Surgical Size Reduction of Zebrafish for the Study of Embryonic Pattern Scaling
Summary
Here, we describe a method for reducing the size of zebrafish embryos without disrupting normal developmental processes. This technique enables the study of pattern scaling and developmental robustness against size change.
Abstract
In the developmental process, embryos exhibit a remarkable ability to match their body pattern to their body size; their body proportion is maintained even in embryos that are larger or smaller, within certain limits. Although this phenomenon of scaling has attracted attention for over a century, understanding the underlying mechanisms has been limited, owing in part to a lack of quantitative description of developmental dynamics in embryos of varied sizes. To overcome this limitation, we developed a new technique to surgically reduce the size of zebrafish embryos, which have great advantages for in vivo live imaging. We demonstrate that after balanced removal of cells and yolk at the blastula stage in separate steps, embryos can quickly recover under the right conditions and develop into smaller but otherwise normal embryos. Since this technique does not require special equipment, it is easily adaptable, and can be used to study a wide range of scaling problems, including robustness of morphogen mediated patterning.
Introduction
Scientists have long known that embryos have a remarkable ability to form constant body proportions although embryo size can vary greatly both under natural and experimental conditions1,2,3. Despite decades of theoretical and experimental studies, this robustness to size variation, termed scaling, and its underlying mechanisms remain unknown in many tissues and organs. In order to directly capture the dynamics of the developing system, we established a reproducible and simple size reduction technique in zebrafish4, which has the great advantage in in vivo live imaging5.
Zebrafish has served as a model vertebrate animal to study multiple disciplines of biology, including developmental biology. In particular, zebrafish is ideal for in vivo live imaging6 because 1) development can proceed normally outside the mother and the egg shell, and 2) the embryos are transparent. In addition, the embryos can withstand some temperature and environmental fluctuations, which allows them to be studied in laboratory conditions. Also, in addition to conventional gene expression perturbation by morpholino and mRNA injection7,8, recent advances in CRISPR/Cas9 technology has made reverse genetics in zebrafish highly efficient9. Furthermore, many classical techniques in embryology, such as cell transplantation or tissue surgery can be applied4,10,11.
Size reduction techniques were originally developed in amphibian and other non-vertebrate animals12. For example, in Xenopus laevis, another popular vertebrate animal model, bisection along the animal-vegetal axis at blastula stage can produce size-reduced embryos12,13. However, in our hands this one-step approach results in dorsalized or ventralized embryos in zebrafish, presumably because dorsal determinants are distributed unevenly and one cannot know their localization from the morphology of embryos. Here we demonstrate an alternative two-step chopping technique for zebrafish that produces normally developing but smaller embryos. With this technique, cells are first removed from the animal pole, a region of naïve cells lacking in organizer activity. To balance the amount of yolk and cells, which is important for epiboly and subsequent morphogenesis, yolk is then removed. Here, we detail this protocol and provide two examples of size invariance in pattern formation; somite formation and ventral neural tube patterning. Combined with quantitative imaging, we utilized the size reduction technique to examine the how the sizes of somites and neural tube are affected in size reduced embryos.
Protocol
All fish-related procedures were carried out with the approval of the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
1. Tool and Reagent Preparation
- Make a wire loop to chop embryos
- Take 20 cm of stainless steel wire that is stiff and non-corrosive with a diameter of 40 μm. Loop the wire through into glass capillary (1.0 mm outer diameter, 0.5 mm inner diameter, no filament), making a small loop at the top (loop length is 1.0 mm)
- Put a little droplet of clear nail polish onto the tip of the glass capillary between the wire loop to hold it in place. Let dry. Make sure not to get any nail polish onto the loop portion, as it may damage the embryos.
- Attach the glass capillary with the loop onto a wooden chopstick (9” bamboo disposable chopsticks broken in half) using lab tape. Leave about 2.5 cm of the glass capillary extending beyond the chopstick, so that the chopstick part of the tool does not dip into the water. Adjust this length to preference.
- Alternatively, make a glass needle to chop embryos
- Pinch one end of glass Pasteur pipette with forceps, while holding the other side with a hand. Heat the thin part of the pipette over a spirit lamp or Bunsen burner.
- Hand-pull the glass pipette. The ideal pipette as a diameter of approximately 30 μm and a gentle curve (radius of curvature = approximately 5 mm). Proper diameter and curvature is obtained with practice and some chance.
