Analysis of Shear Flow-induced Migration of Murine Marginal Zone B Cells In Vitro
Summary
Marginal zone B cells (MZBs) respond to the force of shear flow by re-orienting their migration path up the flow. This protocol shows how to record and analyze the migration using a fluidics unit, pump, microscope imaging system, and free software.
Abstract
Marginal zone B cells (MZBs) are a population of B cells that reside in the mouse splenic marginal zones that envelop follicles. To reach the follicles, MZBs must migrate up the shear force of blood flow. We present here a method for analyzing this flow-induced MZB migration in vitro. First, MZBs are isolated from the mouse spleen. Second, MZBs are settled on integrin ligands in flow chamber slides, exposed to shear flow, and imaged under a microscope while migrating. Third, images of the migrating MZBs are processed using the MTrack2 automatic cell tracking plugin for ImageJ, and the resulting cell tracks are quantified using the Ibidi chemotaxis tool. The migration data reveal how fast the cells move, how often they change direction, whether the shear flow vector affects their migration direction, and which integrin ligands are involved. Although we use MZBs, the method can easily be adapted for analyzing migration of any leukocyte that responds to the force of shear flow.
Introduction
Immune cells are the most motile cells in the human body and often must contend with shear force from blood and lymph flow. However, there are comparatively few studies on shear force-induced migration of leukocytes1,2,3,4,5. We present here a reliable and quantitative protocol to analyze the response of an immune cell to flow in vitro. Performing the assay does not require fabrication of components, and all equipment and consumables are commercially available. The protocol, including cell purification and migration analysis, can be performed in a single day. Finally, although we describe the migration of marginal zone B cells (MZBs), the protocol can be adapted to analyze migration against flow of other types of immune cells. Therefore, it is feasible to use this assay to systematically analyze a broad range of leukocytes with a comprehensive panel of conditions.
MZBs are a population of B cells that, in the mouse, are found only in the spleen and shuttle between the interior of follicles and the marginal zones6,7,8,9. The marginal zone is a layer of immune cells approximately 5–10 cells thick. The cell layer envelops the follicle and consists primarily of MZBs and macrophages but also invariant natural killer T (iNKT) cells, dendritic cells (DCs), and neutrophils, among others10. The cells in the marginal zone are exposed to unidirectional blood flow originating from splenic arteries that terminate in a marginal sinus surrounding the follicle. The blood flows from holes in the marginal sinus through the marginal zone and is then collected in venous sinuses in the red pulp and restored to the circulation11. The free-flow of blood washes over the MZBs and exposes them to antigens carried in the blood. The MZBs carry the antigen into the follicle by shuttling automatically between the marginal zone and inside of the follicle, which is not exposed to blood. Thus, as MZBs shuttle towards the follicle, they must migrate up the shear force of the blood flow12 (Figure 1A).
In this protocol, we describe how to quantitatively determine how immune cells such as MZBs respond to either no flow or high flow in vitro, in order to reveal how they are programmed to migrate in vivo. In the first step, MZBs are purified from a mouse spleen using magnetic beads coupled to antibodies from commercially available kits. The freshly isolated MZBs are introduced into the well of a flow chamber slide, allowed to settle onto integrin ligands, and exposed to the flow of migration buffer using a pump system (Figure 2A). The cells are imaged using a time-lapse video microscopy system. The images are then processed for analysis with a free ImageJ plugin, MTrack213,14, to automatically track the cells. Tracks can then be quantified with the free Ibidi Chemotaxis tool15 to determine various parameters including velocity, straightness, and migration index. These values can be used to determine the effects of migration inhibitors, cell stimulators, chemokines, and other migration-affecting chemicals on the shear-flow induced migration in order to understand the forces controlling immune cell movement in vivo.
Protocol
All experiments involving the use of animals have been previously approved by the Landesverwaltungsamt Halle (Saxony-Anhalt), Germany, in accordance with all guidelines of the medical faculty of the OVGU University of Magdeburg.
1. MZB Cell Purification
- Isolate leukocytes.
- Sacrifice an 8 to 16 week-old mouse and remove the spleen16. Dissociate the spleen in 5 mL of ice-cold cell buffer [phosphate-buffered saline (PBS) + 0.5% fatty acid-free bovine serum albumin (BSA)], by either using a tissue dissociator or gently mashing the spleen through a 70 µm cell strainer mesh into a well on a 6-well plate with the plunger from a 5 mL syringe.
- Transfer the cells in cell buffer into a 15 mL conical tube. Collect additional cells by adding another 5 mL of ice-cold cell buffer to the tissue dissociator tube or the nylon mesh in the well. Transfer the additional 5 mL of cell buffer plus cells to the conical tube. Centrifuge at 300 x g for 10 min at room temperature (RT, 20 °C) and discard the supernatant.
- Perform red blood cell lysis.
