Quantitative Autoradiographic Method for Determination of Regional Rates of Cerebral Protein Synthesis In Vivo
Summary
Protein synthesis is a critical biological process for cells. In brain, it is required for adaptive changes. Measurement of rates of protein synthesis in the intact brain requires careful methodological considerations. Here we present the L-[1-14C]-leucine quantitative autoradiographic method for determination of regional rates of cerebral protein synthesis in vivo.
Abstract
Protein synthesis is required for development and maintenance of neuronal function and is involved in adaptive changes in the nervous system. Moreover, it is thought that dysregulation of protein synthesis in the nervous system may be a core phenotype in some developmental disorders. Accurate measurement of rates of cerebral protein synthesis in animal models is important for understanding these disorders. The method that we have developed was designed to be applied to the study of awake, behaving animals. It is a quantitative autoradiographic method, so it can yield rates in all regions of the brain simultaneously. The method is based on the use of a tracer amino acid, L-[1-14C]-leucine, and a kinetic model of the behavior of L-leucine in the brain. We chose L-[1-14C]-leucine as the tracer because it does not lead to extraneous labeled metabolic products. It is either incorporated into protein or rapidly metabolized to yield 14CO2 which is diluted in a large pool of unlabeled CO2 in the brain. The method and the model also allow for the contribution of unlabeled leucine derived from tissue proteolysis to the tissue precursor pool for protein synthesis. The method has the spatial resolution to determine protein synthesis rates in cell and neuropil layers, as well as hypothalamic and cranial nerve nuclei. To obtain reliable and reproducible quantitative data, it is important to adhere to procedural details. Here we present the detailed procedures of the quantitative autoradiographic L-[1-14C]-leucine method for the determination of regional rates of protein synthesis in vivo.
Introduction
Protein synthesis is an important biological process required for long-term adaptive change in the nervous system1. Inhibiting protein synthesis blocks long-term memory storage in both invertebrates and vertebrates2. Protein synthesis is essential for maintenance of the late phases of some forms of long-term potentiation (LTP) and long-term depression (LTD)3, neuronal survival during development4, and for general maintenance of the neuron and its synaptic connections5. Measurement of rates of brain protein synthesis may be an important tool with which to study adaptive changes as well as neurodevelopmental disorders and disorders related to learning and memory.
We have developed a method to quantify rates of cerebral protein synthesis in vivo in an awake animal that offers inherent advantages over other techniques that estimate rates in ex vivo or in vitro preparations of brain tissue6. Foremost is the applicability to measurements in the intact brain in an awake animal. This is a key consideration because it allows measurements with synaptic structure and function in place and without concerns about post mortem effects. Moreover, the quantitative autoradiographic approach that we employ achieves a high degree of spatial localization. Whereas the energy of 14C is such that we cannot localize the tracer at the subcellular or cellular level, we can measure rates in cell layers and small brain regions such as hypothalamic nuclei, with approximately a 25 µm resolution7.
One challenge of in vivo measurements with radiotracers is to ensure that radiolabel measured is in the product of the reaction of interest rather than unreacted labeled precursor or other extraneous labeled metabolic products6. We chose L-[1-14C]-leucine as the tracer amino acid because it is either incorporated into protein or rapidly metabolized to 14CO2, which is diluted in the large pool of unlabeled CO2 in brain resulting from the high rate of energy metabolism8. Moreover, any 14C not incorporated into protein exists primarily as free [14C]-leucine, which over the 60 min experimental period, is almost entirely cleared from the tissue6. Proteins are then fixed to tissue with formalin and subsequently rinsed with waterto remove any free [14C]-leucine before autoradiography.
Another important consideration is the issue of the dilution of the specific activity of the precursor amino acid pool by unlabeled amino acids derived from tissue proteolysis. We have shown that in adult rat and mouse, about 40% of the precursor leucine pool for protein synthesis in the brain comes from amino acids derived from protein breakdown6. This must be included in the computation of regional rates of cerebral protein synthesis (rCPS) and must be confirmed in studies in which this relationship may change. The theoretical basis and the assumptions of the method have been presented in detail elsewhere6. In this paper, we focus on the procedural issues of the application of this methodology.
This method has been employed for the determination of rCPS in ground squirrels9, sheep10, rhesus monkeys11, rats12,13,14,15,16,17,18,19,20,21, a mouse model of Tuberous Sclerosis complex22, a mouse model of fragile X syndrome23,24,25,26, fragile X premutation mice27, and a mouse model of phenylketonuria28. In this manuscript, we present the procedures for measurement of rCPS with the in vivo autoradiographic L-[1-14C]-leucine method. We present rCPS in brain regions of an awake control mouse. We also demonstrate that in vivo administration of anisomycin, an inhibitor of translation, abolishes protein synthesis in the brain.
Protocol
Note: All animal procedures were approved by the National Institute of Mental Health Animal Care and Use Committee and were performed according with the National Institutes of Health Guidelines on the Care and Use of Animals.
An overview of the protocol is presented in Figure 1.
1. Surgically implant catheters in a femoral vein and artery for administration of the tracer and collection of timed arterial blood samples, respectively. Complete surgery at least 22 h prior to administration of the tracer. Surgery requires about 1 h to complete.