- Prepare methyl cellulose
- Methyl cellulose is used to hold the embryos while chopping. Make 2% methyl cellulose solution in ⅓x Ringer’s solution (116 mM NaCl, 2.9 mM KCl, 1.8 mM CaCl2, and 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.2)). Shake methyl cellulose powder in ⅓x Ringer’s solution at 4 °C overnight.
- Optionally, add ~1.5 mL phenol red (0.5% in DPBS stock solution) to 10 mL 2% methyl cellulose solution, until red, to make the solution visible. Shake it until the color becomes uniform.
2. Preparation of Zebrafish Embryos for Surgical Size Reduction
- Collect embryos
- Place one or two male and one or two female zebrafish in a mating chamber filled with a lot of water. Separate males and females using a plastic divider. Use the AB line for somite imaging (section 4) and a transgenic reporter line of neural tube patterning (nkx2.2:mem-gfp, dbx1b:gfp, or Olig2:dsred) for neural tube imaging. Leave them in the chamber overnight.
NOTE: The choice of adults is important for obtaining eggs that survive surgery well. Typically, young healthy females produce healthy eggs. - On the next morning, transfer the chamber into shallow water and place the chamber with slight tilt. Remove the divider so the fish can mate.
- Collect the eggs by pouring into a tea strainer. Transfer the eggs into a Petri dish with egg water (for 20x egg water, 6 g instant sea salt, 1.5 g CaSO4 and 1 L H2O; use at 1x). For better staging, collect the eggs right after spawning. Place the embryos in a 28.5 °C incubator.
- If necessary, inject morpholino, mRNA, etc., at 1–4 cell stages following a common protocol7,8. Inject mRNA for fluorescent membrane label (mem-mCherry, mem-mBFP1, or mem-mCitrine) for neural tube imaging (section 5).
- Place one or two male and one or two female zebrafish in a mating chamber filled with a lot of water. Separate males and females using a plastic divider. Use the AB line for somite imaging (section 4) and a transgenic reporter line of neural tube patterning (nkx2.2:mem-gfp, dbx1b:gfp, or Olig2:dsred) for neural tube imaging. Leave them in the chamber overnight.
- Dechorionate using pronase
- Around the 128-cell to 256-cell stage, transfer healthy embryos to a 35 mm glass dish filled with egg water. Remove as much egg water as possible from the dish.
- Add 1 mL of 20 mg/mL pronase. Gently swirl embryos around by moving the plate and gently pipette the solution up and down to help dechorionation using a glass pipette.
- When the chorions start to lose elasticity (usually 1-4 min after addition of pronase, depending on the amount of eggs and water), add as much egg water as possible to dilute pronase. Assess elasticity loss by gently touching the chorion with forceps; the chorion should hold the dent without immediately bouncing back. Transfer the eggs to another dish with egg water using a glass pipette (at this point, most of the chorions are not broken).
- Alternatively, gently pour the embryos from the dish into a large 400 mL glass beaker filled with egg water without exposing embryos to air. Then, let the embryos completely settle. Tilt the beaker to let embryos fall to one side. Collect these embryos using a glass pipette and transfer to a new 35 mm glass dish filled with ⅓x Ringer’s solution.
- Wait for several minutes until most of the chorions lose elasticity completely. Remove the remaining chorions from embryos by gently pipetting the eggs. Remove damaged embryos and incubate the embryos in a 28.5 °C incubator.
3. Surgical Size Reduction and Recovery
- Prepare one clean 35 mm glass dish. Spread approximately 0.5 mL of 2% methyl cellulose (in ⅓x Ringer’s solution, with phenol red to help visualize) near the center of the bottom of the larger dish using a plastic spatula. Thinly and evenly spread the methyl cellulose to a thickness of approximately 0.5 mm.
- Pour approximately 30 mL of ⅓x Ringer’s solution to the side of the dish and allow to spread onto the rest of the dish, covering the methyl cellulose.
- Place dechorionated embryos at the 256-cell-1k-cell stage on 2% methyl cellulose. Adjust the orientation of the embryos onto the side to visualize both the cells and the yolk. (Figure 1)
- Chop approximately 30%-40% of cells from the blastoderm near the animal pole by cutting perpendicular to the animal-vegetal axis using the wire loop (or the glass needle) (Figure 1, top panels). After removing the cells, gently tap the ends together to help the remaining cells stick back. Within minutes, the dead cells should slough off when the embryo starts to heal.
