Capture of Environmental DNA (eDNA) from Water Samples by Flocculation
Summary
Environmental DNA (eDNA) is typically captured from water samples using alcohol precipitation, filtration, or flocculation. Presented here is an alternative protocol that uses lanthanum chloride to enable collection of dissolved and particulate-bound nucleic acids from water samples containing eukaryotic cells, prokaryotic cells, and virus.
Abstract
The analysis of environmental DNA (eDNA) has become a widely used approach to problem solving in species management. The detection of cryptic species including invasive and (or) species at risk is the goal, typically accomplished by testing water and sediment for the presence of characteristic DNA signatures. Reliable and efficient procedures for the capture of eDNA are required, especially those that can be performed easily in the field by personnel with limited training and citizen scientists. The capture of eDNA using membrane filtration is widely used currently. This approach has inherent issues that include the choice of filter material and porosity, filter fouling, and time required on site for the process to be performed. Flocculation offers an alternative that can be easily implemented and applied to sampling regimes that strive to cover broad territories in limited time.
Introduction
Before the mid-2000s, the term “environmental DNA” was generally used to refer to DNA obtained from water and soil microbes although some use for the detection of human and animal DNA for purposes of fecal source tracking or non-native species detection had begun1,2,3. Presently, environmental DNA is generally used to describe DNA from cells sloughed from eukaryotic organisms, and the analysis of this DNA has become a widely used management tool. The detection of cryptic species including those that are invasive or species at risk is the goal, typically accomplished by testing water or sediment for the presence of the characteristic DNA signatures of the target species. Studies may focus on a single species of interest using a specific PCR assay (active sampling), or many species may be detected simultaneously using metabarcoding and massively parallel sequencing.
Many improvements in sample collection, sample processing, and data analysis associated with eDNA research have been made during the last two decades. Early studies employed alcohol precipitation to capture DNA from water samples3. While tried and true, the need to analyze large water volumes to increase assay sensitivity make this method unattractive due to the typical requirement for two volumes of ethanol for each volume of water sample.
Later studies have generally used membrane filtration, rationalizing that eDNA is present in a continuum of states from cellular-bound to freely dissolved, but that dissolved DNA is subject to rapid degradation and therefore that cellular-bound DNA represents the most significant fraction. Filter material and pore size optimization is widely discussed and debated as is the optimal method(s) for extraction of captured eDNA from the filter4,5,6.
A third approach to eDNA capture is flocculation, a process that has been used for centuries to clarify liquids including, water, beer, and wine. Several studies have used flocculation to capture virus from natural or potable water sources7,8,9,10,11. These studies have typically used pre-flocculated skim milk or iron chloride as capture agents. Iron chloride methods typically result in downstream analysis molecular problems due to PCR reaction inhibition12,13. Methods using pre-flocculated skim milk require the advance preparation of the flocculant, pH control, and thus far have only been applied to the capture of virus7,8,9.
Recently, flocculation capture of bacteria and virus based on the use of lanthanum (III) chloride has been studied12,13,14,15. The authors demonstrated that viable pathogens could be collected by this method as the binding of lanthanum chloride was effective at low concentration (0.2 mM). Furthermore, lanthanum flocculation is reversible via chelation under mild conditions in contrast to that of iron chloride.
Thus, flocculation using lanthanum chloride to capture biological particulates is attractive given that only a small amount of added concentrated flocculating agent is required, no strict pH control is needed, and sample volumes are scalable from milliliters to gallons. As applied to the capture of eDNA, cellular, sub-cellular (mitochondria and chloroplasts) as well as dissolved DNA are readily recovered. Bacteria and virus are collected simultaneously allowing for pathogen testing and (or) additional ecological studies from a single water sample. A flocculation protocol using lanthanum chloride is presented that enables collection of dissolved and particulate-bound nucleic acids from water samples.
Protocol
1. Decontamination of equipment and flocculation reagent preparation
- Thoroughly wash all bottles, tubing, and other previously used equipment with dishwashing detergent and hot water.
- Spray all bottles, tubing, and other previously used equipment with 6% citric acid solution to remove mineral deposits, let stand 20 minutes, and rinse with tap water.