- Gather necessary materials: sterile surgical instruments (surgical scissors, micro-scissors, forceps, three surgical skin hooks), equipment for isoflurane anesthesia (isoflurane vaporizer, active gas scavenger, sealed anesthesia chamber, anesthesia nose cone), sterile surgery stage, fur clippers, 70% ethanol, betadine, sterile gauze, surgical tape, commercial hand warmers, surgical microscope, sterile 0.9% sodium chloride (saline), sterile heparin 100 USP units/mL in 0.9% sodium chloride (heparinized saline), sterile five 20-cm strips of 6-0 absorbable suture, sterile 25-cm strands of PE-8 and PE-10 polyethylene catheters with one end cut at 45o, sterile 1 cc syringes, sterile 32 gauge needles, cautery equipment, sterile 15-20 cm hollow stainless steel rod (2.5 mm inside diameter, 3 mm outside diameter), local anesthetics (bupivacaine and lidocaine ointment), and an animal enclosure with swivel appendage setup (30 cm spring tether with button, swivel, swivel mount and arm, 20 X 13 cm clear cylindrical container).
- Prepare animal for surgery.
- Assure that proper aseptic and sterile techniques are used as required by your institution.
- Weigh the animal. The animal must be at least 25 g for successful surgery.
- Place the animal inside a sealed plexiglass chamber and connect the chamber to the isoflurane anesthesia apparatus. Set flow rate to 2.5 L/min for males and 3.0 L/min for females of 1.5% isoflurane in O2. After roughly 2 min, ensure the mouse is appropriately sedated by lack of a withdrawal reflex with a toe pinch.
- Once sedated, remove the mouse from the chamber and lay it in a prone position with its face inside the anesthesia nose cone. Set up nose cone to receive gas from the vaporizer and to return gas to the gas scavenger. The scavenger will capture isoflurane in a charcoal filter.
- Use clippers to shave fur between the shoulder blades. Make sure to properly sterilize the shaved region, alternating three times between betadine and ethanol scrubs.
- Flip the mouse over into a supine position keeping the face in the nose cone. Tape down the left leg onto the surgery stage and use clippers to shave fur from the left inner thigh to the upper left abdomen. Make sure to properly sterilize the shaved region, alternating three times between betadine and ethanol scrubs.
- Slide an activated commercially available handwarmer, wrapped in gauze, under the mouse. Tape down the right leg onto the surgery stage.
- Insert catheter into the left femoral vein.
- With the aid of a surgical microscope, use surgical scissors to make a 1 cm incision from the upper medial portion of the left thigh rostrally towards the midline, revealing the femoral artery and vein.
- Retract loose skin with surgical skin hooks to further expose the vesicles.
- Apply sterile 0.9% sodium chloride to exposed area to maintain adequate moisture.
- Use forceps to blunt dissect, separating connective tissue around a small section of the femoral artery and vein. Carefully, separate the artery and vein (Figure 2).
- Use forceps to thread one strand of absorbable suture (Strand A) under both the femoral vein and artery at the most lateral point of the incision. Pull the suture halfway through so the ends are even.
- At a more proximal point to the groin, use forceps to thread a second suture (Strand B) under only the femoral vein. Gently tie a half knot that will be used to restrict blood flow.
- At a point between Strand A and Strand B, use forceps to thread a third suture (Strand C) under only the femoral vein. Gently tie a full knot that will be used to restrict blood flow. Be careful not to tear the vein.
- Gently tug on Strand B to restrict blood flow. Use a hemostat to gently pull Strand B to maintain restricted blood flow.
- Cut a small hole in the restricted area of the femoral vein with microscissors and carefully insert the angled end of the PE-8 tubing (previously flushed with heparin saline) towards Strand B. Once inserted, release Strand B’s tension and guide the catheter further up the vein. Tighten Strand B around the vein containing the catheter.
- Using Strand C, tie an additional knot around the catheter. Make sure this knot does not capture the femoral artery.
- Gently pull back on the syringe barrel to partially fill the tubing with blood to ensure that the catheter has been implanted properly.
- Following the same procedure, insert a PE-10 catheter into the left femoral artery.
- Complete surgical procedure.
- Once both femoral vein and artery catheters have been secured, tie Strand A into a knot around both catheters.
- Cut all excess sutures and remove skin hooks. Flush the arterial catheter with heparinized saline to prevent clotting. Cauterize the ends of both catheters to create a seal.
- Place the mouse in the prone position and make a small incision at the base of the neck and apply saline to the exposed area.
- Insert hollow metal rod subdermally from the neck incision to the femoral incision. Snake the catheters through the hollow rod and out of the neck incision. Remove the hollow rod. Implanting catheters subdermally will prevent mice from damaging the catheters.
- Apply bupivacaine to the sides of the wound. Close the femoral incision with suture. Add lidocaine ointment over closed, sutured wound.
- Snake the catheters through a 30-cm flexible hollow tube (spring tether) and suture the button of the spring tether under the skin. Apply bupivacaine to the sides of the wound. Suture the button of the spring tether under the skin. Add lidocaine ointment over closed, sutured wound.
- Move the mouse into a clear cylindrical container (20 cm high, 13 cm diameter) with a swivel mount and arm to house the animal during the recovery period. Place a hand warmer under the container to keep the animal warm.
- Screw the top of the spring tether to a swivel and secure the swivel to the swivel arm attached to the cylindrical container. Make sure the mouse has full range of motion and the catheters can be accessed.