- Make a small wound to the yolk near the vegetal pole instead of “chopping” the yolk (Figure 1 middle panels). Wound the yolk by nicking the egg membrane with the mounted wire. Yolk will ooze out for few minutes after wounding, and then the wound will heal (Figure 1 bottom panels).
- When the yolk stops oozing out, move the embryo using a pipet outside of the methyl cellulose in the same dish to allow them to better recover.
- Repeat steps 3.4-3.6 for all embryos. Leave the dish stable for 30 min while the embryos recover.
- Transfer the embryos to a new dish with fresh ⅓x Ringer’s solution and put them in 28.5 °C incubator to allow them to completely recover.
- (Optional) If the stage of interest is the early somite stage, embryos can be incubated at 20 °C after being incubated at 28.5 °C until the shield stage, to adjust timing for experimentation and imaging.
4. Live Imaging of Zebrafish Somitogenesis
- Prepare 100 mL of 1% agarose solution by adding 1 g of agarose to 100 mL of egg water, heating until agarose is fully dissolved. Let cool for 5-10 min to a temperature of 62-72 °C.
- Prepare a mount for imaging.
NOTE: This guide was developed for use with an inverted wide-field microscope.- Pour ~15 mL of 1% agarose in to a 100 mm x 15 mm plastic Petri dish.
- Gently place a Dorsal Mount V1 mold template5 onto the bottom of the Petri dish. Keep the mold on the bottom by using a weight, such as 15 mL tube with water.
NOTE: This is so the embryos are as close to the bottom of the Petri dish as possible, when using an inverted microscope. Although the embryos are mounted laterally for somite imaging, here Dorsal Mount V15 is used since it holds the embryos better at early somite stages. - Let cool until agarose is fully solidified. Pour in ~30 mL of egg water with 0.01% tricaine, and carefully remove mold by gently prying off with forceps.
- Mount embryos for somite imaging
- Prepare 1% Low Melting Agarose in ⅓x Ringer’s solution (or egg water) and keep it at 42 °C. Wait until the temperature comes down to 42 °C.
- Under a dissection microscope, place one embryo per well. Pipette approximately 1 μL of Low Melting Agarose in the well to finely adjust the well size to the size of individual embryos that vary in size, especially between those with and without size reduction.
- Quickly orient the embryos before the Low Melting Agarose is solidified so the embryos face completely laterally to the dish. After mounting embryos, gently place the cover slip in the agarose mount to hold the embryos in place. The orientation is particularly important for the long-term somite imaging because the embryos have to be exactly laterally mounted for all the somite boundaries to be clearly imaged at later stages.
- Submerge away from the mounting area and manipulate a 25 mm x 25 mm cover glass such that it is 45 degrees offset from the orientation of the square indent made by the mold. Slide the coverslip over the mold in this orientation until each corner is resting of a different side of the mold.
NOTE: Although this protocol is for an inverted microscope, placing a cover glass on top of the mold is still important so that the embryos do not move while transferring to the microscope.
- Imaging somite formation process
- Prewarm the microscope to 28 °C in an incubator built with foamcore boards and a cabinet heater.
- Place the Petri dish with the mounted embryos onto the microscope stage. Find the embryo mounted in the top-left well of the agarose mount with lower magnification lens, then switch the objective to 10x.
- Set up image acquisition.
NOTE: This instruction is for bright field. - Find conditions for the light power, exposure time, and condenser so that the somite boundary can be clearly seen. Using z-stack settings set the lowest and highest desired imaging plane. Set the total time and time interval. Find and register the xy positions of all the embryos mounted so multiple embryos can be timelapse imaged at once.
- Measure the lengths of PSM and somites using Fiji16.
5. Imaging of Neural Tube Patterning
- Prepare 100 mL of 1% agarose solution by adding 1 g of agarose to 100 mL of egg water, heating until agarose is fully dissolved. Let cool for 5-10 min to 62-72 °C. Adjust tricaine to 0.01%.
- Prepare a dorsal mount for imaging.
NOTE: This guide is developed for use with an upright fluorescence microscope. For inverted microscopes coverslip bottom dishes are necessary and mounting orientation in step 3 is flipped.- Pour in ~15 mL of 1% agarose in to a 100 mm x 15 mm plastic Petri dish. Gently float a Dorsal Mount V1 mold on the surface of the agarose to avoid introducing air bubbles. Let cool until agarose is fully solidified.
