Using In Vitro and In-cell SHAPE to Investigate Small Molecule Induced Pre-mRNA Structural Changes
Summary
Detailed protocols for both in vitro and in-cell selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) experiments to determine the secondary structure of pre-mRNA sequences of interest in the presence of an RNA-targeting small molecule are presented in this article.
Abstract
In the process of drug development of RNA-targeting small molecules, elucidating the structural changes upon their interactions with target RNA sequences is desired. We herein provide a detailed in vitro and in-cell selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) protocol to study the RNA structural change in the presence of an experimental drug for spinal muscular atrophy (SMA), survival of motor neuron (SMN)-C2, and in exon 7 of the pre-mRNA of the SMN2 gene. In in vitro SHAPE, an RNA sequence of 140 nucleotides containing SMN2 exon 7 is transcribed by T7 RNA polymerase, folded in the presence of SMN-C2, and subsequently modified by a mild 2'-OH acylation reagent, 2-methylnicotinic acid imidazolide (NAI). This 2'-OH-NAI adduct is further probed by a 32P-labeled primer extension and resolved by polyacrylamide gel electrophoresis (PAGE). Conversely, 2'-OH acylation in in-cell SHAPE takes place in situ with SMN-C2 bound cellular RNA in living cells. The pre-mRNA sequence of exon 7 in the SMN2 gene, along with SHAPE-induced mutations in the primer extension, was then amplified by PCR and subject to next-generation sequencing. Comparing the two methodologies, in vitro SHAPE is a more cost-effective method and does not require computational power to visualize results. However, the in vitro SHAPE-derived RNA model sometimes deviates from the secondary structure in a cellular context, likely due to the loss of all interactions with RNA-binding proteins. In-cell SHAPE does not need a radioactive material workplace and yields a more accurate RNA secondary structure in the cellular context. Furthermore, in-cell SHAPE is usually applicable for a larger range of RNA sequences (~1,000 nucleotides) by utilizing next-generation sequencing, compared to in vitro SHAPE (~200 nucleotides) that usually relies on PAGE analysis. In case of exon 7 in SMN2 pre-mRNA, the in vitro and in-cell SHAPE derived RNA models are similar to each other.
Introduction
Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) is a method of measuring the kinetics of each nucleotide in an RNA sequence of interest and elucidating the secondary structure at single-nucleotide resolution1. SHAPE methodologies, both in in vitro conditions2,3,4 (purified RNA in a defined buffer system) and in living mammalian cells5,6, have been developed to investigate the secondary structure of medium length RNA sequences (typically <1,000 nucleotides for in-cell SHAPE and <200 nucleotides for in vitro SHAPE). It is particularly useful to evaluate structural changes in receptor RNA upon binding to RNA-interacting small molecule metabolites2,4,7,8 and to study mechanistic actions of RNA-targeting molecules during drug development9,10.
RNA-targeting drug discovery has recently drawn attention in academic laboratories and the pharmaceutical industry11,12 via different approaches and strategies13,14,15,16. Recent examples of RNA-targeting small molecules for clinical use include two structurally distinct experimental drugs, LMI-07017 and RG-791618,19, for spinal muscular atrophy (SMA), which showed promising results in phase II clinical trials20. Both molecules were demonstrated to target survival of motor neuron (SMN) 2 pre-mRNA and regulate the splicing process of the SMN2 gene6,17,21. We previously demonstrated the application of in vitro and in-cell SHAPE in an examination of the target RNA structural changes in the presence of an analog of RG-7916 known as SMN-C26.
In principle, SHAPE measures the 2'-OH acylation rate of each nucleotide of an RNA sequence in the presence of excess amounts of a self-quenching acylation reagent in an unbiased manner. The acylation reagent is not stable in water, with a short half-life of (e.g., T1/2 = 17 s for 1-methyl-7-nitroisatoic anhydride; or 1M7, ~20 min for 2-methylnicotinic acid imidazolide, or NAI)22 and insensitivity to the identity of the bases23. This results in a more favorable acylation of the 2'-OH groups of flexible bases, which can be transformed into an accurate assessment of the dynamics of each nucleotide. Specifically, a nucleotide in a base-pair is usually less reactive than an unpaired one to a 2'-OH modifying reagent, such as NAI and 1M7.
Looking at the source of the RNA template and where 2'-OH acylation takes place, SHAPE can generally be categorized into in vitro and in-cell SHAPE. In vitro SHAPE uses purified T7 transcribed RNA and lacks a cellular context in experimental designs. In in-cell SHAPE, both the RNA template transcription and 2'-OH acylation occur within living cells; therefore, the results can recapitulate the RNA structural model in a cellular context. In-cell SHAPE has been referred to as in vivo SHAPE for the SHAPE carried in living cells in the literature24. Since this experiment is not performed in an animal, we termed this experiment as in-cell SHAPE for accuracy.
The strategies for the primer extension stage of in vitro and in-cell SHAPE are also different. In in vitro SHAPE, reverse transcription stops at the 2'-OH acylation position in the presence of Mg2+. A 32P-labled primer extension therefore appears as a band in polyacrylamide gel electrophoresis (PAGE) and the intensity of the band is proportional to the acylation rate1. In in-cell SHAPE, reverse transcription generates random mutations at the 2'-OH adduct position in the presence of Mn2+. The mutational rate of each nucleotide can be captured by in depth next-generation sequencing, and the SHAPE reactivity at single-nucleotide resolution can then be calculated.
A potential problem for in-cell SHAPE is the low signal-to-noise ratio (i.e., a majority of the 2'-OH groups is unmodified, while the unmodified sequences occupy most of the read in next-generation sequencing). Recently, a method to enrich the 2'-OH modified RNA, referred to as in vivo click SHAPE (icSHAPE), was developed by the Chang laboratory25. This enrichment method may be advantageous in studying weak small molecules such as RNA interactions, especially in a transcriptome-wide interrogation.
Protocol
1. In Vitro SHAPE
NOTE: The protocol is modified from the published protocol1.
- Preparing the RNA template
NOTE: The template for T7 transcription was ordered as a synthetic double-stranded DNA (dsDNA) and amplified by either insertion into an E. coli vector that carries a unique pair of EcoRI/BamHI restriction endonuclease sites, such as pET28a, or by PCR. The protocol for PCR amplification is illustrated below.- Mix the following material: 50 μL of PCR master mix (see Table of Materials), 2 μL for each primer in 10 μM (see Table 1 for sequences), 200 ng of dsDNA template in 2 μL and 3 μL of dimethyl sulfoxide (DMSO), and 41 μL of H2O. Aliquot into two PCR tubes (each tube contains 50 μL of reaction mixture).
- Set up the thermal cycler as follows: stage 1) 95 °C for 30 s; stage 2) 95 °C for 20 s; stage 3) 56 °C for 20 s; stage 4) 72 °C for 20 s; stage 5) loop back to stage 2 for 30 cycles; stage 6) 72 °C for 3 min; and stage 7) infinite holding at 4 °C until the following step.
- Purify the PCR amplicon by extraction of gel slices (see Table of Materials and use the manufacturer’s protocol). Elute the product DNA with 35 μL of Tris-EDTA (TE) buffer, containing 10 mM Tris (pH 8.0) and 1 mM EDTA, to yield a 0.1–0.5 μg/μL template. Normalize the purified DNA template into 0.10 μg/μL.