- Add a solution of 10-fold diluted household bleach (0.5-0.6% final concentration of sodium hypochlorite) to all bottles (approximately 10% of the container volume), shake, let it stand at least 1 h, then drain and rinse with tap water until no trace of bleach remains. Then rinse once with deionized water.
- Dry all bottles.
- Place all collection bottles and caps in a biological safety cabinet and irradiate using ultraviolet germicidal lamps for 2 h rotating each bottle and cap a quarter turn every half hour.
- Place caps onto bottles and screw down loosely. Weigh each bottle and record its empty weight on the bottle.
- Prepare a 100 mM solution of lanthanum (III) chloride heptahydrate by dissolving 3.71 g in 100 mL deionized water. To minimize exposure of the chemical to air, as lanthanum chloride is hygroscopic, seal the bottle with tape between uses.
CAUTION: Lanthanum chloride carries the signal word warning hazard statement(s). H315 causes skin irritation. H319 causes serious eye irritation. H335 may cause respiratory irritation. Use personal protective equipment. See the material’s SDS for further information. - Prepare a 1 M solution of sodium bicarbonate by dissolving 84.01 g sodium bicarbonate in 1 L deionized water. Filter sterilize and store at room temperature.
2. Sample collection and flocculation
- Collect samples from each site plus one field blank (a sample containing drinking water filled on site to control for contamination) by filling appropriately sized bottles. Adjust the number of samples taken according to the experimental design and intended purpose. Do not overfill bottles beyond the shoulder of the bottle to allow for the expansion if samples are to be frozen.
- Record pertinent metadata including date, time, and GPS location. Label sample containers.
- Add 1 M sodium bicarbonate solution to each sample and field blank. Add 10 mL per 500 mL sample.
- Cap the bottles and mix.
- Add 100 mM lanthanum chloride to each sample and field blank. Add 1 mL per 500 mL sample.
- Cap and mix thoroughly. Use electrical tape to secure the cap.
- Store on ice until returning to the field laboratory, then the samples may either be stored overnight refrigerated and standing upright to allow the flocs to settle to the bottom of the container or frozen at -20 °C for shipment to a second laboratory for further processing. Samples must be frozen standing upright to allow for the liquid expansion and to prevent container rupture.
3. Processing of flocs
- Thaw frozen samples at room temperature in a cooler or move samples that were not frozen to a suitable workbench area that includes a peristatic pump and tubing setup for aspiration of sample supernatants. Ensure that the tubing running through the peristaltic pump discharge into a large bucket on the floor. The intake should be equipped with a clean, plastic drinking straw for controlling the intake.
- Use the peristaltic pump to remove the supernatant by using the drinking straw to direct suction first just below the liquid surface and moving the straw intake down with the liquid level. Remove the last of the supernatant by carefully moving the intake to rest on the pontil (raised dimple in the bottom of the bottle). This allows maximum liquid removal while flocs collect around the periphery (see Figure 1).

Figure 1: Collection of flocs settled at unit gravity. Flocs containing cellular material and eDNA are settled at the bottom of the sample bottle and are ready to be recovered after the supernatant is removed. Please click here to view a larger version of this figure.
- Attach a clean drinking straw and repeat the supernatant removal for each sample.
- Use a clean 20 mL pipette and pipette controller to transfer the floc suspension completely to a 50 mL centrifuge tube. Use low volume deionized water rinses (approximately 5 mL) to ensure that any residual flocs are recovered from the sample bottle.
- Adjust all 50 mL tubes to equal volumes with deionized water and collect the flocs by centrifugation at 2,500 x g for 20 min.
- Pour off the supernatant and make tubes stand inverted on a clean paper towel to drain.
- Resuspend flocs in 1 mL of 0.5 M EDTA, pH 8.0 by vortex mixing and then add 4 mL 1x TE solution, pH 8.0 and mix again. Transfer the resuspended flocs to a 15 mL conical centrifuge tube, add 50 µL of 10% linear polyacrylamide and vortex. Add 5 mL 100% isopropanol, mix, and let it stand on ice for 20 min.