- Carefully remove the mold with forceps or a razor blade. Fill the dish with egg water with 0.01% tricaine and cover until use.
- Mount embryos for neural tube imaging.
- Allow transgenic or injected fluorescent size-reduced and control embryos to develop until the 20-25 somite stage (roughly 18-22 h after fertilization). Embryos expressing a fluorescent membrane label (mem-mCherry, mem-mBFP1, or mem-mCitrine) and a transgenic reporter of neural tube patterning (nkx2.2:mem-gfp, dbx1b:gfp, or Olig2:dsred) are used for imaging.
- Replace egg water medium in prepared dorsal mount with 0.01% tricaine working solution. Gently pipette in embryos to be imaged in to wells of the dorsal mount.
- Manipulate embryos into the correct orientation such that the head is facing forward in the mount and the tail is pointing towards the rear with the dorsal portion facing towards the water surface.
- Orient the embryo such that the tail is lying flat and not pointed downward towards the bottom of the dish. It is sometimes helpful to rest the posterior portion of the tail on the side ledge of the mounting well to prevent the tail from sinking and inadvertently imaging the hindbrain. Repeat for each embryo. Make certain with size reduced embryos that they do not fall to deeply into the wells to be imaged.
- Submerge away from the mounting area and manipulate a 25 mm x 25 mm cover glass such that it is 45° offset from the orientation of the square indent made by the mold. Slide the coverslip over the mold in this orientation until each corner is resting of a different side of the mold.
- Slowly rotate the coverglass until it gently falls on to the mounting area. Check to be sure embryos are still mounted in the correct positions and orientations. If too much movement is caused by the falling of the coverglass, remove gently and repeat mounting from step C.
- Image in 3-D the developing spinal cord.
- Gently transport the dish containing mounted embryos to the confocal microscope. For long-term live imaging, be sure to use an incubated microscope. Otherwise, low temperatures are tolerable.
- Under brightfield illumination, navigate to the embryo to image and center the field of view on the preferred anterior-posterior position. To enable a detailed comparison between embryos, this position on each embryo must be consistent.
- Set the imaging parameters to excite and capture signal from the fluorescent proteins being imaged. Parameters can be tuned by eye to create a desired image on the initial sample. To enable measurement consistency, apply these settings to all embryos in the dataset. If many z-stacks are required optimize settings for speed.
- Using z-stack settings, set the lowest and highest desired imaging plane. For optimal image quality in the z-stack, set the z-resolution to the highest that is optimal for the aperture setting. Good images can be generated with 1 μm z-spacing.
- Analyze imaging data by any preferred method.
NOTE: In this case, analysis was performed with custom MATLAB scripts that enable simple segmentation of the neural portions of an image and quantification of imaging signal.
Representative Results
Yolk volume reduction is important for normal morphology
As recently described in Almuedo-Castillo et al.17, size reduction of embryos can be achieved without reducing yolk volume. To compare with and without yolk volume reduction, we performed both two-step chopping (both blastula and yolk) and blastula-only chopping (Figure 2 and Supplemental Movie 1). Two-step chopped embryos showed seemingly normal overall morphology compared to the control (dechorionation only) embryos, other than size difference, throughout the developmental stages (see top and middle panels in Figure 2). On the other hand, blastula-only chopped embryos showed a peculiar morphology, especially at earlier stages. During epiboly, the embryos had a constricted and indented appearance (See bottom panel for 70% epiboly in Figure 2). At the following somite stage, midline structures were found to be flattened (i.e. DV length is relatively shorter than ML length) at many axial levels (See bottom panels for 8 and 20 somite in Figure 2). At later stages, the body structures adjacent to the yolk, such as the mid- and hindbrain, and the first ~10 somites, still showed a relatively flattened shape, possibly due to increased tension from the relatively larger yolk.
Somite size reduction in size reduced embryos
Somites are segmental structures that appear transiently during embryogenesis and give rise to vertebrae and skeletal muscle. From presomitic mesoderm (PSM), somites are formed one by one from the anterior to posterior direction in a periodic manner (e.g. 25 min for zebrafish, 2 h for mice) (Figure 3A). We performed time lapse imaging of somite formation both for control and chopped embryos and measured the size of most newly formed somites (Figure 3B). In both control and chopped embryos, the sizes of somites that were formed at later stages were found to be smaller compared to the ones from earlier stages. Also, throughout the somite formation stages, chopped embryos had smaller somites than the ones in control embryos (Figure 3C).