NOTE: Optionally, the quality of the template may be analyzed on a 1.8% agarose gel electrophoresis with 0.10 μg of DNA. A single band of desired template DNA is expected on the gel. - Set up the T7 transcription reaction (total volume 40 μL) by adding the following materials into a PCR tube: 4 μL of 10x reaction buffer, 4 μL of ATP, 4 μL of CTP, 4 μL of GTP, 4 μL of UTP, 4 μL of T7 enzyme (see Table of Materials), 4 μL of purified PCR amplicon from step 1.1.3 (0.4 μg), and 12 μL of diethyl pyrocarbonate (DEPC)-treated water. Mix by pipetting up and down 4x. Incubate at 37 °C for 4–16 h in a thermal cycler or in a 37 °C incubator.
NOTE: Start to set up step 1.1.6 while the reaction in step 1.1.4 is ongoing. - Add 2 μL of DNase, mix by pipetting up and down 4x, and incubate at 37 °C for 15 min. Mix with equal volume of 2x Tris-borate-EDTA (TBE)-urea sample buffer (see Table of Materials). Heat at 70 °C for 3 min to inactivate.
- In an RNase-free bottle, mix 75 mL of 40% acrylamide/bisacrylamide solution (29:1, see Table of Materials), 180.2 g of urea, and 50 mL of 10x TBE buffer (RNase-free, see Table of Materials). Put the bottle on an orbital shaker (250 rpm) until all crystals of urea are dissolved (may take up to 2 h). Adjust the volume to 500 mL with DEPC-treated water. Filter the solution with a 0.2 μm membrane (see Table of Materials).
NOTE: In this process, avoid using specular and stir-bar to prevent RNase contamination. The solution can be stored at 4 °C for up to 1 year. - Clean two electrophoresis glass plates of a proper size (16.5 cm x 24 cm) with deionized water and 70% EtOH using a piece of wiping paper. Align the plates with a pair of 2.0 mm spacers (the plate with a siliconized surface is facing inward) and seal the edge of the plates with tape. Double-tape the corner of the plate to prevent leaking.
- In a beaker, mix 200 mL of acrylamide solution from step 1.1.6, 2.0 mL of 10% ammonium persulfate solution, and 100 μL of tetramethylethylenediamine (TEMED) with an RNase-free pipette tip by stirring rapidly for 30 s. Tilt the plates on a piece of benchtop cover and pour the solution from the gap of the plates. Tap the plate to lift any bubbles and lay the plate flat on the benchtop cover. Immediately insert the comb and let the gel polymerize for at least 1 h.
NOTE: If the gel is to be used within the next day, cover the top of the gel with a piece of wet paper towel and wrap up with a larger piece of plastic wrap. Place it lying flat at 4 °C. - Remove the tape and clamp the plate in an electrophoresis stand. Remove the comb and add adequate 1x TBE buffer at the top and bottom of the reservoir. Pre-run the gel for 30 min at 20 W.
NOTE: Optionally, if the content of the desired RNA is ~90% or more, a mini-gel can be used in substitution of a preparative gel. - Wash each of the loading wells by pipetting up and down twice with the liquid in the reservoir. Add crude RNA from step 1.1.5 into the wells. Run the gel at 20 W until the xylene cyanol FF dye (light blue) passes about two-thirds of the gel’s length (~60 min).
NOTE: Ensure to load no more than one-third of the height of the well (use multiple wells if necessary). - Immerse the gel in 1x TBE buffer with 1:10,000 dilution of the safe dye (see Table of Materials) in a container large enough for the gel. Put the container on an orbital shaker for 5 min at 80 rpm.
- Under UV light, identify the desired RNA band and swiftly cut a thin band of the gel using a clean razor blade. Place 1–2 gel slices in a 1.7 mL tube.
- Recover the RNA from the gel slice by passive elution. Add adequate elution/precipitation mix (~0.3 mL per slice) to each tube: 0.3 M NaOAc (pH 5.2), 2.5 M LiCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate (SDS) in DEPC-treated water. Rotate the tube that contains the gel splice(s) at 4 °C for 16 h.
NOTE: A typical recovery rate is ~75% in this step. To increase the yield, remove and collect the eluent and add the same volume of elution buffer, then repeat this step. - Combine the eluent in a new 1.7 mL tube. Add 2.5x ice-cold EtOH, invert the tubes six times to mix the liquid, and place the tubes at -80 °C for at least 1 h.
- Centrifuge for 10 min at 10,000 x g and 4 °C. Carefully remove the supernatant by pipetting. Wash the RNA pellet by adding 80% EtOH (1 mL per tube), vortex for 15 s, and centrifuge for 10 min at 10,000 x g and 4 °C.
- Remove the supernatant and air-dry the RNA pellet for 5 min. Add 50 μL of RNase-free water and mix by pipetting up and down 10 times. Normalize the RNA concentration to 0.10 μg/μL. Store the purified RNA at -80 °C.
NOTE: Do not over-dry the RNA pellet. - Optionally, to analyze the quality of the RNA, dilute 1 μL (100 ng) of RNA from step 1.1.16 in 5 μL of water and mix with 5 μL of 2x TBE-urea sample buffer. Then, heat at 70 °C for 3 min and load on a 6% TBE-urea mini-gel.
- Run the gel at 180 V for 1 h. Perform the staining as done in step 1.1.11. Visualize the gel with UV in an imaging system. A single band is expected at the desired length.
- Preparing the 32P-labeled primer
- Set up the reaction by mixing the following materials: 10 μL of γ-32P-ATP (~1.5 mCi, ~25 μM, see Table of Materials), 2 μL of reverse transcription (RT) primer (100 μM, for sequence see Table 1), 2 μL of 10x T4 polynucleotide kinase (PNK) reaction buffer, 1 μL of T4 PNK (see Table of Materials), and 5 μL of RNase-free water. Incubate at 37 °C for 1 h.
CAUTION: Proper protection is need for steps 1.2-1.5 in an authorized workplace for radioactive materials. - Heat-inactivate for 20 min at 65 °C. Add the samples onto a desalting column and centrifuge at 1,000x g for 1 min. Mix the eluent with 25 μL of 2x TBE-urea sample buffer. .
NOTE: Store the labeled crude product at -20 °C if it is not to be immediately purified. - Run a 10% acrylamide gel at 20 W for 1 h.
CAUTION: Load the 32P-labeled primer from step 1.2.1 onto the gel and avoid bleeding the radioactivity into the top reservoir. Do not load more than one-third of the height of the well to ensure a thin band on the gel (use multiple wells if necessary). The liquid in the bottom reservoir can be highly radioactive. - Remove the glass plates of the gel cassette and lay the gel on a piece of plastic wrap. Fold the plastic wrap to cover the top of the gel to make an air-tight sandwich. Place the gel sandwich on a calcium tungstate phosphor screen and expose with an imaging instrument. Image the gel again with regular light to know where the edges of the gel are.
- Use an image processing software to overlay the two images. Align the gel on to the printed image. Use a fresh razor blade to cut the desired bands out of the gel . Place 1 to 2 gel slices in a 1.7 mL tube.
NOTE: Make sure the printed image has the same size as the actual gel. Double check the alignment by imaging the leftover gel again after cutting out the gel slice. - Recover the 32P-labeled primer by following steps 1.1.13 to 1.1.16. Take 1.0 μL of solution into a 0.4 mL tube and dilute the primer with 1x TE buffer to obtain the radioactivity of 1.0 μL at ~100,000 cpm. Store the purified 32P-labeled primer at -80 °C until RNA 2’-OH modification reaction but for no longer than 4 weeks.