- Centrifuge at 6,000 x g for 20 min to collect DNA and cellular particulates. Pour off supernatants and let it stand inverted on clean paper towels to drain. Allow to dry slightly, but do not over dry.
- Begin the floc DNA extraction process using a commercial kit that employs bead beating and spin column binding with a capacity of 25 µg.
- First, resuspend the flocs in 800 µL of the lysis buffer provided with the kit and completelly transfer the suspension to a bead beating tube. Bead beat the floc suspension in short durations with cooling between. Homogenates may be stored frozen at this point or continue with the kit protocol.
- Elute the DNA from the spin column first with 100 µL, then recycle the eluate through the spin tube.
- Finally, elute the residual DNA from the column with an additional 100 µL aliquot of elution buffer to make a total volume of 200 µL. This maximizes the recovery.
- Collect, prepare, and analyze eDNA to demonstrate the performance of this protocol and its performance relative to the widely used filtration method.
- Prepare reactions that contain 1x qPCR master mix, 25 μg/mL bovine thrombin14,16, 0.4 μM forward and reverse primers, and 2 μL of template (1% of recovered sample DNA). Construct standard curves using serial dilutions of purified and quantified PCR amplicons generated using voucher DNA. Alternatively, targets can be synthesized commercially.
- Use a comparative quantification method if no standard is available. For this example, conduct thermocycling using reaction conditions of 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 57 °C for 30 s, and 72 °C for 30 s, although these conditions must be adjusted for other assays. Analyze the data using the manufacturer's software.
Representative Results
Eight 500 mL water samples (450 mL each) taken from an aquarium housing two Eastern painted turtles (Chrysemys picta picta) were used to prepare eDNA using the described protocol. Another eight 500 mL water samples were filtered over 47 mm glass fiber filters of 1.5 µm porosity containing no binder and eDNA was extracted using the same extraction method as used for the flocculation protocol. DNA prepared using both methods was analyzed using quantitative PCR that targets the mitochondrial control region of the target species and uses forward and reverse primers specific for the target organism. The target organism for this demonstration was Chrysemys picta picta and the forward and reverse primers had the sequences (5’-CGTCCATGTAACAGATGGCGT-3’) and (5’-AAACCACCGTATGCCAGGTT-3’) respectively.
Comparative quantification was used first to demonstrate the reproducibility of the protocol as no standard curve was required. Eight water samples prepared using the flocculation protocol were subjected to comparative quantification qPCR and the take-off point (the cycle at which the second derivative is at 20% of the maximum level) for these reactions was found to be 23.5 with a mean PCR reaction efficiency of 1.92 (range 1.89-1.93). Mean relative quantification was 1.003 with a 99% confidence interval of 0.88 – 1.12.
To further demonstrate the robustness of the flocculation protocol, we compared eight 500 mL water samples containing Eastern painted turtle eDNA that was captured using flocculation with eight additional 500 mL water samples from which the eDNA was captured using filtration over 1.5 µm retention glass fiber filters. This type of filter is widely used for the capture of eDNA. Material captured by flocculation and (or) filtration was extracted from the settled flocs and from the filters and analyzed by qPCR as described above except that quantification was performed by comparison to serially diluted standards (see Figure 2). These results support the high performance and reproducibility of the method (see Figure 3).
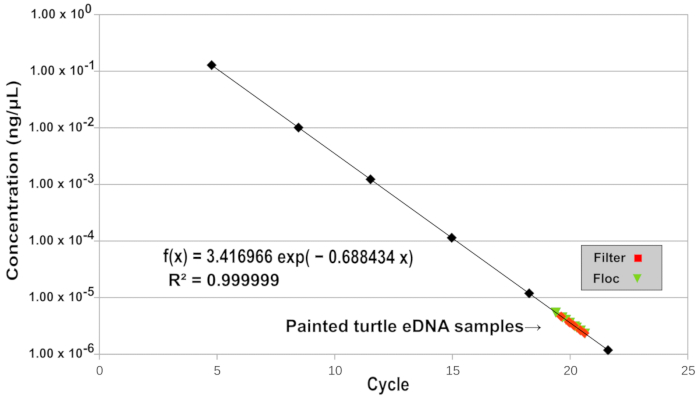
Figure 2: Quantification of Eastern painted turtle eDNA by qPCR. 8 flocculant-captured (green triangles) and 8 filter-captured (red squares) eDNA samples were quantified by qPCR. The black diamonds indicate the diluted standards used for quantification and are connected by a line of best fit. The equation describing the line of best fit and coefficient of determination (R2) are shown. The linearity, efficiency, and reproducibility of the assay are demonstrated. Please click here to view a larger version of this figure.