- Place a slotted lid that does not interfere with the swivel mechanism over the animal enclosure. Refer to Figure 3 for the full setup.
- Allow the mouse to recover. At least 22 h is recommended.
2. Prepare L-[1-14C]leucine solution for injection and 16% (w/v) 5-sulfosalicylic acid (SSA) dihydrate solution for deproteinizing plasma samples. In the SSA solution, also include 0.04 mM norleucine and 1 µCi/mL [H3]leucine as internal standards for amino acid analysis and analysis of tracer concentration in the acid-soluble plasma fractions, respectively. Store the SSA up to two months at 4 ˚C.
- Purchase commercially available L-[1-14C]leucine (50-60 mCi/mmol), which is sold as a solution in 2% ethanol or 0.1 N HCl. Blow dry a known activity of the tracer under a gentle stream of nitrogen and reconstitute in a solution of sterile normal saline made up to a concentration of 100 μCi/mL.
3. Administer L-[1-14C]leucine intravenously and collect arterial blood samples.
- Gather necessary materials: 18 1.5 mL microtubes for deproteinizing plasma samples (add 70 mL of deionized water to each tube), 17 250mL glass vial inserts (15 inserts for collection of arterial blood samples and 2 inserts for collection of dead space blood to be reinjected. To limit the collection of extra and unnecessary blood, small, thin glass vial inserts with tapered bottoms that allow for accessible pipetting of supernatant plasma is recommended), 2 microcapillary tubes (32 X 0.8mm, for hematocrit measurement), 1 heparin and lithium fluoride-coated microcentrifuge tube (to prevent clotting and glycolysis, respectively), hemostats (cover the tips with tygon tubing so that clamps will not damage PE tubing), blood glucose monitor, blood pressure transducer, 1 mL sterile syringes (for saline flushes), and commercially available euthanasia solution (diluted 1:1 in deionized water (for mice)).
- Ensure the animal is in a normal physiological state at the outset of the experiment.
- Clamp the arterial tubing about 2 cm from the end and cut off the tip, creating an opening for blood to flow. Then unclamp tubing and collect dead space blood ((c. 30 mL) to collect any residual saline and/or blood from previous draws), and, in a separate tube, collect a control sample (approx. 30 µL), hematocrit samples (about half of the capillary tube volume), and a glucose sample (approx. 20 µL).
- Measure hematocrit by plugging one end with sealant putty and centrifuge for 1 min at 4500 x g. Measure the ratio of the volume of red cells to the total blood volume. If an animal has a hematocrit below 30%, do not continue the study.
- Use a commercially available blood glucose monitor to measure the glucose level in a drop of blood.
- Centrifuge control sample for 2 min at 18,000 x g to separate plasma. Deproteinize plasma samples as follows: add 5 µL of plasma to 70 µL of deionized water in a 1.5 mL microtube, add 25 µL of the 16% SSA solution and vortex. Place on ice for 30 min before freezing on dry ice.
- Return dead space blood to the animal through the venous line, followed by a heparinized saline flush to prevent excess blood loss.
- Connect the arterial line to a blood pressure transducer to measure mean arterial blood pressure.
- After taking the samples be sure to re-clamp the arterial line and to flush the line with a small (50 mL) volume of heparinized saline.
- Administer tracer intravenously and collect timed arterial blood samples.
- Use a Y-connector to attach one syringe with the tracer (100µCi/kg) and one syringe with 50 mL sterile saline to flush the venous line after injection of tracer. Connect Y-connector to the venous line.
- Initiate the study by simultaneously starting a stop watch and injecting the tracer. Flush the venous line with saline (c. 100mL) immediately following injection.
- Collect blood samples 1-7 continuously throughout the first 2 min of the experiment in the same manner. After collecting the 7th sample, collect 30 µL dead-space blood before each remaining sample. Samples 8-14 are collected at 3, 5, 10, 15, 30, 45, and 60 min, respectively.
- Process blood samples immediately after collection, as was described for the control sample. If there is a delay, place the samples on ice. Carefully reinject dead space blood into the artery via the arterial catheter and flush with heparin saline.
- At some point during the experiment, process three internal standards by adding 25 µL 16% SSA, 0.04 mM norleucine, and 1 mCi/mL [3H]leucine to 75 µL water, vortex and place on ice.
- After collecting the 14th sample at 60 min, inject approximately 0.2 mL of B-euthanasia-D into the venous line to euthanize the animal. Record the time of death.
- Unscrew the animal from the swivel mount and remove from the animal enclosure. Carefully remove the brain, place on aluminum foil, and freeze on dry ice. Do not freeze brains with liquid nitrogen as brains may crack. Store brain, samples, and internal standards at -80 ˚C until ready for processing. Processing can be performed at any point afterwards.
4. Analyze concentrations of leucine and L-[1-14C]leucine in plasma samples.
- Thaw samples and internal standards on ice, vortex, and centrifuge 18,000 x g for 5 min at 2 ˚C. The supernatant fraction will contain the free labeled and unlabeled leucine.
- Transfer 40 µL of the supernatant to a liquid scintillation vial and add scintillation cocktail. Quantify disintegrations per min (DPM) of 3H and 14C by means of liquid scintillation counting and a quench curve designed for simultaneous double-label (3H and 14C) counting.