- Set up the reaction by mixing the following materials: 10 μL of γ-32P-ATP (~1.5 mCi, ~25 μM, see Table of Materials), 2 μL of reverse transcription (RT) primer (100 μM, for sequence see Table 1), 2 μL of 10x T4 polynucleotide kinase (PNK) reaction buffer, 1 μL of T4 PNK (see Table of Materials), and 5 μL of RNase-free water. Incubate at 37 °C for 1 h.
- Small molecule binding and RNA 2’-OH modification
- To prepare four samples, add 8 pmol RNA in 32 μL of 0.5x TE buffer to a 1.7 mL tube, heat at 80 °C for 2 min, and snap-cool on ice for at least 1 min.
NOTE: Use the following equation to calculate the approximate molecular weight of RNA: MW = (# of bases) x 340 Da. - Add 8 μL of 5x folding mix containing 500 mM HEPES (pH 8.0), 20 mM MgCl2, and 500 mM NaCl. Aliquot 9 μL of the RNA mixture into each of the four PCR tubes and label them #1-#4. Add 1 μL of 10% DMSO into the #1 tube, 1 μL of concentrated small molecule solution (10% DMSO) into #2, 1 μL of diluted small molecule solution (10% DMSO) into #3, and 1 μL of water into #4.
- Incubate the PCR tubes at 37 °C in a thermal cycler for 30 min.
- Right before the 2’-OH modification reaction, dilute 1 μL of NAI stock solution (2 M) with 3 μL of DEPC-treated water to yield a 0.5 M working solution. Without delay, add 1 μL of NAI working solution into tubes #1, #2, and #3, and 1 μL of 25% DMSO into #4. Incubate at 37 °C in a thermal cycler for 15 min.
- Add 100 μL of elution/precipitation mix (see step 1.1.13 for recipe), 2 μL of glycogen (15 mg/mL), and transfer all the liquid in each PCR tube into a 1.7 mL tube. Add 0.34 mL of ice-cold 200-proof EtOH (3x volume) into each tube. Mix by vortexing for 5 s and place the tubes in a -80 °C freezer for at least 1 h.
NOTE: During this period, start setting up the sequencing gel to be used in step 1.5.1. - Centrifuge for 15 min at 14,000 x g and 4 °C. Remove the supernatant without disturbing the RNA pellet. Wash the RNA pellet by adding 80% EtOH (0.5 mL per tube), vortex for 15 s, and centrifuge for 15 min at 20,000 x g and 4 °C. Remove the supernatant again.
NOTE: Optionally, use a Geiger counter to ensure the radioactive pellet retaining in the tube. - Air-dry the RNA pellet for 5 min. Add 9 μL of RNase-free water and mix by pipetting up and down 10 times.
NOTE: Do not over-dry the RNA pellet. All precipitation is expected to be dissolved at the end of this step.
- To prepare four samples, add 8 pmol RNA in 32 μL of 0.5x TE buffer to a 1.7 mL tube, heat at 80 °C for 2 min, and snap-cool on ice for at least 1 min.
- Marker preparation and primer extension
- Transfer the modified RNA into 4 new PCR tubes and label them #1-#4 as previously. Add 2 pmol control RNA in 7 μL of DEPC-treated water to 4 additional PCR tubes and label them #5–#8. Add 2 μL of radiolabeled primer (step 1.2.6) in all eight tubes. Heat to anneal at 65 °C for 5 min, then immediately cool down to 4 °C in the thermal cycler.
- Add 4 μL of 5x RT buffer (see Table of Materials), 1 μL of dithiothreitol (DTT, 0.1 M), and 1 μL of dNTP mix (10 mM each) to each of the eight PCR tubes.
- Add 2 μL of DEPC-treated H2O to tubes #1–#4. Add 4 μL of ddATP (5 mM), 4 μL of ddTTP (5 mM), and 4 μL of ddCTP (5 mM) into tubes #5–#7, respectively. Add 1 μL of ddGTP (5 mM) and 3 μL of DEPC-treated water to tube #8. Pipette four times to mix.
- Heat at 52 °C for 1 min in the thermal cycler. Add 1 μL of reverse transcriptase (see Table of Materials), then mix by pipetting up and down four times. Incubate the reaction at 52 °C for 20 min.
- Add 1 μL of 5 M NaOH into each of the tubes to hydrolyze the RNA. Heat the tubes at 95 °C for 5 min.
- Add 5 μL of 1 M HCl and repeat the ethanol precipitation (steps 1.3.5 and 1.3.6), except omit the glycogen and liquid transfer.
NOTE: Omitting step 1.4.6 results in a blurring region in the middle of the gel known as the “salt front”. - Air-dry the DNA pellet for 5 min. Add 10 μL of H2O and 10 μL of 2x TBE-urea sample loading buffer and pipette up and down 10 times to redissolve. Heat at 70 °C for 5 min. Place the tubes on ice before loading onto the gel.
- PAGE analysis
- To prepare an 8% polyacrylamide sequencing gel, follow steps 1.1.6 to 1.1.10 with the following modifications: use 100 mL of 40% acrylamide/bisacrylamide solution (29:1) to prepare an 8% acrylamide solution, and use sequencing gel glass plates (e.g., 46 x 57 cm) with a 0.4 mm spacer.
- Load 10 μL of the sample from step 1.4.7. Run the gel at 65 W for 2 h or until the bottom dye reaches 3/4 of the gel.
NOTE: Store the rest of the samples at -80 °C for repeating if needed. - Remove the top siliconized glass plate by removing the spacer and gently inserting a spatula. Place a piece of large filter paper to cover the gel and press gently to allow the gel to adhere to the filter paper. Starting from the bottom end, lift the filter paper and remove the gel from the glass plate together.
- Place the gel together with the filter paper on a piece of plastic wrap that is large enough to cover the whole gel (filter paper facing up). Transfer the filter paper/gel/plastic wrap sandwich to a gel drier with the filter paper side facing down.
CAUTION: To avoid radioactive material contamination, use 3 to 4 more pieces of large filter paper between the gel sandwich and gel drier. - Dry the gel at 70 °C for 1 h with a vacuum. Transfer the gel sandwich onto a photo-bleached phosphor storage screen cassette. Expose the radioactivity of the gel to the screen for 4–16 h.
- Place the phosphor storage screen on an imaging device and scan with medium resolution (~30 min).
NOTE: If a smile-like artifact is present on the gel image, use correcting software (e.g., SAFA) to process the image to a publishable quality. Keep in mind that the standard marker lane is 1 nucleotide longer than the 2’-OH modification lanes.
2. In-cell SHAPE
- Primer design
NOTE: Unlike in vitro SHAPE, in-cell SHAPE does not need to design an expression cassette. However, an optimal primer set should be used and must be evaluated before SHAPE experiment.- Generate at least three unique primer pairs by a BLAST-based primer design tool (e.g., Primer-BLAST). To study pre-mRNA in human cells, select the human genome (not transcriptome) as a reference database in Primer Pair Specificity Checking Parameters. Pick the size of amplicon in the range of 200–1,000 base-pair (bp).
- Extract the genomic DNA from ~106 cells of interest with a spin column-based method with the treatment of RNase A and protease K (see Table of Materials and use the manufacturer’s protocol). Normalize the purified genomic DNA in 0.1 μg/μL TE buffer.
- Test PCR with the protocol performed in steps 1.1.1 and 1.1.2.
NOTE: The desired primer set should yield a single band on a 1.8% agarose gel.