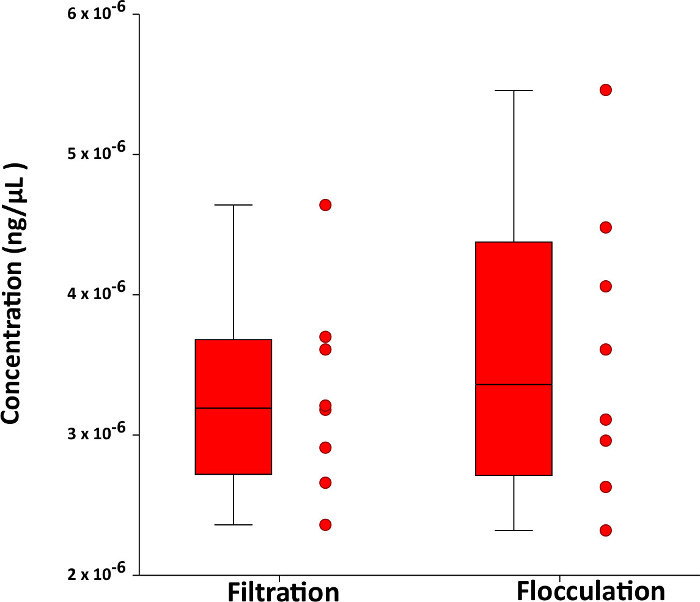
Figure 3: Comparison of flocculant and filter protocols for capture of eDNA. The efficiency of eDNA capture by flocculation is shown to be equivalent to eDNA capture by filtration on glass fiber filters. The boxes are bounded by the 25th an 75th percentiles of the data and error bars indicate the data range. A line inside the boxes shows the median. Red dots adjacent to the boxes show the individual determinations of the replicates. Please click here to view a larger version of this figure.
Discussion
Although this technique is straightforward, some steps are critical to success. The collection and transfer of the flocs must be done completely so as not to lose DNA. Residual material must be gathered using strategic rinses. The described protocol uses a commercial DNA extraction kit for the final processing of flocculated material. Other extraction methods may perform similarly or may be superior in performance, but modifications of this part of the protocol have not been exhaustively tested.
The described protocol is useful for sampling strategies that require taking samples from widely dispersed sites with minimal time differential between samples or when high levels of suspended sediments would clog filters readily. A disadvantage of this protocol is the requirement for the transport water samples from the field to a field station for processing on site or shipping to another laboratory for processing. Filtration methods produce a light weight and compact filter that carries the bound target material that can be transported and shipped easily. Unfortunately, on site filtration requires a variable amount of time to be spent at each sampling site dependent on water conditions. Additionally, filtration requires carrying equipment into the field, decontamination of equipment between samples, and specialized training of personnel and is not generally a process usable for citizen scientists.
The described protocol is simple, and the sample collection portion of the procedure can easily be performed by field personnel with minimal training, or even by citizen scientists. Future applications can be envisioned that include bio-surveillance of pathogens, invasive species, and changes in biodiversity resulting from contaminants or other abiotic factors. Like many protocols, future modifications and improvements are expected that will broaden its usefulness.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was completed with funding from the U.S. Geological Survey.