- Re-suspend the cell pellet in 1 mL of red blood cell (RBC) lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) that is pre-warmed to RT. Add an additional 1.5 mL of RBC lysis buffer and swirl to mix. Incubate at RT (20 °C) for a total of 2 min, including the time required to re-suspend the pellet.
- Add 7.5 mL of ice-cold cell buffer to the cell suspension to stop the lysis. Centrifuge at 300 x g and 4 °C for 10 min and discard the supernatant.
- Re-suspend the cell pellet in 1 mL of cell buffer and add an additional 9 mL of cell buffer. Pipette the 10 mL cell suspension through a 30 µm pre-separation filter into a new 15 mL conical tube to remove cell debris.
- Centrifuge cells at 300 x g and 4 °C for 10 min and discard the supernatant. Re-suspend the cell pellet in 500 µL of cell buffer. Keep cells cold at all subsequent steps of the procedure until incubation in the flow chamber slides.
- Purify MZBs.
- Add 50 µL of biotin-coupled antibodies from a commercially available kit against all undesired cells, including CD43 (on non-B cells), CD4 (on T cells), CD93 (on immature B cells), and Ter119 (on erythrocytes), to the cell suspension in a conical tube. Shake or flick the tube gently to mix, but do not vortex the cells. Lay the conical tube on a bed of ice at an almost-flat angle and rock at 25 rpm for 15 min.
- Add magnetic beads coupled to anti-biotin antibodies in a total volume of 100 µL to the cell suspension. Continue to rock the cell suspension at 25 rpm on ice for 20 min. Wash the cells by adding 5 mL of cell buffer, centrifuging at 300 x g and 4 °C for 10 min, and discarding the supernatant. Re-suspend cells in 1 mL of cell buffer.
- Deplete the cell suspension of the magnetic-bead-coupled cells by retaining them on a magnetic column. Discard labeled cells (all non-B cells and immature B cells). Collect the non-labeled cells (mature B cells) in a 15 mL conical tube that has been pre-coated with a brief application of 5 mL of cell buffer, in order to prevent non-specific adhesion by MZBs.
- Wash the cell suspension with 5 mL of cell buffer. Centrifuge at 300 x g and 4 °C for 10 min and discard the supernatant. Re-suspend cells in 90 µL of cell buffer and add 10 µL of anti-CD23-coupled beads. Rock cell suspension gently on ice for 20 min at 25 rpm.
- Wash the cell suspension with 5 mL of cell buffer. Centrifuge at 300 x g and 4 °C for 10 min and discard the supernatant. Re-suspend cells in 1 mL of cell buffer.
- Deplete the cell suspension of the magnetic-bead-coupled cells by retaining them on a magnetic column. Discard the labeled cells (follicular B cells). Collect the non-labeled cells (MZBs), in a 15 mL conical tube that has been pre-coated with a brief application of 5 mL of cell buffer. Remove 50 µL of the cells for counting and checking purity.
- Stain the sorted MZBs to check for purity.
- Add 10% volume (5 µL) of Fc block to 10,000 cells in 50 µL and incubate at 4 °C for 5 min. Add 50 µL of cell buffer containing 0.3 µL each of B220-FITC (0.15 µg), CD21-PE (0.06 µg), and CD23-APC (0.06 µg) in a total volume of 105 µL (each antibody diluted 1:350), or any other combination of fluorophores for distinguishing between follicular B cells and MZBs. Stain the cells at 4 °C in the dark for 15 min.
- Wash the stained cells once with 1 mL of cell buffer. Centrifuge at 300 x g and 4 °C for 5 min and discard the supernatant. Resuspend in 300 µL of cell buffer and acquire samples using a flow cytometer and standard methods. Gate on live lymphocytes, then on B220+ cells, and finally on CD23low CD21high cells (Figure 1B).
NOTE: The procedure typically yields 500,000 MZBs per spleen from a C57B/6 mouse.
- Prepare MZBs for flow migration.
- Wash purified MZBs once in 1–2 mL of cold migration buffer [Hanks’ balanced salt solution (HBSS) containing cations, 10 mM Hepes, 0.2% fatty acid-free BSA] to ensure that the cation-containing migration buffer is not diluted by residual cation-free PBS buffer. Centrifuge at 300 x g and 4 °C for 10 min and discard the supernatant. Re-suspend MZBs in cold migration buffer to a concentration of 50,000 MZBs per 30 µL (equivalent to the amount loaded in one well of a flow chamber slide).
NOTE: The freshly isolated MZBs should be used as soon as possible. However, they are still capable of migrating for up to 8 h or more after the start of the experiment for as long as they are kept on ice. Optimal temperatures should be tested for other cell types. For example, it may be better to keep cultured T cells at 37 °C until the start of migration.
