Isolation of Mouse Kidney-Resident CD8+ T cells for Flow Cytometry Analysis
Summary
Following viral infection, kidney harbors a relatively large number of CD8+ T cells and offers an opportunity to study non-mucosal TRM cells. Here, we describe a protocol to isolate mouse kidney lymphocytes for flow cytometry analysis.
Abstract
Tissue-resident memory T cell (TRM) is a rapidly expanding field of immunology research. Isolating T cells from various non-lymphoid tissues is one of the key steps to investigate TRMs. There are slight variations in lymphocyte isolation protocols for different organs. Kidney is an essential non-lymphoid organ with numerous immune cell infiltration especially after pathogen exposure or autoimmune activation. In recent years, multiple labs including our own have started characterizing kidney resident CD8+ T cells in various physiological and pathological settings in both mouse and human. Due to the abundance of T lymphocytes, kidney represents an attractive model organ to study TRMs in non-mucosal or non-barrier tissues. Here, we will describe a protocol commonly used in TRM-focused labs to isolate CD8+ T cells from mouse kidneys following systemic viral infection. Briefly, using an acute lymphocytic choriomeningitis virus (LCMV) infection model in C57BL/6 mice, we demonstrate intravascular CD8+ T cell labeling, enzymatic digestion, and density gradient centrifugation to isolate and enrich lymphocytes from mouse kidneys to make samples ready for the subsequent flow cytometry analysis.
Introduction
Tissue-resident memory (TRM) T cells represent one of the most abundant T cell populations in adult human and infected mice. TRM cells provide the first line of immune defense and are critically involved in various physiological and pathological processes1,2,3,4,5. Comparing with circulating T cells, TRM cells carry distinct surface markers with unique transcription programs6,7,8. Expanding our knowledge of TRM biology is the key to understanding T cell responses which is essential for future development of T cell-based vaccines and immunotherapies.
In addition to commonly shared molecular markers of TRMs across all non-lymphoid tissues, accumulating evidence suggest that tissue-specific features are a central component of TRM biology9. Kidney harbors many immune cells including TRM cells after infection and offers a great opportunity to study TRM cell biology in a non-mucosal tissue. Acute LCMV (lymphocytic choriomeningitis virus) infection via intraperitoneal route is a well-established systemic infection model to study antigen-specific T cell responses in mice. The infection is usually resolved in 7-10 days in wild type mice and leave a large number of LCMV-specific memory T cells in a variety of tissues, including the kidney10. P14 TCR (T cell receptor) transgenic mice carry CD8+ T cells recognize one of the immune-dominant epitopes of LCMV presented by MHC-I (class I major histocompatibility complex) molecule H2-Db in C57BL/6 mice. Combining congenically marked P14 T cell adoptive transfer and LCMV infection in mice, CD8+ effector and memory T cells are tracked, including TRM differentiation and homeostasis.
In some barrier tissues, such as intestinal intraepithelial lymphocytes (IEL) compartment and salivary glands, established lymphocyte isolation procedure yields high percentage of TRM cells with minimal blood borne T cell contamination in mouse LCMV model11. However, in non-barrier tissues, such as the kidney, dense vascular network contains a large number of circulating CD8+ T cells. It has been well documented that even successful perfusion cannot efficiently remove all circulating CD8+ T cells. To overcome this technical hurdle, intravascular antibody staining has been established as one of the most commonly used techniques in TRM labs12. In brief, 3-5 minutes before euthanasia, 3 µg/mouse anti-CD8 antibody (to label CD8+ T cells) is delivered intravenously. Intact blood vessel wall prevents rapid diffusion of the antibody within this short period of time (i.e., 3-5 minutes) and only intravascular cells are labeled. Following standard lymphocyte isolation protocol, intravascular versus extravascular cells can be easily distinguished using flow cytometry.
Here, we will describe a standard protocol commonly used in TRM labs to perform intravascular labeling, lymphocyte isolation and flow cytometry analysis of kidney CD8+ T cells using a C57BL/6 mouse that has received CD45.1+ P14 T cells and LCMV infection 30 days before13. Same protocol can be used to study both effector and memory T cells in the kidney.