Any use of trade, firm, or products names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Materials
| 0.5M EDTA, pH 8.0 | Available from several commercial sources | ||
| 1 x TE Buffer, pH 8.0 | Available from several commercial sources | ||
| 15 ml Conical Centrifuge Tubes | Single use | ||
| 16 oz. PET Clear Square Bottles | Reusable with cleaning and decontamination | ||
| 38/400 Polypropylene Cap with Pressure Sensitive Liner | Leave caps loosely attached until use. Cap tightly to seal pressure sensitive liner. | ||
| 50 mL Conical Centrifuge Tubes | Single use | ||
| 6% Citric Acid Solution | Use to remove mineral deposits from reusable sample bottles. | ||
| Fecal/Soil Microbe Kit | Use inhibitor removal resin or column | ||
| Lanthanum (III) chloride heptahydrate | 100 mM Stock Solution | ||
| Peristaltic DC Pump | Any peristaltic pump will do, or can be syphoned | ||
| Polyacryl Carrier | 10% linear polyacrylamide | ||
| Sodium Bicarbonate | 1 M, filter sterilized and stored at room temperaruture |
Referencias
- Mathes, M. V., et al. . Presumptive Sources of Fecal Contamination in Four Tributaries to the New River Gorge National River, West Virginia, 2004. , (2007).
- Caldwell, J. M., Raley, M. E., Levine, J. F. Mitochondrial Multiplex Real-Time PCR as a Source Tracking Method in Fecal-Contaminated Effluents. Environmental Science & Technology. 41 (9), 3277-3283 (2007).
- Ficetola, G. F., Miaud, C., Pompanon, F., Taberlet, P. Species detection using environmental DNA from water samples. Biology Letters. 4 (4), 423-425 (2008).
- Djurhuus, A., et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Frontiers in Marine Science. 4 (314), (2017).
- Hinlo, R., Gleeson, D., Lintermans, M., Furlan, E. Methods to maximise recovery of environmental DNA from water samples. PLOS One. 12 (6), 0179251 (2017).
- Majaneva, M., et al. Environmental DNA filtration techniques affect recovered biodiversity. Scientific reports. 8 (1), 4682 (2018).
- Aguado, D., et al. A Point-of-Use Method for the Detection of Viruses in Water Samples. Journal of Visualized Experiments. (147), e58463 (2019).
- Bofill-Mas, S., et al. Cost-effective Method for Microbial Source Tracking Using Specific Human and Animal Viruses. Journal of Visualized Experiments. (58), e2820 (2011).
- Gonzales-Gustavson, E., et al. Characterization of the efficiency and uncertainty of skimmed milk flocculation for the simultaneous concentration and quantification of water-borne viruses, bacteria and protozoa. Journal of Microbiological Methods. 134, 46-53 (2017).
- Payment, P., Fortin, S., Trudel, M. Ferric chloride flocculation for nonflocculating beef extract preparations. Applied Environmental Microbiology. 47 (3), 591-592 (1984).
- Poulos, B. T., John, S. G., Sullivan, M. B. Iron Chloride Flocculation of Bacteriophages from Seawater. Methods Molecular Biology. 1681, 49-57 (2018).
- Zhang, Y., Riley, L. K., Lin, M., Hu, Z. Lanthanum-based concentration and microrespirometric detection of microbes in water. Water Research. 44 (11), 3385-3392 (2010).
- Zhang, Y., Riley, L. K., Lin, M., Hu, Z. Determination of low-density Escherichia coli and Helicobacter pylori suspensions in water. Water Research. 46 (7), 2140-2148 (2012).
- Schill, W. B., Galbraith, H. S. Detecting the undetectable: Characterization, optimization, and validation of an eDNA detection assay for the federally endangered dwarf wedgemussel, Alasmidonta heterodon (Bivalvia: Unionoida). Aquatic Conservation: Marine and Freshwater Ecosystems. 29 (4), 603-611 (2019).
- Zhang, Y., Riley, L. K., Lin, M., Purdy, G. A., Hu, Z. Development of a virus concentration method using lanthanum-based chemical flocculation coupled with modified membrane filtration procedures. Journal of Virology Methods. 190 (1-2), 41-48 (2013).
- Zhang, Y., et al. Bovine thrombin enhances the efficiency and specificity of polymerase chain reaction. BioTechniques. 57 (6), 289-294 (2014).

.