- Wash purified MZBs once in 1–2 mL of cold migration buffer [Hanks’ balanced salt solution (HBSS) containing cations, 10 mM Hepes, 0.2% fatty acid-free BSA] to ensure that the cation-containing migration buffer is not diluted by residual cation-free PBS buffer. Centrifuge at 300 x g and 4 °C for 10 min and discard the supernatant. Re-suspend MZBs in cold migration buffer to a concentration of 50,000 MZBs per 30 µL (equivalent to the amount loaded in one well of a flow chamber slide).
2. Flow Experiment
- Coat slides with integrin ligand.
- On the day before the experiment, thaw an aliquot of ICAM-1 (400 µg/mL). Dilute ICAM-1 by a factor of 1:80 in PBS to reach a final concentration of 5 µg/mL. Add 30 µL of ICAM-1 to one chamber on a flow slide for every migration movie required. Incubate slide in a humid chamber at 4 °C overnight.
NOTE: Each slide contains 6 chambers and up to 8 movies in one session can be easily recorded.
- On the day before the experiment, thaw an aliquot of ICAM-1 (400 µg/mL). Dilute ICAM-1 by a factor of 1:80 in PBS to reach a final concentration of 5 µg/mL. Add 30 µL of ICAM-1 to one chamber on a flow slide for every migration movie required. Incubate slide in a humid chamber at 4 °C overnight.
- Block the coated wells.
- On the day of the experiment, wash the chamber by adding 100 µL of PBS to one well and withdrawing 100 µL from the other well of the chamber. Add 100 µL of blocking buffer (2% fatty acid-free BSA in PBS) to a chamber and incubate the slide in a humid chamber at RT for 1.5 h.
- Wash the chamber by adding 100 µL of PBS to one well, withdrawing it from the other well, then adding 100 µL of migration buffer to the chamber. Store the slides in a humid chamber at RT until the experiment.
- Prepare the fluidics unit.
- Plug in a fluid pump, attach the fluidics unit, and turn on the pump software. Wash the tubing once with 5–10 mL of migration buffer, then add 11.7 mL of migration buffer equally to both reservoirs and remove the air bubbles.
- Clamp the tubing about 10 cm from the connector piece. Place the fluidic unit in the heated incubation chamber on the microscope.
- Set the slide choice to “µ-slide VI 0.4” and the tubing to “white” in the software. Set the desired shear stress (e.g., 4 dyn cm-2). Set the time of imaging to 30 min.
- Set the tubing calibration factor by determining the flow rate through the tubing in mL/min. Measure the time required for 2 mL of the buffer to flow from one reservoir of the other and divide by 2. Make at least 4 measurements for accuracy. Use the actual flow rate to set the desired flow rate and shear stress by entering the values into the pump software.
- Prepare the imaging system.
- Pre-warm the microscope incubation chamber, fluidics unit, and migration buffer aliquots to 37 °C (Figure 2B). Test the fluidic pump by ensuring that the migration buffer flows back and forth in the reservoirs and that there are no air bubbles in the system.
- Load the cells on the slide.
- Clean the bottom of the slide with an ethanol-dampened tissue. Add 30 µL of the cell suspension into one well of the chamber and withdraw 30 µL from the other well. Incubate the slide with a cover on to prevent evaporation at 37 °C for 30 min to allow the cells to attach.
- Slowly add the pre-warmed migration buffer to each well of the flow chamber slide until a positive meniscus rises out of the well and remove any air bubbles with the pipette tip.
- Attach the slide to the fluidics unit.
- Clamp the slide to the microscope stage. Remove the unmarked side of the fluidic tubing from the connector piece and fill up the end with migration buffer until a positive meniscus without air bubbles rises out of the end.
- Flip the end of the tubing over and insert it into the top well of the flow chamber, mopping up spilled buffer with a tissue. While keeping the tubing clamped, repeat the procedure for the marked end of the tubing.
- Image the cells.
- Switch the imaging system to “Live” to set the focus on the cells. Select a field of view that is in the middle of the slide. De-focus the cells slightly to enhance the black outline of the cell and produce a white interior, which can then be thresholded to a round black cell shape on a white background for use with an automated tracking program.
- Start the imaging sequence program and record 1 image every 5 s for 30 min using the 10x dry objective. Remove the clamp from the tubing and turn the pump on, then non-adherent cells will wash off and adherent cells will begin to migrate. Image for 30 min or longer using a 10x objective or higher, as desired. Label and save the movie.
- Detach the fluidic unit from the slide.
- At the end of the imaging program, reset the pump for the next experiment: re-attach the clamp to the tubing, remove the tubing from the slide, re-attach the connector piece, open the clamp, and use the manual control of the pump to push several mL of buffer from one reservoir to the other to remove the air bubbles. Re-attach the clamp and lay the clamped tubing on the microscope stage for the next movie.
NOTE: The protocol can be paused indefinitely here. If using an inhibitor or modifier of cell migration, depending on its mode of action, add it to either the cell suspension before settling in the migration chamber or to the migration buffer in the fluidic unit.