- To quantify plasma leucine concentrations, use an HPLC system with a sodium cation exchange column and post-column derivatization with o-phthaldehyde and fluorometric detection.
- Set HPLC to the following specifications: fluorometer excitation of 330 nm and emission of 465 nm. The mobile phase consists of sodium eluant, pH 7.40, and sodium eluant + 5% sulfolane, pH 3.15. Set the buffer flow rate of 0.400 mL/min and the derivatization instrument flow rate to 0.300 mL/min. Set the column temperature to 48 ˚C and reactor temperature to 45 ˚C.
- Calibrate the system with a range of amino acid concentrations (including norleucine) between 30 and 500 pmol/10mL. The calibration curve is linear. The amino acid concentrations of the tested 10 mL injection samples fall within the ranges of this calibration curve.
5. Perform quantitative autoradiography.
- Prepare brain sections 20 µm in thickness for autoradiography. Section brain by means of a cryostat at -20 ˚C.
- Thaw mount serial brain sections on gelatin-coated slides. Air dry.
- Wash slides in five changes of 10% formalin for 30 min per change, followed by a continuous flow of deionized water for 1 h. Cover the slides loosely with foil to avoid dust and allow to dry for 24 h.
- Arrange slides in an X-ray film cassette (cassettes that fit 20X25 cm mammography films are recommended) along with a set of [14C]methylmethacrylate standards, which were previously calibrated against tissue of known 14C concentrations as described29. Standards can be commercially purchased but ensure that they cover a range of 2-300 mCi/g of tissue and are calibrated against 20 µm tissue thickness. Under red safelight, place a piece of mammography film, emulsion side down, on top of the sections.
- Seal the cassettes and place in a black changing bag and store in a cabinet for 40-45 days.
- Develop films according to manufacturer’s directions. Note: Automated film development is not recommended because the background may be uneven and can affect quantification.
6. Analyze images.
Note: A commercially available program for image analysis coupled with a CCD camera and a fluorescent light box with even illumination is recommended. The relative optical densities in the illuminated film are detected by the CCD camera.
- Construct a calibration curve of optical density (OD) v. tissue 14C concentration based on the ODs of the set of calibrated standards on the film. Fit these data (including the blank or background) to a polynomial equation. Either a second or third degree polynomial equation fits very well.
- To analyze specific brain regions, locate the region of interest (ROI) in six to eight sections by comparison with a brain atlas. Record the ODs of the pixels within a ROI in all sections and, based on the calibration curve, compute the tissue 14C concentration in each pixel. Compute the average tissue 14C concentration in the ROI.
7. Computation of rCPS. Compute rCPS in each ROI by means of the following equation:
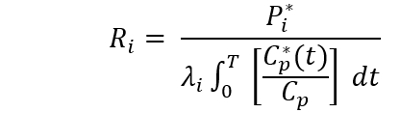
Where P*(T) is the weighted average tissue concentration of 14C in the ROI, Cp(t) and C*p(t) are the arterial plasma concentrations of unlabeled and labeled leucine at time, t, T is the time that the animal died (about 60 min), and λ is the fraction of leucine in the tissue precursor pool that comes from the plasma. Evaluation of λ is carried out in a separate experiment 6. λ has been evaluated in WT, Fmr1 knockout, Tsc+/-, and PKU mice 6,22,25,28. If an experiment involves either genetic or pharmacological changes that might affect rates of protein synthesis, degradation, or metabolism of leucine, λ should be evaluated under the new conditions.
Representative Results
Here we show a representative experiment demonstrating the effects of prior administration of a protein synthesis inhibitor on rCPS. Anisomycin in normal saline was administered to an adult C57/BL6 male wild-type mouse subcutaneously (100 mg/kg) 30 min prior to initiation of rCPS determination. Effects of anisomycin treatment compared to a vehicle-treated control animal show that rCPS is almost undetectable in the anisomycin-treated mouse (Figure 4). These data represent a validation that the in vivo autoradiographic L-[1-14C]-leucine method measures rates of protein synthesis in brain.
We present a figure of L-[1-14C]-leucine autoradiograms at four levels of the brain to demonstrate the resolution of the method (Figure 5). Illustrated are the cell layers in the olfactory bulb (Figure 5A and B), the hippocampus (Figure 5C), and the cerebellum (Figure 5G). Nuclei in the hypothalamus (Figure 5D), the pons (Figure 5E and F), and the brain stem (Figure 5H) are also clearly seen in the autoradiograms. We also show the quantitative regional rates of protein synthesis in the frontal cortex (5.88 nmol/g/min) (Figure 6A) and dorsal hippocampus (5.35 nmol/g/min) (Figure 6B) of a typical control animal.
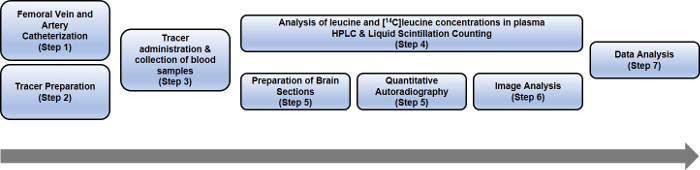
Figure 1: Schematic representing the steps of the entire rCPS protocol. Please click here to view a larger version of this figure.