Protocol
All animal experiments performed following this protocol must be approved by the respective institutional Animal Care and Use Committee (IACUC). All procedures described here have been approved by IACUC UT Health San Antonio.
1. Adoptive transfer of P14 T cells into C57BL/6 recipients and LCMV infection
- Use P14 mice at 6-12 weeks of age. Ensure that the sex of donor P14 TCR transgenic mouse should match the sex of C57BL/6 recipient mice. Otherwise, cells such as male T cells transferring into female mice will result in immune-mediated rejection of circulating donor T cells in around 12 days14.
- Purify naïve CD8+ T cells from the spleen and lymph nodes of a P14 TCR transgenic mouse using mouse CD8+ T cell purification kit following the manufacturer’s protocol.
- Briefly, dissect spleen and lymph nodes, mash the samples to generate single cell suspension.
- To enrich for naïve T cells, add biotin-anti-CD44 antibody during the first antibody incubation step13.
NOTE: Negative selection commercial kit used in this step contains two major components for two steps of incubation (i.e., biotin-antibody mixture incubation and streptavidin-magnetic bead incubation).
- Use a hemocytometer to count the cells and inject 1,000 to 10,000 purified P14 T cells in 200 µL of PBS into each sex matched C57BL/6 recipient via the tail vein.
NOTE: Detailed tail vein injection protocol is described in step 2. - On the next day, all C57BL/6 recipients receive 2 x 105 pfu LCMV Armstrong diluted in 200 µL of PBS via an intraperitoneal route.
2. Intravascular labeling of CD8+ T cells
NOTE: Depending on the research interests, different time points can be chosen following infection. An example of day 30 post infection (day 30 after step 1.4) is demonstrated, which is often considered as an early memory time point for CD8 T cell response.
- Dilute biotin anti-CD8α antibody to 15 µg/mL in PBS. Ensure each mouse receive 200 μL of PBS containing 3 µg antibody.
NOTE: Biotin anti-CD8 antibody is used here so that it will be more flexible to design the final FACS staining panel. Fluorescent dye-labeled antibody can be used directly. However, it is important to use different clones of monoclonal antibodies for intravenous labeling versus FACS staining. - Properly heat the tail vein of mice with an overhead heat lamp for 5-10 min to dilate the veins.
NOTE: Extra care must be taken to prevent overheating the animals. Reduce the heat or remove the heat lamp if the mice stopped to move around. - Draw 200 µL of prediluted anti-CD8α antibody mix into a 100 U insulin syringe (28G) and remove any air bubbles by moving the piston up and down. Bend the needle to create a 150° angle between the needle and syringe, bevel up, so the needle will be parallel to the vein.
- Restrain the mouse with a rodent restrainer of appropriate size and spray the tail with 70% ethanol to make the vein clearly visible.
NOTE: The duration of the restraint should be kept to a minimum. - Hold the tail at the distal end with thumb and middle fingers of the non-dominant hand. Place the index finger underneath the site where the needle will be inserted.
- Hold the syringe with the dominant hand and insert the needle into the vein in parallel towards the direction of the heart.
NOTE: When placing correctly, the needle should move smoothly into the vein. - Slowly inject 200 µL of anti-CD8α antibody via the tail vein.
NOTE: If there is a resistance or a white area is seen above the needle on the tail, the needle is not inside the vein. Start the initial injection close to the tip of the tail. When necessary, move forward towards the bottom of the tail for subsequent injections. - Remove the needle and gently apply compression until bleeding stops.
- Return the mouse to the cage, and euthanize the mouse after 3-5 min.
NOTE: The mice should be euthanized via isoflurane chamber followed by cervical dislocation. Other methods, such as CO2 inhalation will take up to 5 min and may introduce unnecessary variation of intravenous antibody labeling time. - Dissect the kidney using scissors, transfer the whole kidney to 1.5 mL of microcentrifuge tubes and leave the samples on ice until further processing.
NOTE: Intact whole kidney can be safely stored on ice for a few hours before subsequent processing and digestion.
3. Enzymatic digestion of the kidney
- Prepare 6 well plates containing 3mL of the digestion solution (RPMI/2% FCS/1 mg/ml Collagenase B) for each mouse, store on ice.