- At the end of the imaging program, reset the pump for the next experiment: re-attach the clamp to the tubing, remove the tubing from the slide, re-attach the connector piece, open the clamp, and use the manual control of the pump to push several mL of buffer from one reservoir to the other to remove the air bubbles. Re-attach the clamp and lay the clamped tubing on the microscope stage for the next movie.
3. Migration Track Analysis
NOTE: Cells can be tracked automatically using the MTrack2 plugin or by hand using the Manual Tracking plugin17. Automatic tracking works well with MZBs because these cells are mainly round and remain this way while migrating, making it easy to threshold the image of the cells to black objects on a white background. Automatic tracking is more difficult if other cell types are used, such as cultured, activated CD8+ T cells, because these cells stretch out during migration and become somewhat transparent, making it difficult to define the edges. In this case, either (1) the cells can be stained with an intra-vital fluorescent dye to produce images that can be thresholded to show black objects on a white background, or (2) other programs to outline the cells such as image segmentation and/or edge detection can be used. Manual tracking is a useful option when producing a high-contrast image of cell outlines is not possible.
- Perform manual tracking using the plugin.
- Install the “Manual Tracking” plugin in ImageJ and open the movie in ImageJ. Under the “Manual Tracking” menu, select “add new track”. Click the center of a cell as the movie advances automatically frame-by-frame. When the cell has been selected in every frame, select “add new track” to start following a new cell.
- After the tracks have been recorded, copy the output results into a data spreadsheet. Change the format for decimal display for all spreadsheet cells to zero places after the decimal. Save the file as a “tab stop-separated” .txt file for subsequent analysis.
NOTE: Typically, 50–100 cells are tracked, which takes about 1 h to complete for a 20 min movie of about 240 frames.
- Perform automatic tracking using the MTrack2 plugin. Download and install the MTrack2 kt plugin according to instructions in the file (see Supplemental Coding File).
- Threshold the images from the cell migration movie. Open the image stack in ImageJ. Process the images in ImageJ to convert the video from color to high-contrast images showing the cells as black objects on a white background (Figure 3A).
- Sharpen the images twice, despeckle the images, and convert the images to 8 bit. Select the “Image | adjust brightness/contrast” sub-menu and put the contrast slider at maximum contrast, then the cells will appear as white objects on a black background. Select the “Image | adjust threshold” sub-menu to ensure that the cells appear as black objects on a white background.
- Run the automatic cell tracking plugin “MTrack2 kt” (see Supplemental Coding File). Set the following parameters on the options screen.
- Set the particle size minimum at 1 pixel and particle size maximum at 30 pixels for MZBs.
- For velocity (maximum distance between 2 particles on successive frames of the movie), if the cell density on the slide is high, set this value low to avoid having the track “jump” from one cell to another. For MZBs, set this value at 10 pixels to capture outliers, although MZBs will generally not exceed 2 pixels on successive frames that are 5 s apart.
- For the minimum number of frames that a particle has to be present to be tracked, set this value at the number of frames in the movie for MZBs (e.g., 240) for a 20 min movie with 12 frames/min. However, if the cells are fast enough to leave the field of view before the end of imaging, then set the minimum number of frames at less than the duration of the movie. For example, with fast-moving T cells, set the minimum number at the equivalent of 5-10 min of imaging.
NOTE: Record a macro of these steps (thresholding and MTrack2-kt) to automate this part of the procedure and save time (see Supplemental Coding File). Using the macro to automatically track a movie in ImageJ takes less than a minute.
- Copy the output results into a data spreadsheet. Change the format for decimal display for all spreadsheet cells to zero places after the decimal. Add an empty row at the top of the cell tracks in the spreadsheet and delete the “D” column. Save the file as a “tab stop-separated” .txt file for subsequent analysis in the Ibidi Chemotaxis tool.
NOTE: The MTrack2 plugin outputs the cell tracks in rows of 75 parallel columns. For analysis in the Ibidi chemotaxis tool, the cell tracks must be in a single column. To convert the output, the MTrack2 plugin was modified to output either in the original format or as a single column (Figure 3B) (see Supplemental Coding File “MTrack2 kt”, which is written in Java). Copy the .java file to the ImageJ plugins folder. In ImageJ, select “Plugins > Compile and Run". Restart ImageJ, and the modified version of MTrack2 should appear in the Plugins menu.
- Threshold the images from the cell migration movie. Open the image stack in ImageJ. Process the images in ImageJ to convert the video from color to high-contrast images showing the cells as black objects on a white background (Figure 3A).
- To analyze cell tracks with the Ibidi Chemotaxis tool, download the Ibidi Chemotaxis and Migration Tool v2.0 (“stand alone”) and open the program.
- Select “Import data” and navigate to the .txt cell track files. Multiple files can be selected at the same time. Imported files appear in the “Datasets” pane in red. Select all of them and enter the correct values in the “Initialisation” menu below to apply the following settings.
- Enter either the exact number or a range of the number of slices (the number of frames in the movie).