- Cell preparation and 2’-OH modification
NOTE: To increase the abundance of the pre-mRNA of interest, a minigene with the correct sequence under cytomegalovirus (CMV) promoter for constitutive expression is transfected into the host cells.- Pre-warm the culture medium containing 10% fetal bovine serum (FBS) and 1x antibiotic mixture in Dulbecco's Modified Eagle Medium (DMEM) at 37 °C. Seed 293T cells at 50% confluency 24 h prior to the transfection in culture medium. Change the medium into 2% FBS in DMEM without antibiotics ~1 h prior to the transfection.
- Add 10 μg of minigene plasmid into 100 μL of reduced serum media (see Table of Materials) in 1 well of a 96-well plate. Pipette the solution up and down four times to mix.
- Add 40 μL of transfection reagent (see Table of Materials) into 100 μL of reduced serum media in another well of the plate. Pipette up and down four times to mix. Incubate at room temperature for 5 min.
- Add the entire plasmid solution to the transfection reagent suspension. Pipette up and down four times to mix. Incubate at room temperature for 30 min.
- Add the entire DNA/ transfection reagent mixture dropwise on the surface of the medium throughout the dish. Incubate at 37 °C for 6–12 h.
- Aspirate the medium and gently wash once with 5 mL of phosphate-buffered saline (PBS, pH 7.4, without CaCl2 and MgCl2, see Table of Materials). Trypsinize the cells with 3 mL of trypsin solution. Incubate at room temperature for 5 min.
- Knock the dish gently to dissociate the cells from the bottom of the dish. Neutralize trypsin with 6 mL of culture medium. Centrifuge at 300 x g for 4 min and remove the medium.
- Resuspend the ~106 cells for each sample in 199 μL of reaction medium (1% FBS in DMEM) and transfer the cell suspension into a 96-well plate. Incubate the cells with 1 μL of compound working solution or 1 μL of DMSO in all three controls for 30 min at 37 °C.
NOTE: Three control samples should be included: DMSO control with NAI, denatured RNA with NAI, and NAI negative control. - Add 10 μL of 2 M NAI to each sample, except for the denaturing control (DC) RNA and NAI negative controls. Pipette up and down four times to mix. Incubate at 37 °C for 15 min. Gently shake the plates to re-suspend the cells every 5 min.
- Transfer the cells into 1.7 mL tubes and centrifuge at 400 x g for 2 min without delay. Remove the supernatant without disturbing the cell pellet.
- Add 0.4 mL of guanidinium thiocyanate (see Table of Materials). Vortex for 15 s to homogenize. Incubate the samples at room temperature for 5 min.
NOTE: The samples can be stored at -20 °C until proceeding to the next step.
- RNA extraction
- Add 0.2 mL of chloroform to each of the guanidinium thiocyanate homogenized samples. Vortex for 15 s. Incubate at room temperature for 2 min.
- Centrifuge for 5 min at 12,000 x g and 4 °C. The top colorless aqueous layer contains RNA. Transfer ~380 μL of aqueous solution into a new 1.7 mL tube.
CAUTION: Do not disturb the interphase to avoid contamination. - Add 1.5x volume of EtOH (~570 μL). Vortex for 15 s to mix.
- Transfer 600 μL of RNA mixture into an RNA isolation spin-column (see Table of Materials). Centrifuge at 12,000 x g for 15 s. Discard the eluent. Repeat this step to pass through all liquid into the same spin column.
- Wash the column with 0.35 mL of RW1 buffer (see Table of Materials) by spinning for 15 s. Add 80 μL of RNase-free DNase I in RDD buffer (see Table of Materials). Incubate at room temperature for 20 min.
NOTE: It is critical to remove all DNA contamination. - Subsequently wash the column with 0.35 mL of RW1 buffer and 0.6 mL of 2x RPE buffer (see Table of Materials) by spinning for 15 s at each step. Dry the column by centrifuging the spin column with an empty collection tube at 12,000 x g for 1 min.
- Add 32 μL of RNase-free water to the center of the column. Incubate for 1 min. Centrifuge at 12,000 x g for 30 s to obtain ~30 μL of purified RNA solution. Put the samples at -20 °C while processing the denatured sample.
- Denatured RNA control preparation
- Place 30 µL RNA sample for DC in a 1.7 mL plastic tube on ice. Add 50 μL of formamide and 15 μL of DC buffer containing 300 mM HEPES (pH 8.0) and 25 mM EDTA into the tube.
- Heat to denature the RNA at 95 °C for 1 min. Quickly add 5 μL of NAI stock solution (2 M), then flick twice to mix and put the tube back on to the heating block. Incubate at 95 °C for an additional 1 min then immediately put the tube on ice.
- Add 0.4 mL of guanidinium thiocyanate. Repeat steps 2.3.1–2.3.4.
- Wash the column with 0.6 mL of 2x RPE buffer (see Table of Materials) by spinning for 15 s at each step. Dry the column by centrifuging the spin column with an empty collection tube at 12,000 x g for 1 min.
- Add 32 μL of RNase-free water to the center of the column. Incubate for 1 min. Centrifuge at 12,000 x g for 30 s to obtain ~30 μL of recovered DC RNA solution.
- Primer extension
- Normalize the RNA solution from 2.3.7 and 2.4.5 into 0.5 μg/μL with RNase-free water. Freshly prepare 30 mM MnCl2 solution from MnCl2∙4H2O powder.
- Transfer 9 μL of RNA solution for each sample into a PCR strip. Add 1 μL of 10 mM dNTP and 1 μL of 2 μM gene-specific primer (GSP) in each tube. Pipette up and down four times to mix. Incubate at 65 °C for 5 min in a thermal cycler and put on ice for >1 min.
NOTE: Alternatively, 1 μL of a random nonamer can be used in substitution of GSP. - In each PCR tube, add a mixture of 2 μL of 10x RT buffer (see Table of Materials), 4 μL of 30 mM MnCl2, and 2 μL of 100 mM DTT. Pipette up and down 4x to mix. Incubate at 42 °C for 2 min.
- Add 1 μL of reverse transcriptase (see Table of Materials). Pipette up and down 4x to mix. Incubate at 42 °C in a thermal cycler for 3 h.
- Library preparation and next-generation sequencing
- Set up a PCR reaction by adding 1 μL of DNA polymerase, 5 μL of 10x DNA polymerase buffer (see Table of Materials), 2 μL of DMSO, 1.5 μL for each of the primers (10 μM), 34 μL of H2O, and 5 μL of cDNA from step 2.5.4.
- Set up the thermal cycler as follows: stage 1) 95 °C for 2 min; stage 2) 95 °C for 30 s; stage 3) 60 °C for 30 s; stage 4) 68 °C for 30 s; stage 5) loop back to stage 2 for 30 cycles; stage 6) 68 °C for 3 min; and stage 7) infinite holding at 4 °C until the following step.
- Purify the amplicon by using 1.8% agarose gel.
NOTE: The PCR band should appear as a single band on the gel. - Construct the library by using standard fragmentation and ligation methods established in the literature6,26.
- Sequence on a next-generation sequencing equipment using 1 x 75 single-end reads to generate approximately 10 million reads per sample.
- Analyze the results using ShapeMapper and SuperFold software packages developed by the Weeks group26.
Representative Results
We previously demonstrated that an RNA splicing modulator, SMN-C2, interacts with AGGAAG motif on exon 7 of the SMN2 gene's pre-mRNA, and used SHAPE to assess the RNA structural changes in the presence of SMN-C26. The binding site of SMN-C2 is distinct from the FDA-approved antisense oligonucleotide (ASO) for SMA, nusinersen, which binds and blocks the intronic splicing silencer (ISS) on intron 727,28 (Figure 1A). Most known splicing regulators of SMN2 exon 7 are within the ~100 nucleotide range of exon 7 in the pre-mRNA29; therefore, a 140 and 276 nucleotide-long RNA sequences were used for in vitro and in-cell SHAPE, representatively, which covers exon 7 and the adjacent intron region (Figure 1A).