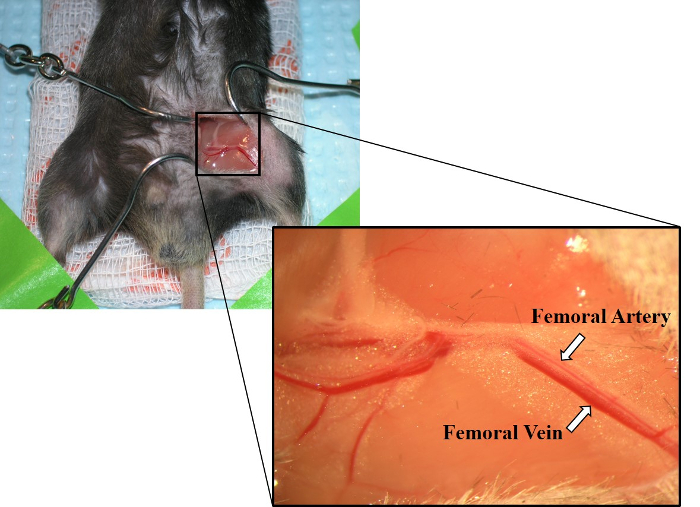
Figure 2: Image of exposed femoral artery and femoral vein. Laying parallel to one another, the femoral artery is shown above the femoral vein. The femoral vein also has a deeper red color than the femoral artery. Please click here to view a larger version of this figure.

Figure 3: Image of recommended animal enclosure set-up for rCPS experiment. It utilizes a clear cylindrical animal enclosure with swivel appendage connected to a spring tether. Please click here to view a larger version of this figure.
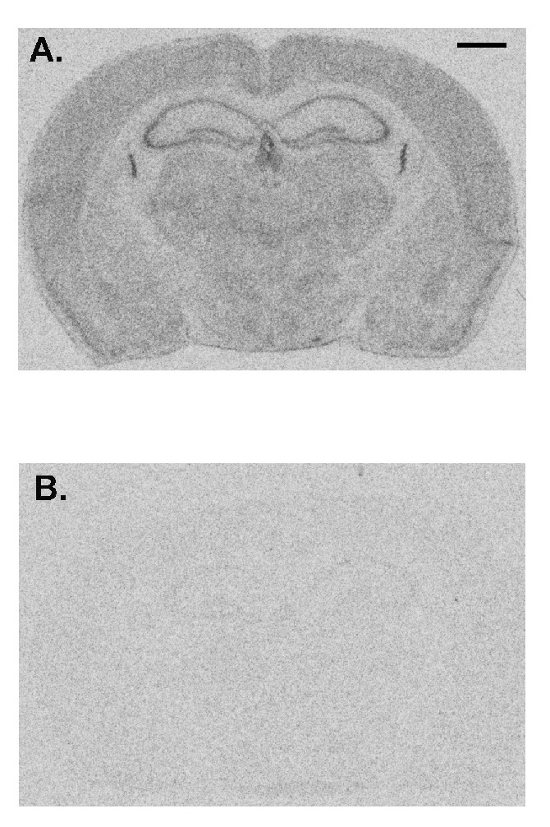
Figure 4: Representative images from a vehicle-treated animal (A) compared with an animal treated with anisomycin (100 mg/kg, subcutaneously) 30 min prior to administration of tracer (B). Rates of protein synthesis are proportional to the level of darkness in the image. Anisomycin drastically reduces the measured rates of protein synthesis indicating the specificity of this method. The scale bar in the upper right of A represents 1 mm and applies to both images. Please click here to view a larger version of this figure.
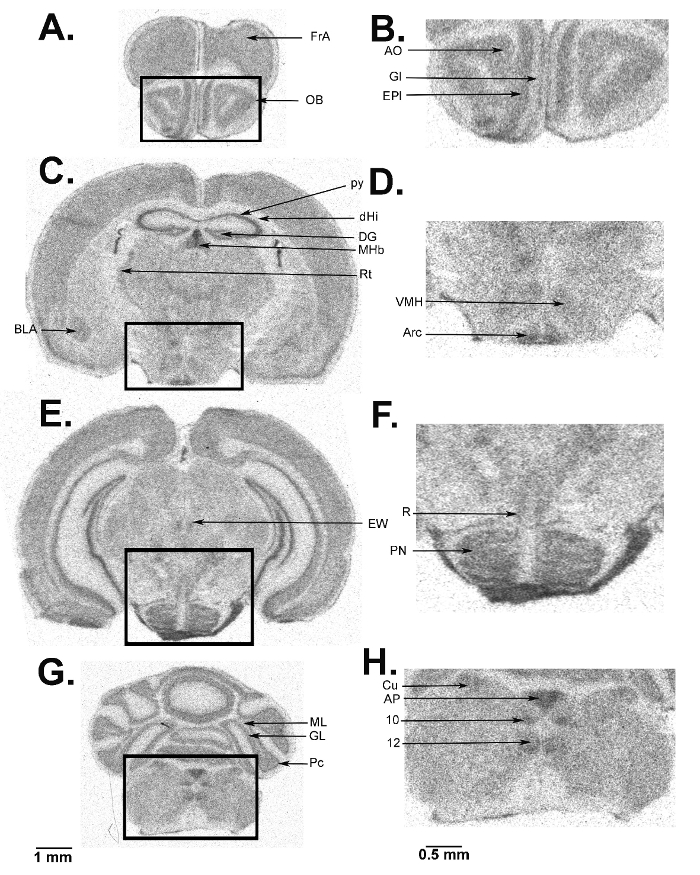
Figure 5: Digitized autoradiograms from an awake behaving mouse at the level of the olfactory bulb (A, B), hypothalamus (C, D), pons (E, F), cerebellum (G), and brain stem (G, H). The darker regions have higher rCPS. The scale bar in panel G applies to panels A, C, E, and G. Autoradiograms on the right (B, D, F, and H) are enlarged images from the areas designated on the images on the left and the scale bar in panel H applies to panels B, D, F, and H. Abbreviations are as follows: FrA, frontal association cortex; OB, olfactory bulb; AO, anterior olfactory nucleus; Gl, glomerular layer; EPl, external plexiform layer; BLA, basolateral amygdala; py, pyramidal cell layer; dHi, dorsal hippocampus; DG, dentate gyrus; MHb, medial habenula; Rt, thalamic reticular nucleus; VMH, ventral medial hypothalamic nucleus; Arc, arcuate nucleus; EW, Edinger-Westphal nucleus; R, red nucleus; PN, pontine nucleus; ML, molecular layer; GL, granular layer; Pc, Purkinje cell layer; Cu, cuneate nucleus; AP, area postrema; 10, dorsal motor nucleus of the vagus; 12, hypoglossal nucleus. Please click here to view a larger version of this figure.