- In the Calibration menu (X/Y calibration), enter the size of a pixel. Locate this information in the properties file of the movie. For MZBs, a 10x objective was used, giving a pixel size of 1.14 µm. In “Calibration menu | Time interval”, enter the length of time between frames in the movie.
- Apply the settings. Select “Plot data” to view the track plots and confirm that the track files will open. From this point, many options are available which are described in the documentation for the program, including marking the tracks with different colors based on different properties and displaying the individual or average values for velocity, forward migration index, directness, and more (Figure 3B).
NOTE: The MZBs take about 10 min to detect and respond to the flow, presumably by polarizing their integrin complexes to the flow-forward point of the cell. A graph of migration index over time will show an initial drop below zero, corresponding to the first couple of minutes that the cell is exposed to flow, followed by a gradual rise upwards until the forward migration index values are positive. Thus, it may be optimal to exclude these first minutes in the final calculations, unless this process is of interest.
Representative Results
We used the protocol outlined above to compare migration of MZBs on ICAM-1-coated slides without flow (0 dyn/cm2) and exposed to shear flow (4 dyn/cm2). Cells were tracked automatically with MTrack2, and the resulting track files were overlaid on the cell migration movies of no flow (0 dyn/cm2) and (4 dyn/cm2) to show the distribution and shape of the tracks (Figure 4A). Cell tracks were then imported into the Ibidi Chemotaxis tool (ICT) to generate track plots of each movie (Figure 4B). The average migration index (called "FMIy" in the ICT), velocity, straightness (called "directness" in the ICT), and straight-line distance (called "Euclidean distance" in the ICT) of cell tracks from 4 movies each for both conditions were calculated in the Chemotaxis tool using the "measured values" command. These average values were then copied into GraphPad Prism for generating graphs and calculating statistical significance (Figure 4C).
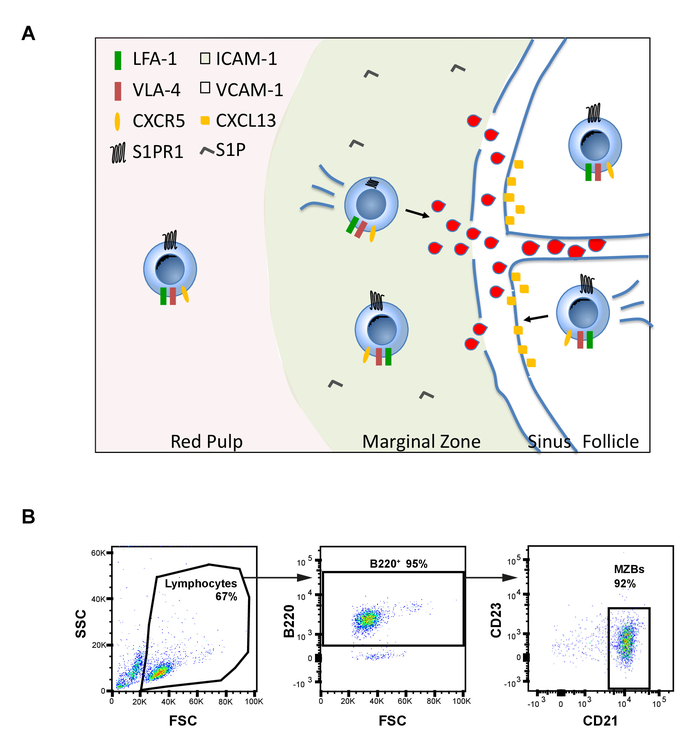
Figure 1: MZB shuttling. (A) Model of MZB shuttling between the marginal zone and the follicle in the spleen. MZBs require internalization of S1PR1, a receptor for S1P, and functional CXCR5, a receptor for the CXCL13 chemokine, in order to enter the follicle. Additionally, to reach the follicle, MZBs must migrate against the force of blood flow emanating from pores in the marginal sinus that envelops the follicle. If a MZB loses adhesion to ICAM-1, the ligand for LFA-1 integrin, it is pushed into the red pulp by the force of the flow, where increased amounts of VCAM-1, the ligand for VLA-4, would not support migration. (B) Flow cytometry gating strategy to test purity of sorted MZBs using antibodies against B220, CD23, and CD21. Please click here to view a larger version of this figure.