In this representative in vitro SHAPE analysis, the RNA sequence of interest is embedded into an optimized cassette developed by the Weeks laboratory, which is compatible for most RNA sequences1. Occasionally, the sequence of interest interacts or interferes with this cassette. In these cases, a modified cassette can be used with the following three characteristics: i) a 3'-end specific primer binding site with a more efficient hybridization affinity for the primer than any part of the RNA sequence, ii) a highly structured hairpin loop located directly upstream of the primer binding site that will show specific and reproducible SHAPE signal (this will act as both an internal control for the experiment and method to align the signal from experiment to experiment), and iii) a 5'-end hairpin structure element, which indicates the end of the SHAPE signal. The SHAPE cassette is further flanked with self-cleaving ribozyme sequences at both 5'- and 3'-ends in order to generate a homogenous RNA30. We found that hammerhead and hepatitis delta virus ribozymes are compatible to the SHAPE cassette and usually give high yield of the desired RNA. The resulting template RNA has a 3'-end of 2',3'-cyclic phosphate and a 5'-end of hydroxyl group, which do not interfere with primer extension. A 140-nucleoside long sequence covering exon 7 of SMN2 and adjacent region in the pre-mRNA was synthesized within SHAPE and ribozyme cassette as illustrated in Scheme 1.
In general, the sequence of interest ligated in the expression cassette should be long enough to cover the potential secondary or higher order of structure. In the case of SMN2 exon 7, a 140-nucleotide region contains two stem-loop structures6,31. The structure of the region of interest varies case-by-case and should be evaluated by trial-and-error.
Polyacrylamide sequencing gel with 32P-labeled primer extension products was chosen to visualize the in vitro SHAPE profile in this representative experiment. An alternative visualization method is to use capillary electrophoresis with a fluorescently labeled DNA primer32. In polyacrylamide sequencing gels, ~20 nucleosides near the 5'-end and ~10 nucleotides near the 3'-end of the RNA sequence of interest will not be visualized quantitatively, due to occasionally stops at the initiation steps of reverse transcription and intense bands on PAGE gels for full-length transcripts1.
An alternative way for preparative gel recovery of an RNA template (steps 1.1.6 to 1.1.12) is to overload a mini-gel if the yield of desired RNA template is >90%. It is advised to remove the excess NTP from the RNA product by desalting the column and eluting the RNA in 50 µL of TE buffer. Measure the concentration of the RNA and load 5.0 µg in each well. During the purification step of the T7 transcribed RNA template, stop the PAGE when the xylene cyanol FF dye (light blue) passed two-thirds of the 6% denatured TBE-urea gel. The self-cleavage RNA fragment (<80 nucleosides) passed through the gel, which left the desired RNA as the only major band in the gel upon staining (Figure 1B).
SMN-C2 (Figure 1C) was synthesized according to the published procedure33 and dissolved in 10 mM DMSO stock solution. The stock solution is further diluted into 500 and 50 µM in 10% DMSO solution to achieve final concentrations of 50 and 5 µM, respectively. Snap-cooled RNA refolded to its equilibrium stage in the presence of DMSO or SMN-C2 within 30 min at 37 °C. A longer incubation time did not change the outcome of the experiment. Two experimental samples (50 and 5 µM SMN-C2), two controls (DMSO and NAI- controls), and four markers (A, T, G, C) were treated for primer extension. After exposing the gel to phosphor storage screen, a successful SHAPE experiment will show: i) a single and most-intense band at the top of the gel and ii) bands throughout the gel at single-nucleotide resolution without smear (Figure 1D). A common problem in PAGE analysis is that a smear region known as the "salt front" may appear in the middle of the gel (Figure 1E). This is probably due to high concentration of salt, DMSO, or other unwanted substance in the loading sample that can be removed by ethanol precipitation.
In in vitro SHAPE, a pure RNA template is the key for a successful experiment. An impure RNA template is usually the cause of undesired results. If PAGE analysis clearly shows a pattern of 2 sets of markers, it indicates that the RNA template is not homogenous and needs to be repurified by preparative TBE-urea gel.
For in-cell SHAPE, a key to a successful experiment is to design an optimized primer set for amplification. With 0.10 µg of genomic DNA-free cDNA template, a single band should be observed within 25 PCR cycles in agarose gel analysis. The repetitive intron sequences should therefore be avoided. To study the structural impact of SMN-C2 on SMN2 pre-mRNA, three primer sets (Table 2) was tested, and all were satisfactory (Figure 2A).
A low copy number of a target RNA sequence is generally a problem for in-cell SHAPE. To enrich the RNA of interest, a minigene that contains the RNA sequence of interest under a strong CMV promoter was transfected into 293T cells. Because the splicing pattern of the SMN2 minigene recapitulates that of endogenous SMN2 with or without SMN-C219,34, we envision that the structure of SMN-C2 interacting RNA in the overexpressed SMN2 pre-mRNA likely was the same as the endogenous gene product. While the EC50 of SMN-C3 in the splicing assay was ~0.1 µM6,19, a concentration at the higher end (20 µM) was used to ensure the binding state of the small molecule and target RNA sequence.
Upon isolation of the RNA, both gene-specific and random primers can be used for extension. In the case of exon 7 of SMN2, we found that SMN2E7-338-RV (Table 2) yields a higher copy of desired cDNA than a random nonamer, evidenced by a more intense band in the PCR amplification with SMN2E7-276 primer set (Table 2) after 25 cycles. In the 2'-OH modification step, a 91 µM final NAI concentration was used for an incubation period of 15 min. If a different cell type or medium is used, the incubation time must be re-optimized. If the incubation time is too long, agarose gel analysis of the amplicon will sometimes fail to show a band.
A python-based program, ShapeMapper, developed by the Weeks laboratory35 was used to analyze the data generated by next-generation sequencing [for raw data of SMN-C2 treated in-cell SHAPE of SMN2 exon 7 pre-mRNA, refer to the Sequence Read Archive (SRA) database]. Throughout the amplicon, the SHAPE reactivity did not significantly change (>1), which indicated that the secondary structure remains in the presence of SMN-C2 (Figure 2B). This is also confirmed by the arc plots generated by SuperFold35. The connection lines indicate the possible base pairing based on SHAPE activity of each nucleotide. SMN-C2- and DMSO-treated RNA modeling is essentially the same (Figure 2C). Differential in-cell SHAPE reactivity was calculated for each nucleotide (Figure 2B), and the most reactivity change occurred at TSL-1 (5'-end of exon 7) but not TSL-2 (3'-end of exon 7). This result agrees with the in vitro SHAPE; although, the base pairing shifted from in vitro SHAPE RNA modeling in the in-cell SHAPE analyses (Figure 2D).
A common problem of in-cell SHAPE is the low mutation rate throughout the amplicon. This is usually due to genomic DNA contamination. DNA does not contain 2'-OH group; therefore, no acylation product is formed with NAI. PCR amplification thus merely reflects the low mutation rate of the DNA polymerase. Isolation of RNA by guanidinium thiocyanate followed by on-column DNase digestion is sufficient to remove all DNA in most cases. If DNA contamination persists, repeat step 2.3 for RNA isolation.