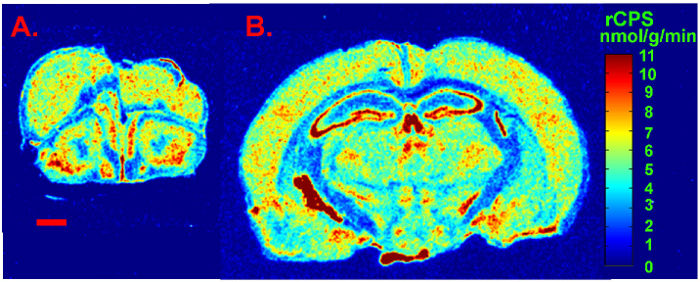
Figure 6: Digitized autoradiograms from an awake behaving control mouse at the level of the frontal cortex (A) and dorsal hippocampus (B). Rates of cerebral protein synthesis are color coded in the images according to the color bar shown on the right. The scale bar in the lower left of A represents 1 mm and applies to both images. Please click here to view a larger version of this figure.
Discussion
We present a quantitative method for determination of regional rates of cerebral protein synthesis (rCPS) in vivo in experimental animals. This method has considerable advantages over existing methods: 1. Measurements are made in the awake behaving animal, so they reflect ongoing processes in the functioning brain. 2. Measurements are made by means of quantitative autoradiography affording the ability to determine rCPS in all regions and subregions of the brain simultaneously. 3. The kinetic model of the method takes into account the possibility of recycling of unlabeled amino acids derived from tissue protein degradation and its effect on the precursor pool for protein synthesis6.
The primary limitation of this method is that it is time-consuming and demanding. Whereas it is tempting to employ simpler and higher throughput methods, the limitations of data obtained must be acknowledged.
Because of the complexity of measuring rCPS in an intact mouse, problems with maintenance of a normal physiological state, collecting adequate blood samples, and avoiding possibly interfering conditions may be encountered. Surgical implantation of the venous and arterial catheters is challenging. As with any surgical procedure, especially with the handling of delicate vasculature, there is an inherent risk for mortality of the animal. For us, it is rare (about 1%). During the ensuing 22 h recovery period, occasionally (about 4%) an animal will pull a catheter out. During the measurement, it is important that catheters are patent and that animals are in a normal physiological state. In our recent experience, arterial blood cannot be collected in about 2% of animals and about 1% of animals had a low hematocrit (< 40%) or low arterial blood pressure (< 85 mm Hg), suggesting blood loss during surgery and/or recovery.
In the preparation of brain sections for autoradiography, it is important to ensure that section thickness is 20 µm because that is the section thickness to which [14C]methylmethacrylate standards have been calibrated. Use care to ensure good quality sections, i.e., without tears, folds, or bubbles as these imperfections will interfere with the autoradiographic analysis. We develop autoradiographic films by hand rather than in an automated film processor because we find that background optical density can be uneven following automated processing, and this can affect the quantification.
In the equation for rCPS, we include a factor, lambda (λ), that is the fraction of leucine that comes from the arterial plasma, the remainder comes from the recycling of amino acids derived from tissue protein degradation6. We have evaluated λ in separate experiments in WT and Fmr1 KO (fragile X model) C57Bl/6J mice and shown that its value is 0.603. The value of λ may vary depending on species, genetic background, or the presence of a genetic mutation. Therefore, if designing protein synthesis experiments for other models, one will need to evaluate λ before an accurate measurement can be obtained.