NOTE: Stock collagenase B solution at higher concentration (e.g., 20x) can be prepared and stored at or below -20°C. Working solution should be prepared fresh. Please see Table 1 for the recipes for all solution/buffer used in the current protocol. - Add 300 μL of digestion solution into the 1.5 mL microcentrifuge sample tubes and mince the kidney samples with a straight spring scissor.
NOTE: The kidney tissue should be minced into even pieces with sizes no larger than 1.5 mm in diameters. No decapsulation step is required. - Transfer the minced kidney samples to 6 well plates containing the digestion solution
NOTE: 6 well plates are used for convenience only when there are multiple samples to be processed. - Incubate the samples at 37 °C with gentle rocking (around 60 rpm) for 45 min.
- After digestion, mash the tissue with the plunger flange of a 3 mL syringe directly inside the dish, and transfer into 15 mL conical tubes.
NOTE: Usually filtration through a cell strainer is not required at this step because density gradient centrifugation is performed next to purify live lymphocytes. - Spin down the samples at 500 x g for 5 min at 4 °C.
- Resuspend the pellet in 3 mL of RPMI/10% FCS.
4. Density gradient centrifugation to enrich lymphocytes from the digested kidney
- Spin down the sample at 500 x g for 5 min at 4 °C.
- Remove the supernatant and resuspend the cell pellet with 5 mL of 44% Percoll/RPMI mix (44% Percoll + 56% RPMI without FBS).
- Put the tip of a 3 mL pipette containing 3 mL of 67% Percoll/PBS (67% Percoll + 33% PBS) directly to the bottom of each tube, and slowly release the solution so that the heavy solution (67% Percoll/PBS) forms a distinct layer at the bottom and the light solution (44% Percoll/RPMI) is raised up. Together, cocktail layers will form.
NOTE: For newly arrived density gradient medium from the vendor, add 1/10 volume of sterile 10x PBS to the bottle, mix well and consider the PBS-balanced density gradient medium as 100% solution. - Spin the samples at 900 x g for 20 min at room temperature with reduced accelerator and brake setting.
NOTE: For centrifuges with accelerator and brake settings (on a 1-9 scale (1 means minimum and 9 means maximum)), use 6 for accelerator and 4 for brake. - Carefully remove the tubes from the centrifuge without disturbing the layers;
NOTE: A clear lymphocyte layer is identified between pink color RPMI layer and colorless PBS layer. - Remove the top layer with a transfer pipette without touching the lymphocyte layer.
- Transfer the lymphocyte layer to a new 15 mL tube, fill the tube with PBS/5% FCS and mix by inverting the tube 4-6x slowly to ensure sufficient mixing.
NOTE: This step is important to wash out any residual Percoll. - Spin down the samples at 500 x g for 5 min at 4 °C.
- Remove the supernatant and re-suspend the cell pellet with 500 μL of complete RPMI medium. The cells are ready for flow cytometry staining.
5. Flow cytometry staining and analysis
NOTE: Please follow standard flow cytometry staining protocol for T lymphocytes.
- Take around 1 x 106 cells for each FACS staining.
NOTE: Use a U-bottom 96 well plate for FACS staining. However, if only a limited number of samples are involved, 5 mL FACS tubes can also be used. - Spin down the cells at 500 x g for 5 min at 4 °C. Discard the supernatant.
- Resuspend each sample in 50 µL of FACS buffer containing 10 µg/mL of 2.4G2 FcR blocker (an anti-CD16/32 monoclonal antibody to block the interaction between added FACS antibody with FcR). Incubate on ice for 15 min.
- Without further centrifugation, add 50 µL of surface staining antibody mixture for each sample so that the final staining volume is 100 µL/each sample. Incubate on ice for 30 min in the dark.
NOTE: Include fluorescently labeled streptavidin (2 µg/mL or follow manufacturer’s recommendation) in the antibody mixture to label CD8α+ intravascular CD8+ T cells and use fluorescent labeled anti-CD8β antibody to label all CD8 T cells. Antibody mixture is made by adding desired amounts of interested antibodies into the FACS buffer. - Wash the samples with PBS twice. For each wash, fill the wells of 96 well plate with cold PBS, spin at 500 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the cells in 100 µL/well of diluted live/death dye. Incubate on ice for 30 min in the dark.