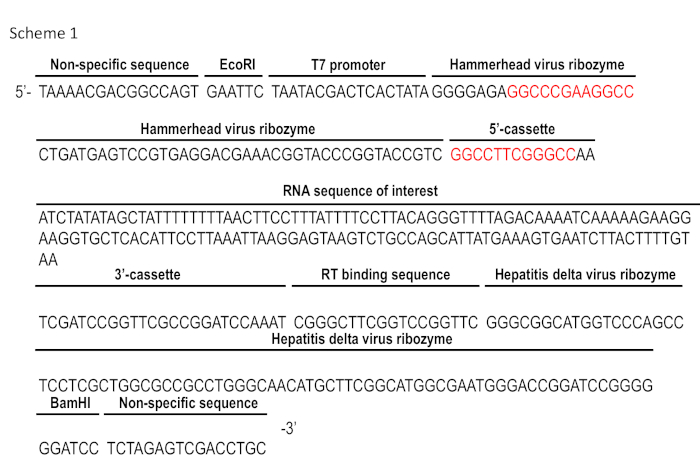
Scheme 1: DNA sequence for the template of RNA transcription. The 12 bp sequence within the Hammerhead virus ribozyme sequence in red is the reverse complement of the first 12 bp of the 5'-cassette. The RNA sequence of interest presented herein is a 140 bp pre-mRNA sequence contains exon 7 of human SMN2 gene. Please click here to view a larger version of this figure.
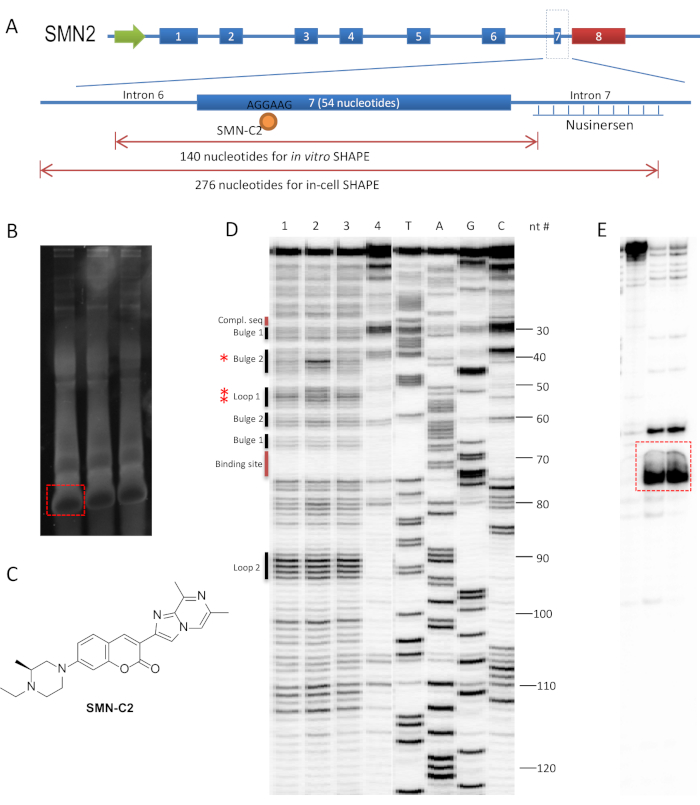
Figure 1: Experimental design and result of in vitro SHAPE for SMN2 exon 7 pre-mRNA in the presence of SMN-C2. (A) Overview of the sequence of interest for in vitro and in-cell SHAPE studies of the SMN2 gene. SMN-C2 binds to the AGGAAG motif on exon 7, a distinct location from the nusinersen binding site. (B) Purification of the T7 transcription product. Each of the three lanes contains 5.0 µg of crude RNA. The TBE-urea gel was stained with SYBR-Safe (1:10,000) for 5 min in 1x TBE buffer. Red dashed box indicates the edge of excision for RNA recovery. (C) The structure of SMN-C2. (D) In vitro SHAPE experiment with NAI and a 140 nucleotide-long RNA template containing exon 7. 1 = DMSO; 2 = SMN-C2 (50 µM); 3 = SMN-C2 (5 µM); 4 = lacking of NAI; 5-8 = ladders generated by addition of ddATP, ddTTP, ddCTP, and ddGTP during primer extension. PAGE was carried out on a TBE-urea sequencing gel at 60 W for 3 h. Red asterisks indicate increased band intensity with 50 µM SMN-C26. (E) An example of a "salt front" region in the dashed red box from a separate experiment. Please click here to view a larger version of this figure.
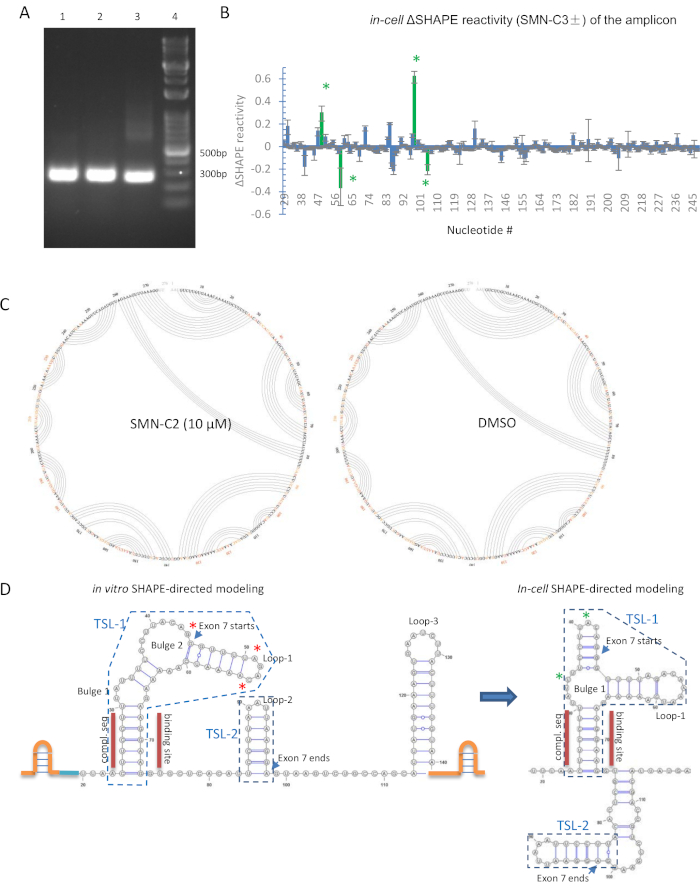
Figure 2: In-cell SHAPE derived RNA modeling for SMN2 exon 7 pre-mRNA. (A) PCR amplification with all three primer sets (Table 2) yielded a single band in agarose gel analysis. 1 = SMN2E7-338, 2 = SMNE7-276, 3 = SMN2E7-251, 4 = DNA ladder. (B) Differential in-cell SHAPE reactivity in SMN2 minigene-transfected 293T cells for 10 µM SMN-C2 and DMSO in TSL1. SHAPE reactivity at single-nucleotide resolution. Its standard deviation was calculated by ShapeMapper software35. Green asterisks indicate significant SHAPE reactivity change induced by 10 µM SMN-C2. The numbering of the nucleotides in the 276 bp amplicon shown on x-axis. Error bars were estimated by Shape-Mapper software26. (C) Arc plot generated by SuperFold35 for the most plausible RNA secondary structure modeling by in-cell SHAPE data. (D) In vitro and in-cell SHAPE-directed modeling of exon 7 and adjacent regions. For in vitro RNA model, SHAPE stabilizing cassette (orange) and nucleotides 1-19 (blue) are shown in sketch. For in-cell RNA model, nucleotide numbering is aligned with in vitro SHAPE template. Nucleotides 1-18 and 120-140 were omitted. Significant reactivity changes are indicated in red and green asterisks for in vitro and in-cell SHAPE, respectively. The secondary structures that were previously named TSL1 and TSL231 are enclosed in blue boxes. Please click here to view a larger version of this figure.