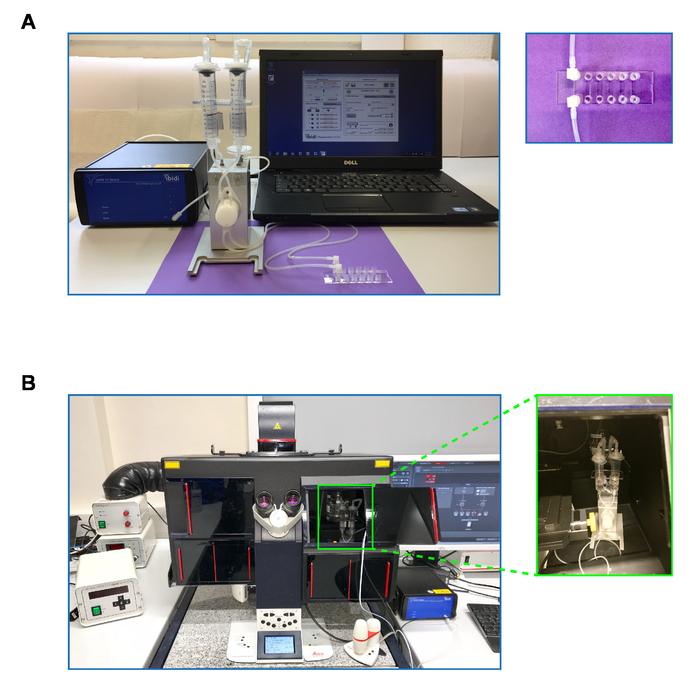
Figure 2: Setup of the fluidics system, pump, microscope, and incubation chamber. (A) Image of the pump system (Ibidi18) consisting of a pump with connected fluidics unit and laptop running the software to control the pump. Higher magnification image: the flow chamber slide with 6 flow chambers, the first of which is attached to the tubing of the fluidics unit. (B) Typical setup for measuring in vitro migration of MZBs against flow. A microscope equipped with a heating chamber (black box) containing the fluidics unit connected to a pump (blue square object to the lower right of the microscope) outside the microscope. Higher magnification image: the fluidics unit inside the microscope heating chamber. Please click here to view a larger version of this figure.
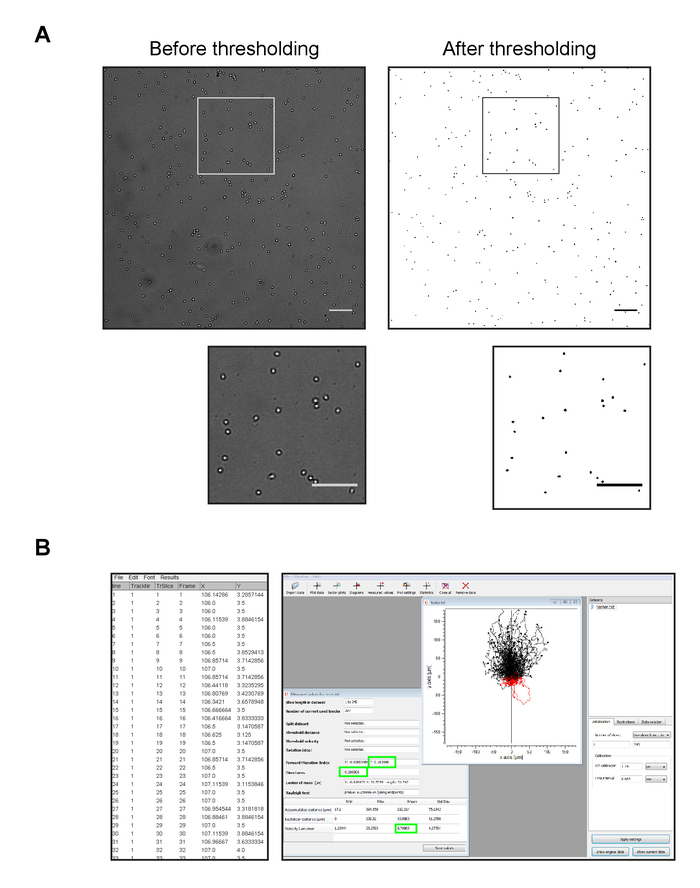
Figure 3: Quantification of imaging data. (A) Representative images of a frame from a movie of migrating MZBs before (left) and after (right) thresholding the image to convert the cells to black objects on a white background. Scale bars in both low and high magnification images = 100 µm. (B) Left panel: image of typical results output from the MTrack2 (single column output) plugin. Right panel: image of the Ibidi Chemotaxis and Migration 2.0 tool after input of MTrack2 results showing a track plot of migrating MZBs and a "measured values" display window showing the averages of parameters including velocity, migration index, and directness (green boxes), among others. Calibration settings shown include the pixel resolution of 1.14 µm per pixel and a time interval of 5 s (0.083 min) per movie frame. Please click here to view a larger version of this figure.