NOTE: Even with density gradient medium spin, enzymatic digestion step often induces significant cell death in tissue-resident T cells due to ARTC2.2/P2RX7 signaling15. Live/death dye staining is highly recommended before fixation. Anti-ARTC2 nanobody can be administrated into the mice before euthanasia to prevent cell isolation-induced cell death. - Wash the samples with FACS buffer twice. For each wash, fill the wells of 96-well plate with cold FACS buffer, spin at 500 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the cells in 100 μL/well fixing buffer. Incubate at 37 °C for 10 min.
NOTE: Fixing buffer is made freshly by diluting formaldehyde with PBS to a final concentration of 2% formaldehyde. - Wash the samples with FACS buffer twice. For each wash, fill the wells of 96 well plate with cold FACS buffer, spin at 500 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the cells in 250 μL/well FACS buffer. Seal the plate, store at 4 °C and protect from light until FACS analysis.
NOTE: Before FACS analysis, filter the samples through a 70 μM nylon mesh to remove possible cell clusters that may clog the FACS machine.
Representative Results
The protocol described here is summarized in a flow chart (Figure 1A). At day 30 post LCMV infection, we performed intravascular labeling of CD8+ T cells. 5 minutes later, both kidneys of the animal were dissected, minced and subjected to collagenase digestion. Lymphocytes were further purified from the digested samples via Percoll centrifugation and analyzed by flow cytometry. As shown in Figure 1B, even after density centrifugation-mediated lymphocyte enrichment, it was very common to see a large portion of non-lymphocytes in the final product. However, after gating on live lymphocytes, it was easy to identify CD8+ T lymphocytes. We could distinguish intravascular vs extravascular CD8+ T cells. Due to highly correlative expression pattern of CD8α and CD8β on CD8+ T cells, it is expected that intravascular CD8+ T cells exhibit a diagonal pattern on CD8α-CD8β FACS profile. As expected, only extravascular CD8+ T cell efficiently acquired TRM phenotypes, such as upregulation of CD69 and downregulation of Ly6C (Figure 1B). In our experiments, we were interested in donor P14 T cells with congenic marker CD45.1. Using the same protocol, endogenous host derived CD8+ T cells can be examined as well. In contrast to infected mice, the vast majority of CD8+ T cells isolated from young naïve mice housed in SPF facility were labeled with i.v. CD8α antibody and, therefore, belonged to the intravascular compartment (Figure 1C).
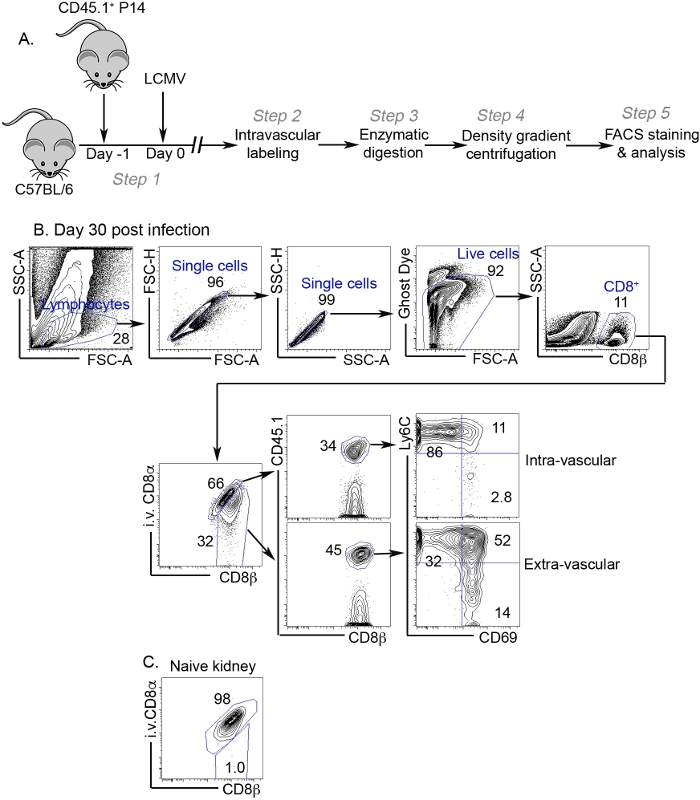
Figure 1: Representative results.