| Primer name | 5'-3' Sequence |
| Ribozyme-FW | TAAAACGACGGCCAGTGAAT |
| Ribozyme-RV | GCAGGTCGACTCTAGAGGAT |
| RT primer | GAACCGGACCGAAGCCCG |
Table 1: The primer sequences used in the representative experiment.
| Name | Sequence (5'->3') |
| SMN2E7-338-FW | AAAGACTATCAACTTAATTTCTGA |
| SMN2E7-338-RV | TGTTTTACATTAACCTTTCAACT |
| SMN2E7-276-FW | AATGTCTTGTGAAACAAAATGCT |
| SMN2E7-276-RV | AACCTTTCAACTTTCTAACATCT |
| SMN2E7-251-FW | TGAAACAAAATGCTTTTTAACATCC |
| SMN2E7-251-RV | TCAACTTTCTAACATCTGAACTTTT |
Table 2: The primer sets for amplification of the pre-mRNA sequence of interest. All three primer sets yield a single amplicon in a PCR reaction with genomic DNA-free cDNA template.
Discussion
In in vitro SHAPE, it is critical to use high quality homogeneous RNA template. T7 transcription, however, often yields heterogeneous sequences36. Especially, sequences with ±1 nucleotide at the 3'-terminus with non-negligible yields36 are usually difficult to be removed by polyacrylamide gel purification. Heterogeneous RNA template can result in more than one set of the signal in the sequencing gel profiling of the primer extension product, which sometimes makes it difficult to interpret the result. The ribozyme at both 5'- and 3'-ends of the RNA template expression cassette will make both ends homogenous.
For both in vitro and in-cell SHAPE, the incubation time of 2'-OH modification reagents is another critical factor. It was suggested by the Weeks group that at least five times the half-life of the water-quenching 2'-OH acylation reagent should be used1. In our hands, incubating with NAI (T1/2 = ~20 min) for >30 min in both in vitro and in-cell SHAPE results in an overreacted result. As shown in Figure 1D, a good in vitro SHAPE profiling should have >50% total signal on top of the gel as full-length transcript, ensuring that most of the modified RNA is only acylated once. Overreaction will render the double-acylated product non-negligible and the profiling biased to the enriched short length extension products. In in-cell SHAPE, the amplicon for the library construction should appear as a single band in agarose gel analysis (step 2.6.3). Smear of the band indicates an overreaction, and the incubate time should be reduced. In the representative experiments, a 15 min incubation time at 37 °C was optimal for in vitro and in-cell SHAPE. This should be used as a starting point for NAI modification in similar applications.
There are other 2'-OH modification reagents22 widely used for SHAPE, such as 1M7. Compared to NAI, 1M7 has a better reactivity to 2'-OH group and shorter half-life in water22. In our hands, 1M7 formed massive amount of yellow precipitation in cell culture media in an in-cell SHAPE experiment, which complicated the RNA isolation. For a comparison reason, both in vitro and in-cell SHAPE used NAI as the 2'-OH modification reagent for a study of SMN-C2 and SMN2 pre-mRNA interactions. If only in vitro SHAPE is required, 1M7 is an alternative option as demonstrated by various studies in riboswitch structural determination7,8.
In general, in-cell SHAPE over in vitro SHAPE is preferred, especially if the molecule is presumably acting in the nucleus of a eukaryotic cell. RNA-binding proteins in the nucleus are abundant, and it is almost impossible to recapitulate the cellular context under in vitro conditions.
In the past decade, SHAPE becomes the standard method for studying the secondary structure of RNA. Compared to traditional RNA footprinting with RNase37, it is more suitable to study small molecule-RNA interactions, as these interaction can sometimes be weak and insensitive to RNase challenges.
The major limitation of using SHAPE to study small molecule-induced RNA structural changes is that its results do not reveal the binding site. In in vitro and in-cell SHAPE for SMN-C2 bound pre-mRNA, the SHAPE reactivity did not change at the putative binding site (Figure 1D, Figure 2B); rather, the reactivity at 2 to 3 remote sites at the loop or budge region were altered. SMN-C2 presumably binds to an RNA double-helix region (Figure 1D, Figure 2D), which usually has a low SHAPE reactivity. Therefore, further stabilizing the structure to decrease SHAPE reactivity would probably not be observable. To generate a putative binding site, other methods such as ChemCLIP38 should be used, in which a crosslinking chemical probe is involved.
A common alternative approach to map RNA secondary or higher structure and nucleotide dynamics is NMR spectrometry39,40. It has been demonstrated that RNA dynamics derived from quantitative NMR analysis strongly correlate with SHAPE activity39. In the context of small molecule binding, chemical shift perturbations can reveal the interacting nucleotides21.
As RNA-targeting small molecules become a new modality of drug development11, we envision that SHAPE will be established as a standard methodology to evaluate the structural impacts of target RNA in the presence of small molecules. In the future, a transcriptome-wide interrogation method for RNA-targeting small molecule drugs is desired.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was made possible by the NIH R01 grant (NS094721, K.A.J.).
Materials
| DNA oligonucleotide | IDT | gBlock for > 200 bp DNA synthesis | |
| Phusion Green Hot Start II High-Fidelity PCR Master Mix | Thermo Fisher | F566S | |
| NucleoSpin gel and PCR clean-up kit | Takara | 740609.50 | |
| MegaScript T7 transcription kit | Thermo Fisher | AM1333 | Contains 10X reaction buffer, T7 enzyme, NTP and Turbo DNase |
| DEPC-treated water | Thermo Fisher | 750023 | |
| 2X TBE-urea sample buffer | Thermo Fisher | LC6876 | |
| 40% acrylamide/ bisacrylamide solution (29:1) | Bio-Rad | 1610146 | |
| 10X TBE buffer | Thermo Fisher | 15581044 | |
| Nalgene Rapid-Flow™ Filter Unit | Thermo Fisher | 166-0045 | |
| Kimwipe | Kimberly-Clark | 34133 | |
| TEMED | Thermo Fisher | 17919 | |
| SYBR-Safe dye | Thermo Fisher | S33102 | |
| 6 % TBE-urea mini-gel | Thermo Fisher | EC6865BOX | |
| ChemiDoc | Bio-Rad | ||
| T4 PNK | NEB | M0201S | |
| γ-32P-ATP | Perkin Elmer | NEG035C005MC | |
| Hyperscreen™ Intensifying Screen | GE Healthcare | RPN1669 | calcium tungstate phosphor screen |
| phosphor storage screen | Molecular Dynamics | BAS-IP MS 3543 E | |
| Amersham Typhoon | GE Healthcare | ||
| NAI (2M) | EMD Millipore | 03-310 | |
| GlycoBlue | Thermo Fisher | AM9515 | |
| SuperScript IV Reverse Transcriptase | Thermo Fisher | 18090010 | Contains 5X RT buffer, SuperScript IV |
| dNTP mix (10 mM) | Thermo Fisher | R0192 | |
| ddNTP set (5mM) | Sigma | GE27-2045-01 | |
| large filter paper | Whatman | 1001-917 | |
| Gel dryer | Hoefer | GD 2000 | |
| QIAamp DNA Blood Mini Kit | Qiagen | 51104 | Also contains RNase A and protease K |
| SMN2 minigene34 | Addgene | 72287 | |
| Heat inactivated FBS | Thermo Fisher | 10438026 | |
| Pen-Strep | Thermo Fisher | 15140122 | |
| Opti-MEM I | Thermo Fisher | 31985062 | |
| FuGene HD | Promega | E2311 | |
| TrpLE | Thermo Fisher | 12605010 | |
| DPBS without Ca/ Mg | Thermo Fisher | 14190250 | |
| TRIzol | Thermo Fisher | 15596018 | |
| RNeasy mini column | Qiagen | 74104 | Also contains RW1, RPE buffer |
| RNase-Free DNase Set | Qiagen | 79254 | Contains DNase I and RDD buffer |
| Deionized formamide | Thermo Fisher | AM9342 | |
| MnCl2•4H2O | Sigma-Aldrich | M3634 | |
| random nonamer | Sigma-Aldrich | R7647 | |
| SuperScript First-Strand Synthesis System | Thermo Fisher | 11904-018 | Contains 10X RT buffer, SuperScript II reverse transcriptase |
| AccuPrime pfx DNA polymerse | Thermo Fisher | 12344024 | |
| NextSeq500 | Illumina | ||
| NucAway column | Thermo Fisher | AM10070 | for desalting purpose |
Referencias
- Wilkinson, K. A., Merino, E. J., Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols. 1, 1610-1616 (2006).