Neural tube heights are reduced following size reduction
To see the effect of embryo size reduction on neural tube size, we performed our two-step chopping technique on mem-mCherry injected embryos and imaged their spinal cords at 20 hpf using our confocal imaging system (Figure 4A,B). In this dataset, neural tube heights were reduced following size reduction by 12.4% ± 3.2%, as measured manually using custom image analysis code (Figure 4C). Taken together, these data show that size reduction reduces neural tube height. This technique can be used to measure the effects of size reduction on neural patterning.
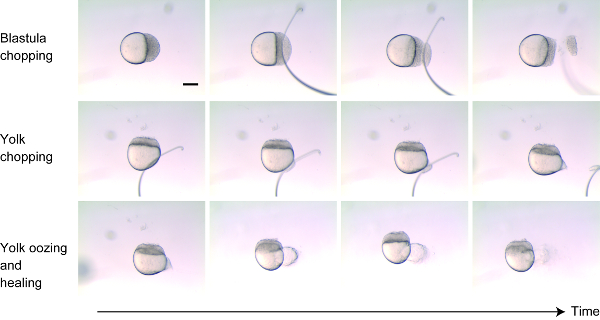
Figure 1: Size reduction technique. Approximately 30%-40% of the cells were cut from the animal pole (top panels). The membrane surrounding the yolk was carefully wounded so that the yolk oozed out (middle panels). For the following few minutes, yolk oozed out and then the wounds on both blastoderm and yolk healed up (bottom panels). Scale bar = 200 μm. Please click here to view a larger version of this figure.
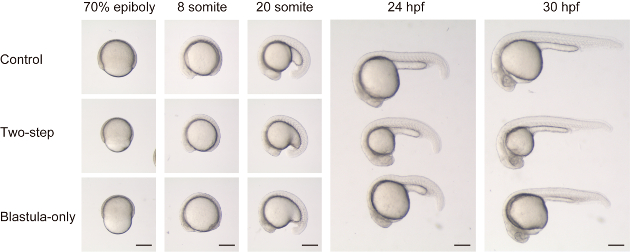
Figure 2: Comparison between two methods of size reduction. Control embryos (top panels, top embryos for 24 hpf and 30 hpf), size reduced embryos with two-step chopping (blastula and yolk, middle panels, middle embryos for 24 hpf and 30 hpf) and size reduced embryos with blastula-only chopping (bottom panels, bottom embryos for 24 hpf and 30 hpf) are compared along developmental stages. Note that in blastula-only chopped embryos, the blastoderm volume is much smaller compared to the yolk (at 70% epiboly). As a result, the embryo has a disproportionately flattened shape at somite stages (i.e., DV axis is relatively shorter compared to AP axis in blastula-only chopped embryos, compared to the control or a two-step chopped one). Scale ba = 200 μm. Please click here to view a larger version of this figure.
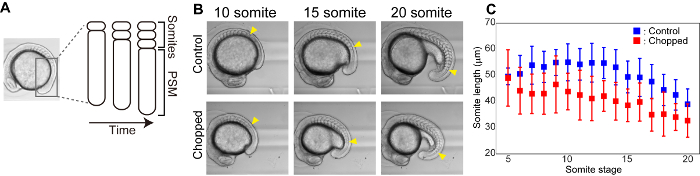
Figure 3: Size reduction reduces the length of somites. (A) Schematic illustration of somite formation. (B) Bright field images of control and chopped embryos over time. Yellow arrowheads indicate the most newly formed somite at each somite stage. (C) Somite length (in anterior-posterior axis) measurements over time for both control and chopped embryos. Error bars represent standard deviation. Please click here to view a larger version of this figure.