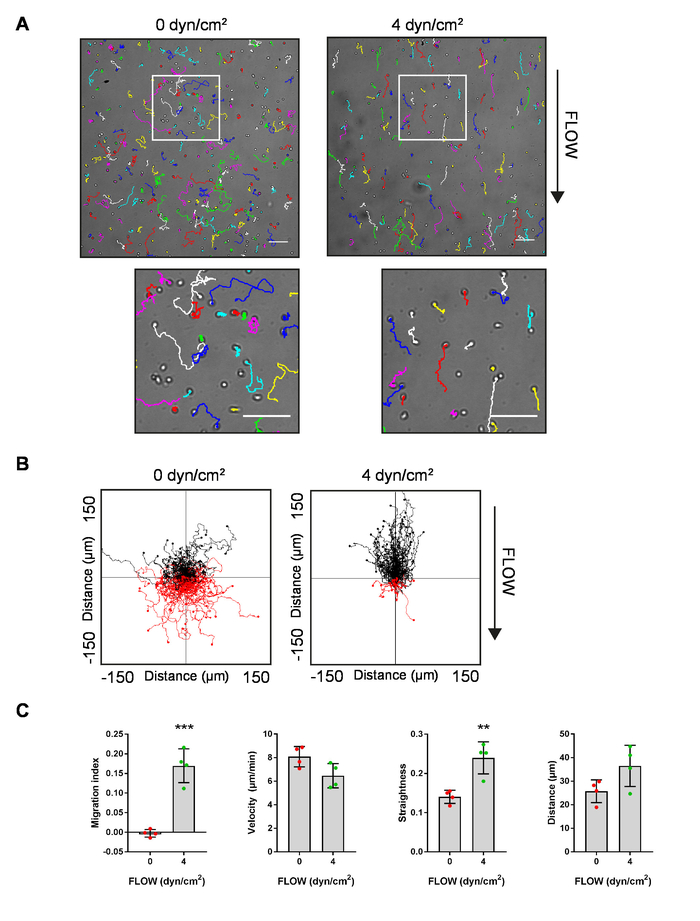
Figure 4: Example of results using the protocol to measure MZB shuttling. (A) Representative stills of migrating MZBs with an overlay of tracks from the ImageJ plugin "MTrack2 kt". Note: the track colors were arbitrarily set by the "Manual Tracking" plugin for ImageJ. Scale bars in both low and high magnification insets = 100 µm. (B) Representative track plots of migrating MZBs from the Ibidi Chemotaxis and Migration 2.0 tool. Red lines and black lines represent cell tracks that terminate below or above the horizontal axis, respectively. In (A) and (B): left, MZBs migrating with no flow (0 dyn/cm2); right, MZBs migrating to a 4 dyn/cm2 flow. (C) Migration index (FMIy), velocity, straightness (directness), and distance for MZBs migrating with no flow (0 dyn/cm2) or flow (4 dyn/cm2) (n = 4 mice in 4 separate experiments). Error bars = mean ± SD; Student's t test, **p < 0.01, ***p < 0.001. Please click here to view a larger version of this figure.
Supplemental File 1: MTrack2_kt.java. Please click here to download this file.
Discussion
We describe here a method for analyzing the migration of cells that detect the force of shear flow and respond by altering their migration. An analysis of MZBs showed that MZBs migrate spontaneously on ICAM-1 and in the presence of flow, will migrate up the flow. In our previous work, we showed that MZBs do not migrate up the flow on VCAM-1 but instead remain fixed in place. The murine splenic marginal zone contains mainly ICAM-1, while the red pulp contains both ICAM-1 and VCAM-1. From these data, it could be inferred that MZBs would migrate up the flow while in the marginal zone but not in the red pulp. The analysis was validated in vivo using MZBs with defective adhesion that were mislocalized to the splenic red pulp by the force of the flow12. For these reasons, MZBs represent a good positive control for testing the flow migration system described here, even if the system is used to study a different cell type.
The most critical aspect of the procedure is ensuring consistency in cell handling on the slide. Because the quantifiable aspects of cell migration depend on the degree to which the cells adhere to the slide, any decrease of cellular adhesion would make reproducible results difficult. Decreased cellular adhesion could result from inconsistency in multiple factors, including temperatures of the migration buffer or the heated incubation box (cold reduces adhesion), levels of cations in the cell suspension (cations affect integrin binding), slide handling (tapping or flexing the slide could dislodge cells), and cell density (collisions between cells could affect migration parameters). Other sensitive points during the procedure include not pipetting the buffer into the slide wells with too much force prior to attaching the tubing (as this could reduce cell adhesion) and keeping the time required for protocol steps consistent in every experiment (cell settling time could affect adhesion). In summary, any possible influence on cell adhesion must be kept consistent throughout repetitions to produce reliable cell migration data.
This method is easy to set up since it uses commercially available equipment and supplies and none of the steps require advanced instruction. For analysis of MZB, as well as other cell types such as cultured CD8+ T cells, coating the slides with various integrin ligands is sufficient to observe the migration up the flow. Studying flow-induced migration response to various integrin ligands leads to direct identification of active integrins on the cell surface, possibly revealing mechanisms relevant to in vivo functions as with MZB migration up the flow on ICAM-1 but not VCAM-1. However, it is also possible to add an endothelial cell layer to the flow chambers. One example of immune cell migration that is affected by shear flow is T cell extravasation through endothelial layers3. This procedure was used to unravel the activation of T cell adhesion via integrins, selectins, and chemokines, and to model lymphocyte migration through the blood-brain barrier. The only limitation to the cells that can be analyzed with this method is that they must be able to sense the force of flow as a directional signal.