- Trausch, J. J., et al. Structural basis for diversity in the SAM clan of riboswitches. Proceedings of the National Academy of Sciences. 111, 6624-6629 (2014).
- Souliere, M. F., et al. Tuning a riboswitch response through structural extension of a pseudoknot. Proceedings of the National Academy of Sciences. 110, E3256-E3264. , E3256-E3264 (2013).
- Rice, G. M., Busan, S., Karabiber, F., Favorov, O. V., Weeks, K. M. SHAPE Analysis of Small RNAs and Riboswitches. Methods in Enzymology. , 165-187 (2014).
- Spitale, R. C., et al. RNA SHAPE analysis in living cells. Nature Chemical Biology. 9, 18-20 (2013).
- Wang, J., Schultz, P. G., Johnson, K. A. Mechanistic studies of a small-molecule modulator of SMN2 splicing. Proceedings of the National Academy of Sciences. 115, E4604-E4612 (2018).
- Price, I. R., Gaballa, A., Ding, F., Helmann, J. D., Ke, A. Mn 2+ -Sensing Mechanisms of yybP-ykoY Orphan Riboswitches. Molecular Cell. 57, 1110-1123 (2015).
- Hennelly, S. P., Sanbonmatsu, K. Y. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Research. 39, 2416-2431 (2011).
- Abulwerdi, F. A., et al. Development of Small Molecules with a Noncanonical Binding Mode to HIV-1 Trans Activation Response (TAR) RNA. Journal of Medicinal Chemistry. 59, 11148-11160 (2016).
- Shortridge, M. D., et al. A Macrocyclic Peptide Ligand Binds the Oncogenic MicroRNA-21 Precursor and Suppresses Dicer Processing. ACS Chemical Biology. 12, 1611-1620 (2017).
- Warner, K. D., Hajdin, C. E., Weeks, K. M. Principles for targeting RNA with drug-like small molecules. Nature Reviews Drug Discovery. 17, 547-558 (2018).
- Mullard, A. Small molecules against RNA targets attract big backers. Nature Reviews Drug Discovery. 16, 813-815 (2017).
- Shortridge, M. D., Varani, G. Structure based approaches for targeting non-coding RNAs with small molecules. Current Opinion in Structural Biology. 30, 79-88 (2015).
- Gallego, J., Varani, G. Targeting RNA with Small-Molecule Drugs: Therapeutic Promise and Chemical Challenges. Accounts of Chemical Research. 34, 836-843 (2001).
- Rizvi, N. F., Smith, G. F. RNA as a small molecule druggable target. Bioorganic & Medicinal Chemistry Letters. 27, 5083-5088 (2017).
- Connelly, C. M., Moon, M. H., Schneekloth, J. S. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chemical Biology. 23, 1077-1090 (2016).
- Palacino, J., et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nature Chemical Biology. 11, 511-517 (2015).
- Ratni, H., et al. Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 ( SMN2 ) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA). Journal of Medicinal Chemistry. 61, 6501-6517 (2018).
- Naryshkin, N., et al. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 345, (2014).
- Chiriboga, C., et al. Preliminary Evidence for Pharmacodynamics Effects of RG7916 in JEWELFISH, a Study in Patients with Spinal Muscular Atrophy who Previously Participated in a Study with Another SMN2-Splicing Targeting Therapy (S46.003). Neurology. 90, (2018).
- Sivaramakrishnan, M., et al. Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers. Nature Communications. 8, (2017).
- Lee, B., et al. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA. 23, 169-174 (2017).
- Weeks, K. M., Mauger, D. M. Exploring RNA Structural Codes with SHAPE Chemistry. Accounts of Chemical Research. 44, 1280-1291 (2011).
- Smola, M. J., et al. SHAPE reveals transcript-wide interactions , complex structural domains , and protein interactions across the Xist lncRNA in living cells. Proceedings of the National Academy of Sciences. 113, 10322-10327 (2016).
- Flynn, R. A., et al. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nature Protocols. 11, 273-290 (2016).
- Smola, M. J., Rice, G. M., Busan, S., Siegfried, N. A., Weeks, K. M. Selective 2 ′ -hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct , versatile and accurate RNA structure analysis. Nature Protocols. 10, 1643-1669 (2015).
- Hua, Y., et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes & Development. 24, 1634-1644 (2010).
- Hua, Y., et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 478, 123-126 (2011).
- Wee, C. D., Havens, M. A., Jodelka, F. M., Hastings, M. L. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PloS One. 9, e115205 (2014).
- Hammond, J. A., Rambo, R. P., Filbin, M. E., Kieft, J. S. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 15, 294-307 (2009).
- Singh, N. N., Singh, R. N., Androphy, E. J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Research. 35, 371-389 (2007).
- Deigan, K. E., Li, T. W., Mathews, D. H., Weeks, K. M. Accurate SHAPE-directed RNA structure determination. Proceedings of the National Academy of Sciences. 106, 97-102 (2009).
- Qi, H., et al. . Preparation of pyridopyrimidine derivatives and related compounds for treating spinal muscular atrophy. , (2013).
- Lorson, C. L., Hahnen, E., Androphy, E. J., Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Academy of Sciences of the United States of America. 96, 6307-6311 (1999).
- Siegfried, N. A., Busan, S., Rice, G. M., Nelson, J. A. E., Weeks, K. M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nature Methods. 11, 959 (2014).
- Milligan, J. F., Groebe, D. R., Witherell, G. W., Uhlenbeck, O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Research. 15, 8783-8798 (1987).
- Nilsen, T. W. RNase Footprinting to Map Sites of RNA-Protein Interactions. Cold Spring Harbor Protocols. , (2014).
- Velagapudi, S. P., et al. Design of a small molecule against an oncogenic noncoding RNA. Proceedings of the National Academy of Sciences. , 5898-5903 (2016).
- Barnwal, R. P., Yang, F., Varani, G. Applications of NMR to structure determination of RNAs large and small. Archives of Biochemistry and Biophysics. 628, 42-56 (2017).
- Scott, L. G., Hennig, M. RNA structure determination by NMR. Methods in Molecular Biology. 452, 29-61 (2008).