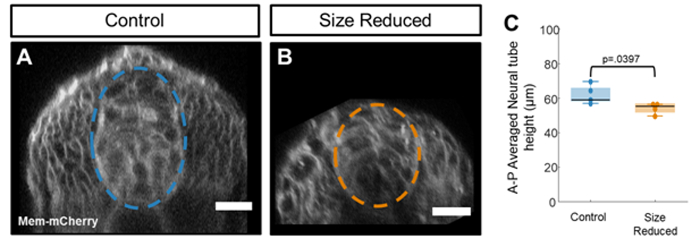
Figure 4: Size reduction reduces the height of the neural tube. (A-B) Example images of normal sized (A) and size reduced (B) tg(ptch2:kaede) embryos which were injected at the single cell stage with mem-mCherry mRNA. Scale bar = 20 μm. (C) Neural tube heights extracted from manual segmentation of the neural tube in each z-stack. Statistically significant differences are observed in average neural height when values are compared using an unpaired t-test (p = 0.0397). Please click here to view a larger version of this figure.

Supplemental Movie 1: Comparison between two-step chopping versus blastula-only chopping. Top row = control embryos, middle row = size reduced embryos with two step chopping, bottom row = size reduced embryos with blastula only chopping. Movies were taken every 3 min for 12 h. Scale bar = 1 mm. Please click here to view this video. (Right-click to download.)
Discussion
Historically, among vertebrate animals, size reduction has been mainly performed using amphibian embryos, by bisecting the embryos along animal-vegetal axis at a blastula stage12. However, there are mainly two differences between frog and zebrafish embryos when we bisect embryos. First, at the stage when zebrafish embryos become tolerant of bisecting (blastula stage), the organizer is located in a restricted area of blastula margin18,19,20,21. Because one cannot tell the position of the organizer from the morphology of embryos, randomly cutting the embryo along the animal-vegetal axis produces dorsalized or ventralized embryos. Second, unlike frog embryos, zebrafish embryos go through a process called epiboly, where cells move towards the vegetal pole around a separated yolk until it is completely surrounded by cells. If only a portion of blastoderm is removed, fewer cells remain to engulf a yolk of relatively larger volume, and as a result, morphology appears affected after epiboly. Therefore, we employ two-step chopping in which we chop blastulae near the animal pole, to avoid cutting off the organizer, and wound the yolk membrane, to make the yolk size proportional to the blastula.
In addition to two-step chopping, we found the medium in which the size reduction surgery is performed is critical for recovery of embryos following the surgery. Among several media we tried (Egg water, Egg water + Albumin, Danieau buffer, L15, L15 + FBS, ⅓x Ringer, 1x Ringer), only ⅓x Ringer and 1x Ringer yielded high survival rates; in other media, embryos failed to recover from the wounds.
An important troubleshooting tip for low survival rate is to use healthy embryos from healthy and young parental fish. We noted that even when the control non-size reduced embryos show almost 100% survival rate, when size reduced, embryos from older fish tend to show lower survival rate. Also, note that the survival rate tends to decrease when the size reduction is combined with additional perturbations, such as morpholino injection.
The simplicity of the size reduction technique described here allows researchers to apply this technique without specialized equipment or intensive training. Further, since the size reduced embryos remain smaller until later stages of development (once they start eating, their size seems to catch up with the control fish), this technique can be applied to study scaling of many tissues and organs. Therefore, this technique makes it possible to combine size reduction and quantitative in vivo live imaging to study scaling and size control of various systems.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The work was supported by the PRESTO program of the Japan Science and Technology Agency (JPMJPR11AA) and a National Institutes of Health grant (R01GM107733).
Materials
| 60 mm PYREX Petri dish | CORNING | 3160-60 | |
| Agarose | affymetrix | 75817 | For making a mount for live imaging |
| Agarose, low gelling temperature Type VII-A | SIGMA-ALDRICH | A0701-25G | |
| CaCl2 | EMD | CX0130-1 | For 1/3 Ringer's solution |
| CaSO4 | For egg water | ||
| Cover slip (25 mm x 25 mm, Thickness 1) | CORNING | 2845-25 | |
| Disposable Spatula | VWR | 80081-188 | |
| Foam board | ELMER'S | 951300 | For microscope incubator |
| Forcept (No 55) | FST | 11255-20 | |
| Glass pipette | VWR | 14673-043 | |
| HEPES | SIGMA Life Science | H4034 | For 1/3 Ringer's solution |
| INCUKIT XL for Cabinet Incubators | INCUBATOR Warehouse.com | For microscope incubator | |
| Instant sea salt | Instant Ocean | 138510 | For egg water |
| KCl | SIGMA-ALDRICH | P4504 | For 1/3 Ringer's solution |
| Methyl cellulose | SIGMA-ALDRICH | M0387-100G | |
| NaCl | SIGMA-ALDRICH | S7653 | For 1/3 Ringer's solution |
| Petri dish | Falcon | 351029 | For making a mount for live imaging |
| Phenol red | SIGMA Life Science | P0290 | |
| Pipette pump | BEL-ART PRODUCTS | F37898 | |
| Pronase | EMD Millipore Corp | 53702-250KU | |
| Tricaine-S (MS222) | WESTERN CHEMICAL INC | NC0135573 | |
| Ultra thin bright annealed 316L dia. 0.035 mm Stainless Steel Weaving Wires | Sandra | The wire we used was obtained ~20 years ago and we could not find exactly the same one. This product has the same material and diameter as the one we use. |
Referencias
- Cooke, J. Scale of body pattern adjusts to available cell number in amphibian embryos. Nature. 290, 775-778 (1981).