(A) Flowchart of the protocol. (B) At day 30 post infection, kidney lymphocytes were analyzed by flow cytometry. Representative FACS profiles and gating strategy are shown. The numbers indicate the percentage of gated subsets in their parental population. (C) Representative FACS profile of kidney CD8+ T cells isolated from a naïve mouse. Please click here to view a larger version of this figure.
| Reagent | Recipe | NOTE |
| 100% Percoll | 9 volumes of Percoll + 1 volume of 10xPBS | |
| 44% Percoll/RPMI | 44% Vol of 100% Percoll + 56% Vol serum free RPMI | Made fresh from 100% Percoll |
| 67% Percoll/PBS | 67% Vol of 100% Percoll + 33% Vol PBS | Made fresh from 100% Percoll |
| Digestion buffer | RPMI + 2% FCS + 1mg/ml collagenase B | Made fresh from collagenase stock |
| PBS/5% FCS washing buffer | PBS + 5% FCS | |
| FACS buffer | PBS + 2% FCS + 0.02% NaN3 | Stored at 4°C |
| Fixing buffer | PBS + 2% formaldehyde | Stored at RT and protected from light |
Table 1: List of recipes for all solution and buffer used in current protocol.
Discussion
As tissue specific immunity is a rapid expanding area of research, accumulating evidence suggest that immune cells, especially lymphocytes population can be identified in almost all organs in adult human or infected or immunized mice. LCMV mouse infection model is a well-established model to study antigen-specific T cell response, effector and memory T cell differentiation including TRM biology across multiple tissues. Here, we described a protocol to analyze CD8+ T cells in the kidney. This protocol is largely adapted from publications focused on TRM12. Presumably due to high level P2RX7 expression and enzymatic digestion induced cell death15, lymphocytes yield from the current protocol is best suited for immediate phenotypic analysis. However, with established protocol to inhibit P2RX7 induced cell death16, it is conceivable that the current protocol can be easily modified (e.g., with the addition of inhibitors to block P2RX7 signaling) to increase the cell survival and be suited for other long-term functional assays. Proper control groups should be included to ensure that P2RX7 inhibitors do not interfere with interested functional assays.
Kidney CD8+ TRM cells are largely generated in response to infection or environmental antigens. We have use intravascular labeling protocol to directly analyze kidney CD8+ T cells isolated from 5-6-week-old naïve C57BL/6 mice. Consistent with published results17, there is almost no extravascular CD8+ T cells in these “clean” mice (Figure 1C). An elegant study has demonstrated that the majority of memory CD8+ T cells identified in non-lymphoid tissues are TRM10. In contrast, using a similar infection system, we often detect a significant population of CD69– cells (most likely represent TEM cells or CD69– TRM cells) even in the extravascular compartment. The discrepancy may be due to the facts that 1) different techniques are used, i.e., microscope vs flow cytometry; 2) early vs late memory time points are focused and 3) CD69 is not a perfect marker to identify TRM. Enzymatic digestion and flow cytometry will significantly under-estimate the total number of TRM cells. However, as a convenient and non-labor-intensive protocol, it is still commonly accepted in most TRM studies.
The gold standard to definitively identify TRM cells is to perform parabiosis experiment. Although the protocol described here provides a convenient way to identify kidney-resident T cells, the results from this protocol only prove that at the time when the mice are euthanized, the T cells are residing outside the blood vessels in the kidney. This protocol alone does not provide any information about the migratory history of T cells.
In addition to infections, any kidney targeting immune responses, including autoimmune responses may induce the differentiation of a significant subset of kidney-resident T cells can be studied using a similar protocol. Further, as described before12, this protocol can be easily modified to use anti-CD45 antibody (to label all hematopoietic cells) to study other kidney-resident immune cells. Together, we demonstrated a relatively convenient way to isolate and analyze CD8+ T cells from the kidney, which can be adapted to various models including infection and autoimmunity.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work is supported by NIH grants AI125701 and AI139721, Cancer Research Institute CLIP program and American Cancer Society grant RSG-18-222-01-LIB to N.Z. We thank Karla Gorena and Sebastian Montagnino from Flow Cytometry Facility. Data generated in the Flow Cytometry Shared Resource Facility were supported by the University of Texas Health Science Center at San Antonio (UTHSCSA), NIH/NCI grant P30 CA054174-20 (Clinical and Translational Research Center [CTRC] at UTHSCSA), and UL1 TR001120 (Clinical and Translational Science Award).