Our work in genetic mouse models of neurodevelopmental disorders demonstrates that this methodology reveals changes in rCPS in these models and in some cases responses to pharmacological treatments22,23,25,26. It is also conceivable that the rCPS measurement may also monitor degenerative changes in brain in conditions such as models of Alzheimer's disease, Parkinson's disease, fragile X tremor ataxia syndrome, traumatic brain injury, etc. In these models, it might be possible to track early degenerative changes and possibly also responses to early interventions. The rCPS method can be used together with immunohistochemistry in parallel sections to further examine specific brain changes25. In summary, the quantitative autoradiographic L-[1-14C]-leucine method is ideal for accurate determination of rCPS values in vivo. It offers considerable advantages in terms of accuracy and applicability to in vivo conditions over existing methods.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Zengyan Xia for the genotyping of the mice, Tom Burlin for the processing of amino acids and films, and Mei Qin for performing some of the rCPS experiments. This research was supported by the Intramural Research Program of the NIMH, ZIA MH00889. RMS was also supported by an Autism Speaks Postdoctoral Fellowship 8679 and a FRAXA Postdoctoral Fellowship.
Materials
| Mice | The Jackson Laboratory | 003024 | Fmr1 knockout breeding pairs |
| Anisomycin | Tocris Bioscience | 1290 | |
| Microhematocrit Tubes | Drummond Scientific | 1-000-3200-H | capillary tubes |
| Critoseal Capillary Tube Sealant | Leica Microsystems | 39215003 | sealant putty |
| Glass vial inserts | Agilent | 5183-2089 | used to collect blood samples |
| Digi-Med Blood Pressure Analyzer | Micro-Med Inc. | BPA-400 | blood pressure analyzer |
| Bayer Breeze 2 Blood Glucose Monitoring System | Bayer Breeze | 9570A | glucose meter |
| Gastight syringe | Hamilton Co. | 1710 | tuberculin glass syringe |
| HeatMax HotHands-2 Hand Warmers | HeatMax | Model HH2 | warming pads |
| Heparin Lock Flush Solution | Fresenius Kabi USA, LLC | 504505 | heparin saline |
| Clear animal container | Instech | MTANK/W | animal enclosure |
| Spring tether | Instech | PS62 | catheter tube/rodent attachment |
| Swivel | Instech | 375/25 | hooks to spring tether |
| Swivel arm and mount | Instech | SMCLA | hooks to swivel and animal enclosure |
| Tether button | Instech | VAB62BS/22 | attaches to bottom of spring tether |
| Stainless steel tube | Made in-house | N/A | used to snake catheters through mouse |
| Matrx VIP 3000 | Matrx | 91305430 | isoflurane vaporizer |
| Isoflurane | Stoelting Co. | 50207 | isoflurane/halothane adsorber |
| Clippers | Oster Finisher | Model 59 | |
| Surgical skin hooks | Made in-house (??) | N/A (??) | |
| 0.9% Sodium Chloride Saline | APP Pharmaceuticals LLC | 918610 | |
| Forceps | Fine Science Tools | 11274-20 | |
| Surgical scissors | Fine Science Tools | 14058-11 | |
| Microscissors | Fine Science Tools | 15000-00 | |
| UNIFY silk surgical sutures | AD Surgical | #S-S618R13 | 6-0 USP, non-absorbable |
| PE-8 polyethylene tubing | SAI Infusion Technologies | PE-8-25 | |
| Syringe | Becton Dickinson and Co. | 309659 | 1cc/mL |
| PE-10 polyethylene tubing | Clay Adams | 427400 | |
| MCID Analysis | Imaging Research Inc. | Version 7.0 | optical density analysis |
| Gelatin-coated slides (75x25mm) | FD Neurotechnologies | PO101 | |
| Cryostat | Leica | CM1850 | |
| Super RX-N medical x-ray film | Fuji | 47410-19291 | |
| Hypercassettes (8×10 in) | Amersham Pharmacia Biotech | 11649 | |
| [1-14C]leucine | Moravek | MC404E | |
| Microcentrifuge tube | Sarstedt Aktiengesellschaft & Co. | 72.692.005 | used to deproteinize blood samples |
| Glass pasteur pipette | Wheaton | 357335 | |
| Glass wool | Sigma-Aldrich | 18421 | |
| Nitrogen | NIH Supply Center | 6830009737285 | |
| Scintillation fluid | CytoScint | 882453 | |
| Liquid scintilllation counter | Packard Tri-Carb | 2250CA | |
| Amino acid analyzer | Pickering Laboratories | Pinnacle PCX | |
| HPLC unit | Agilent Technologies | 1260 Infinity | include 1260 Bio-Inert Pump |
| Surgical microscope | Wild Heerbrugg | M650 | |
| Sulfosalicylic acid | Sigma-Aldrich | MKBS1634V | 5-sulfosalicylic acid dihydrate |
| Norleucine | Sigma | N8513 | |
| 1.0 N HCl | Sigma-Aldrich | H9892 | |
| [H3]leucine | Moraevk | MC672 | |
| Falcon tube | Thermo Scientific | 339652 | 50 mL conical centrifuge tubes |
| Stopwatch | Heuer Microsplit | Model 1000 | 1/100 min |
| Euthanasia Solution | Vet One | H6438 | |
| Northern Light Precision Illuminator | Imaging Research Inc. | Model B95 | fluorescent light box |
| Micro-NIKKOR 55mm f/2.8 | Nikon | 1442 | CDD camera |
Referencias
- West, A. E., et al. Calcium regulation of neuronal gene expression. Proceedings of the National Academy of Sciences of the United States of America. 98, 11024-11031 (2001).
- Siegel, G., Agranoff, B., Albers, R. W., Fisher, S., Uhler, M. . Basic Neurochemistry. , (1999).