- Driesch, H. Entwicklungsmechanische Studien: I. Der Werthe der beiden ersten Furchungszellen in der Echinogdermenentwicklung. Experimentelle Erzeugung von Theil- und Doppelbildungen. Zeitschrift fur wissenschaftliche Zoologie. , (1892).
- Morgan, T. H. Half embryos and whole embryos from one of the first two blastomeres. Anatomischer Anzeiger. 10, 623-638 (1895).
- Ishimatsu, K., et al. Size-reduced embryos reveal a gradient scaling-based mechanism for zebrafish somite formation. Development. 145, (2018).
- Megason, S. G. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods in Molecular Biology. 546, 317-332 (2009).
- Graeden, E., Sive, H. Live imaging of the zebrafish embryonic brain by confocal microscopy. Journal of Visualized Experiments. (26), e1217 (2009).
- Rosen, J. N., Sweeney, M. F., Mably, J. D. Microinjection of zebrafish embryos to analyze gene function. Journal of Visualized Experiments. (25), e1115 (2009).
- Yuan, S., Sun, Z. Microinjection of mRNA and morpholino antisense oligonucleotides in zebrafish embryos. Journal of Visualized Experiments. (27), e1113 (2009).
- Sorlien, E. L., Witucki, M. A., Ogas, J. Efficient Production and Identification of CRISPR/Cas9-generated Gene Knockouts in the Model System Danio rerio. Journal of Visualized Experiments. (138), e56969 (2018).
- Kemp, H. A., Carmany-Rampey, A., Moens, C. Generating chimeric zebrafish embryos by transplantation. Journal of Visualized Experiments. (29), e1394 (2009).
- Mizuno, T., Shinya, M., Takeda, H. Cell and tissue transplantation in zebrafish embryos. Methods in Molecular Biology. 127, 15-28 (1999).
- Cooke, J. Control of somite number during morphogenesis of a vertebrate, Xenopus laevis. Nature. 254, 196-199 (1975).
- Inomata, H., Shibata, T., Haraguchi, T., Sasai, Y. Scaling of dorsal-ventral patterning by embryo size-dependent degradation of Spemann’s organizer signals. Cell. 153, 1296-1311 (2013).
- Gomez, C., et al. Control of segment number in vertebrate embryos. Nature. 454, 335-339 (2008).
- Lauschke, V. M., Tsiairis, C. D., Francois, P., Aulehla, A. Scaling of embryonic patterning based on phase-gradient encoding. Nature. 493, 101-105 (2013).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9, 676-682 (2012).
- Almuedo-Castillo, M., et al. Scale-invariant patterning by size-dependent inhibition of Nodal signalling. Nature Cell Biology. 20, 1032-1042 (2018).
- Koos, D. S., Ho, R. K. The nieuwkoid gene characterizes and mediates a Nieuwkoop-center-like activity in the zebrafish. Current Biology. 8, 1199-1206 (1998).
- Yamanaka, Y., et al. A novel homeobox gene, dharma, can induce the organizer in a non-cell-autonomous manner. Genes and Development. 12, 2345-2353 (1998).
- Jesuthasan, S., Stahle, U. Dynamic microtubules and specification of the zebrafish embryonic axis. Current Biology. 7, 31-42 (1997).
- Schier, A. F., Talbot, W. S. The zebrafish organizer. Current Opinion in Genetics and Development. 8, 464-471 (1998).