Materials
| 1.5 ml microcentrifuge tubes | Fisherbrand | 05-408-130 | |
| 15 ml Conical Tubes | Corning | 431470 | |
| 3 ml syringe | BD | 309657 | |
| 37C incubator | VWR | ||
| Biotin a-CD8a antibody(Clone 53-6.7) | Tonbo Biosciences | 30-0081-U025 | |
| Calf Serum | GE Healthcare Life Sciences | SH30073.03 | |
| Collegenase B | Millipore Sigma | 11088807001 | |
| Disposable Graduated Transfer Pipettes | Fisherbrand | 13-711-9AM | |
| Insulin Syringe | BD | 329424 | |
| Micro Dissecting Spring Scissors | Roboz Surgical | RS 5692 | |
| Mojosort Mouse CD8 Naïve T Cells Isolation Kit | Biolegend | 480043 | |
| overhead heat lamp | Amazon | B00333PZZG | |
| PBS | |||
| Percoll | GE Healthcare Life Sciences | 17089101 | |
| rocker | VWR | ||
| RPMI | GE Healthcare Life Sciences | SH30096.01 | |
| Solid Brass Mouse Restrainer | Braintree Scientific, Inc | SAI-MBR | |
| Swing Bucket Centrifuge with refrigerator | Thermofisher | ||
| Tissue Culture 6-well plate | Corning | 3516 |
Referencias
- Mueller, S. N., Gebhardt, T., Carbone, F. R., Heath, W. R. Memory T cell subsets, migration patterns, and tissue residence. Annual Review of Immunology. 31, 137-161 (2013).
- Mueller, S. N., Mackay, L. K. Tissue-resident memory T cells: local specialists in immune defence. Nature Reviews: Immunology. 16, 79-89 (2016).
- Park, C. O., Kupper, T. S. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nature Medicine. 21, 688-697 (2015).
- Schenkel, J. M., et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346, 98-101 (2014).
- Thome, J. J., Farber, D. L. Emerging concepts in tissue-resident T cells: lessons from humans. Trends in Immunology. 36, 428-435 (2015).
- Mackay, L. K., et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352, 459-463 (2016).
- Mackay, L. K., et al. T-box Transcription Factors Combine with the Cytokines TGF-beta and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 43, 1101-1111 (2015).
- Milner, J. J., et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature. 552, 253-257 (2017).
- Liu, Y., Ma, C., Zhang, N. Tissue-Specific Control of Tissue-Resident Memory T Cells. Critical Reviews in Immunology. 38, 79-103 (2018).
- Steinert, E. M., et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 161, 737-749 (2015).
- Qiu, Z., Sheridan, B. S. Isolating Lymphocytes from the Mouse Small Intestinal Immune System. Journal of Visualized Experiments. (132), e57281 (2018).
- Anderson, K. G., et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nature Protocols. 9, 209-222 (2014).
- Ma, C., Mishra, S., Demel, E. L., Liu, Y., Zhang, N. TGF-beta Controls the Formation of Kidney-Resident T Cells via Promoting Effector T Cell Extravasation. Journal of Immunology. 198, 749-756 (2017).
- Gebhardt, T., et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 477, 216-219 (2011).
- Borges da Silva, H., Wang, H., Qian, L. J., Hogquist, K. A., Jameson, S. C. ARTC2.2/P2RX7 Signaling during Cell Isolation Distorts Function and Quantification of Tissue-Resident CD8(+) T Cell and Invariant NKT Subsets. Journal of Immunology. 202, 2153-2163 (2019).
- Fernandez-Ruiz, D., et al. Liver-Resident Memory CD8(+) T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity. 45, 889-902 (2016).
- Beura, L. K., et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 532, 512-516 (2016).