- Nguyen, P. V., Abel, T., Kandel, E. R. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 265, 1104-1107 (1994).
- Mao, Z., Bonni, A., Xia, F., Nadal-Vicens, M., Greenberg, M. E. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 286, 785-790 (1999).
- Pfeiffer, B. E., Huber, K. M. Current advances in local protein synthesis and synaptic plasticity. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 26, 7147-7150 (2006).
- Smith, C. B., Deibler, G. E., Eng, N., Schmidt, K., Sokoloff, L. Measurement of local cerebral protein synthesis in vivo: influence of recycling of amino acids derived from protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 85, 9341-9345 (1988).
- Schmidt, K. C., Smith, C. B. Resolution, sensitivity and precision with autoradiography and small animal positron emission tomography: implications for functional brain imaging in animal research. Nuclear Medicine and Biology. 32, 719-725 (2005).
- Banker, G., Cotman, C. W. Characteristics of different amino acids as protein precursors in mouse brain: advantages of certain carboxyl-labeled amino acids. Archives of Biochemistry and Biophysics. 142, 565-573 (1971).
- Frerichs, K. U., et al. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proceedings of the National Academy of Sciences of the United States of America. 95, 14511-14516 (1998).
- Abrams, R. M., Burchfield, D. J., Sun, Y., Smith, C. B. Rates of local cerebral protein synthesis in fetal and neonatal sheep. The American Journal of Physiology. 272, R1235-R1244 (1997).
- Nakanishi, H., et al. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. The European Journal of Neuroscience. 9, 271-279 (1997).
- Sun, Y., Deibler, G. E., Sokoloff, L., Smith, C. B. Determination of regional rates of cerebral protein synthesis adjusted for regional differences in recycling of leucine derived from protein degradation into the precursor pool in conscious adult rats. Journal of Neurochemistry. 59, 863-873 (1992).
- Scammell, T. E., Schwartz, W. J., Smith, C. B. No evidence for a circadian rhythm of protein synthesis in the rat suprachiasmatic nuclei. Brain Research. 494, 155-158 (1989).
- Smith, C. B., Eintrei, C., Kang, J., Sun, Y. Effects of thiopental anesthesia on local rates of cerebral protein synthesis in rats. The American Journal of Physiology. 274, E852-E859 (1998).
- Sun, Y., Deibler, G. E., Smith, C. B. Effects of axotomy on protein synthesis in the rat hypoglossal nucleus: examination of the influence of local recycling of leucine derived from protein degradation into the precursor pool. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 13, 1006-1012 (1993).
- Smith, C. B., Yu, W. H. Rates of protein synthesis in the regenerating hypoglossal nucleus: effects of testosterone treatment. Neurochemical Research. 19, 623-629 (1994).
- Orzi, F., Sun, Y., Pettigrew, K., Sokoloff, L., Smith, C. B. Effects of acute and delayed effects of prior chronic cocaine administration on regional rates of cerebral protein synthesis in rats. The Journal of Pharmacology and Experimental Therapeutics. 272, 892-900 (1995).
- Nadel, J., et al. Voluntary exercise regionally augments rates of cerebral protein synthesis. Brain Research. 1537, 125-131 (2013).
- Sun, Y., et al. Rates of local cerebral protein synthesis in the rat during normal postnatal development. The American Journal of Physiology. 268, R549-R561 (1995).
- Smith, C. B., Sun, Y., Sokoloff, L. Effects of aging on regional rates of cerebral protein synthesis in the Sprague-Dawley rat: examination of the influence of recycling of amino acids derived from protein degradation into the precursor pool. Neurochemistry International. 27, 407-416 (1995).
- Ingvar, M. C., Maeder, P., Sokoloff, L., Smith, C. B. The effects of aging on local rates of cerebral protein synthesis in rats. Monographs in Neural Sciences. 11, 47-50 (1984).
- Sare, R. M., Huang, T., Burlin, T., Loutaev, I., Smith, C. B. Decreased rates of cerebral protein synthesis measured in vivo in a mouse model of Tuberous Sclerosis Complex: unexpected consequences of reduced tuberin. Journal of Neurochemistry. 145, 417-425 (2018).
- Liu, Z. H., Huang, T., Smith, C. B. Lithium reverses increased rates of cerebral protein synthesis in a mouse model of fragile X syndrome. Neurobiology of Disease. 45, 1145-1152 (2012).
- Qin, M., et al. Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 33, 499-507 (2013).
- Qin, M., Kang, J., Burlin, T. V., Jiang, C., Smith, C. B. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 25, 5087-5095 (2005).
- Qin, M., et al. R-Baclofen Reverses a Social Behavior Deficit and Elevated Protein Synthesis in a Mouse Model of Fragile X Syndrome. The International Journal of Neuropsychopharmacology. 18, pyv034 (2015).
- Qin, M., et al. Cerebral protein synthesis in a knockin mouse model of the fragile X premutation. ASN Neuro. 6, (2014).
- Smith, C. B., Kang, J. Cerebral protein synthesis in a genetic mouse model of phenylketonuria. Proceedings of the National Academy of Sciences of the United States of America. 97, 11014-11019 (2000).
- Reivich, M., Jehle, J., Sokoloff, L., Kety, S. S. Measurement of regional cerebral blood flow with antipyrine-14C in awake cats. Journal of Applied Physiology. 27, 296-300 (1969).