Although the method outlined here is used to characterize cellular behavior such as velocity and turning, it can also be extended to analysis of the molecular aspects of migration. Molecular complexes relevant to flow-induced migration, including integrins such as LFA-1 and their cytoskeletal adaptors, would be located within 200 nM of the surface of the slide, amenable to visualization with TIRF microscopy. This addition to the method would be ideal for studying cells from mice with mutations in integrin- or cytoskeleton-related proteins. As many immune cells migrate at various stages in their development through blood or lymph flows, the in vitro migration described can be used to systematically test many kinds of cultured or primary leukocytes for response to flow and reveal how the cells are programmed to migrate in an immune response in vivo. In conclusion, the assay described here provides a reliable and straightforward method for analyzing flow-induced migration of MZBs that can also be extended to other cell types.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from the "Deutsche Forschungsgemeinschaft" SFB 854/TP11 to K.-D.F.
Materials
| VWR Cell Strainer, 70 µm | VWR | 10199-656 |
| Pre-Separation Filters, 30 µm | Miltenyi | 130-095-823 |
| MZB and FOB cell isolation kit | Miltenyi | 130-100-366 |
| B220 CD45R, clone RA3-6B2, FITC | Biolegend | 103206 |
| CD21 / CD 35, clone 7G6, APC | BD Biosciences | 558658 |
| CD23, clone B3B4, PE | Biolegend | 101608 |
| HBSS | Biochrom | L2035 |
| D-PBS 1x | Gibco by Life Technologies | 14190-094 |
| BSA albumin fraction V, fatty acid-free | Roth | "0052.3" |
| ICAM-1 | R&D Systems | 796-IC-050 |
| Ibidi µ-slides VI 0.4, hydrophobic, uncoated | Ibidi | 80601 |
| Perfusion set, white, 50 cm, 0.8 mm | Ibidi | 10963 |
| Ibidi Pump system | Ibidi | 10902 |
Referencias
- Alon, R., Ley, K. Cells on the run: shear-regulated integrin activation in leukocyte rolling and arrest on endothelial cells. Current Opinions in Cell Biology. 20 (5), 525-532 (2008).
- Dominguez, G. A., Anderson, N. R., Hammer, D. A. The direction of migration of T-lymphocytes under flow depends upon which adhesion receptors are engaged. Integrative Biology (Cambridge). 7 (3), 345-355 (2015).
- Steiner, O., et al. Differential roles for endothelial ICAM-1, ICAM-2, and VCAM-1 in shear-resistant T cell arrest, polarization, and directed crawling on blood-brain barrier endothelium. Journal of Immunology. 185 (8), 4846-4855 (2010).
- Valignat, M. P., Theodoly, O., Gucciardi, A., Hogg, N., Lellouch, A. C. T lymphocytes orient against the direction of fluid flow during LFA-1-mediated migration. Biophysical Journal. 104 (2), 322-331 (2013).
- Woolf, E., et al. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nature Immunology. 8 (10), 1076-1085 (2007).
- Cinamon, G., et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nature Immunology. 5 (7), 713-720 (2004).
- Cinamon, G., Zachariah, M. A., Lam, O. M., Foss, F. W., Cyster, J. G. Follicular shuttling of marginal zone B cells facilitates antigen transport. Nature Immunology. 9 (1), 54-62 (2008).
- Cyster, J. G., Schwab, S. R. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annual Reviews in Immunology. 30, 69-94 (2012).
- Schwab, S. R., Cyster, J. G. Finding a way out: lymphocyte egress from lymphoid organs. Nature Immunology. 8 (12), 1295-1301 (2007).
- Cerutti, A., Cols, M., Puga, I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nature Reviews Immunology. 13 (2), 118-132 (2013).
- Mebius, R. E., Kraal, G. Structure and function of the spleen. Nature Reviews Immunology. 5 (8), 606-616 (2005).
- Tedford, K., et al. The opposing forces of shear flow and sphingosine-1-phosphate control marginal zone B cell shuttling. Nature Communications. 8 (1), 2261 (2017).
- ImageJ. MTrack2 Available from: https://imagej.net/MTrack2 (2018)
- . MTrack2 Available from: https://valelab4.ucsf.edu/~nstuurman/IJplugins/MTrack2.html (2018)
- . Chemotaxis and Migration Tool Available from: https://ibidi.com/chemotaxis-analysis/171-chemotaxis-and-migration-tool.html (2018)
- Reeves, J. P., Reeves, P. A. Removal of lymphoid organs. Current Protocols in Immunology. , (2001).
- . Manual Tracking Available from: https://imagej.nih.gov/ij/plugins/manual-tracking.html (2018)
- . ibidi Pump System Available from: https://ibidi.com/perfusion-system/112-ibidi-pump-system.html (2018)

