memory T-cell differentiation: implications for vaccine development. Nature Reviews Immunology. 2 (4), 251-262 (2002).
- Iborra, S., Gonzalez-Granado, J. M. In Vitro Differentiation of Naive CD4(+) T Cells: A Tool for Understanding the Development of Atherosclerosis. Methods in Molecular Biology. 1339, 177-189 (2015).
- Toribio-Fernandez, R., et al. Lamin A/C augments Th1 differentiation and response against vaccinia virus and Leishmania major. Cell Death & Disease. 9 (1), 9 (2018).
- Lee, T., Shevchenko, I., Sprouse, M. L., Bettini, M., Bettini, M. L. Retroviral Transduction of Bone Marrow Progenitor Cells to Generate T-cell Receptor Retrogenic Mice. Journal of Visualized Experiments. (113), (2016).
- Cruz-Adalia, A., et al. Conventional CD4(+) T cells present bacterial antigens to induce cytotoxic and memory CD8(+) T cell responses. Nature Communications. 8 (1), 1591 (2017).
- Quah, B. J., Wijesundara, D. K., Ranasinghe, C., Parish, C. R. The use of fluorescent target arrays for assessment of T cell responses in vivo. Journal of Visualized Experiments. (88), e51627 (2014).
- Cruz-Adalia, A., Ramirez-Santiago, G., Torres-Torresano, M., Garcia-Ferreras, R., Veiga Chacon, E. T Cells Capture Bacteria by Transinfection from Dendritic Cells. Journal of Visualized Experiments. (107), e52976 (2016).
- Cossarizza, A., et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. European Journal of Immunology. 47 (10), 1584-1797 (2017).
- Quah, B. J., Parish, C. R. New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE-like fluorescent dyes. Journal of Immunological Methods. 379 (1-2), 1-14 (2012).
- Yardeni, T., Eckhaus, M., Morris, H. D., Huizing, M., Hoogstraten-Miller, S. Retro-orbital injections in mice. Lab Animals (NY). 40 (5), 155-160 (2011).
- Warth, S. C., Heissmeyer, V. Adenoviral transduction of naive CD4 T cells to study Treg differentiation. Journal of Visualized Experiments. (78), (2013).
- Liou, H. L., Myers, J. T., Barkauskas, D. S., Huang, A. Y. Intravital imaging of the mouse popliteal lymph node. Journal of Visualized Experiments. (60), (2012).

