An Efficient Method for Directed Hepatocyte-Like Cell Induction from Human Embryonic Stem Cells
Summary
This manuscript describes a detailed protocol for differentiation of human embryonic stem cells (hESCs) into functional hepatocyte-like cells (HLCs) by continuously supplementing Activin A and CHIR99021 during hESC differentiation into definitive endoderm (DE).
Abstract
The potential functions of hepatocyte-like cells (HLCs) derived from human embryonic stem cells (hESCs) hold great promise for disease modeling and drug screening applications. Provided here is an efficient and reproducible method for differentiation of hESCs into functional HLCs. The establishment of an endoderm lineage is a key step in the differentiation to HLCs. By our method, we regulate the key signaling pathways by continuously supplementing Activin A and CHIR99021 during hESC differentiation into definitive endoderm (DE), followed by generation of hepatic progenitor cells, and finally HLCs with typical hepatocyte morphology in a stagewise method with completely defined reagents. The hESC-derived HLCs produced by this method express stage-specific markers (including albumin, HNF4α nuclear receptor, and sodium taurocholate cotransporting polypeptide (NTCP)), and show special characteristics related to mature and functional hepatocytes (including indocyanine green staining, glycogen storage, hematoxylin-eosin staining and CYP3 activity), and can provide a platform for the development of HLC-based applications in the study of liver diseases.
Introduction
The liver is a highly metabolic organ that plays several roles, including deoxidation, storing glycogen, and secretion and synthesis of proteins1. Various pathogens, drugs and heredity can cause pathological changes in the liver and affect its functions2,3. Hepatocytes, as the main functional unit of the liver, play an important role in artificial liver support systems and drug toxicity elimination. However, the resource of primary human hepatocytes is limited in cell-based therapy, as well as in liver disease research. Therefore, developing new sources of functional human hepatocytes is an important research direction in the field of regenerative medicine. Since 1998 when hESCs have been established4, hESCs have been widely considered by researchers because of their superior differentiation potential (they can differentiate into various tissues in a suitable environment) and high degree of self-renewability, and thus provide ideal source cells for bioartificial livers, hepatocyte transplantation and even liver tissue engineering5.
Currently, the hepatic differentiation efficiency can be greatly increased by enriching the endoderm6. In the lineage differentiation of stem cells into endoderm, levels of the transforming growth factor β (TGF-β) signaling and WNT signaling pathways are the key factors in the node of the endoderm formation stage. Activation of a high-level of TGF-β and WNT signaling can promote the development of endoderm7,8. Activin A is a cytokine belonging to the TGF-β superfamily. Therefore, Activin A is widely used in endoderm induction of human induced pluripotent stem cells (hiPSCs) and hESCs9,10. GSK3 is a serine-threonine protein kinase. Researchers have found that CHIR99021, a specific inhibitor of GSK3β, can stimulate typical WNT signals, and can promote stem cell differentiation under certain conditions, suggesting that CHIR99021 has potential for inducing stem cell differentiation into endoderm 11,12,13.
Here we report an efficient and reproducible method for effectively inducing the differentiation of hESCs into functional HLCs. The sequential addition of Activin A and CHIR99021 produced about 89.7±0.8% SOX17 (DE marker)-positive cells. After being further maturated in vitro, these cells expressed hepatic specific markers and exerted hepatocyte-like morphology (based on hematoxylin-eosin staining (H & E)) and functions, such as uptake of indocyanine green (ICG), glycogen storage and CYP3 activity. The results show that hESCs can be successfully differentiated into mature functional HLCs by this method and can provide a basis for liver disease-related research and in vitro drug screening.
Protocol
1. Stem cell maintenance
NOTE: The cell maintenance protocol described below applies to the hES03 cell line maintained in an adherent monolayer. For all the following protocols in this manuscript, cells should be handled under a biological safety cabinet.
- Prepare 1x mTesR stem cell culture medium by diluting 5x supplementary medium to mTesR basic medium.
- Prepare 30x hESC-qualified Matrigel medium by diluting 5 mL of hESC-qualified Matrigel with 5 mL of DMEM/F12 on the ice. Store at -20 °C.
- Prepare 1x hESC-qualified Matrigel medium by diluting 33.3 µL of 30x hESC-qualified Matrigel with 1 mL of DMEM/F12.
- Coat sterile 6 well, tissue culture-treated plates with 1 mL of 1x hESC-qualified Matrigel in each well in advance and store at 4 °C overnight. Leave at room temperature (RT) for at least 30 min before use.
- Thaw the cryopreserved hESCs in a 37 °C water bath for 3 min without shaking. Then immediately transfer the cells by pipetting into a 15 mL centrifuge tube containing 4 mL prewarmed 37 °C mTesR medium, pipet up and down gently 2 times.
- Centrifuge the hESCs at 200 x g for 2 min at RT.
- Aspirate the supernatant and gently resuspend the cells in 1 mL of mTesR medium.
- Aspirate the DMEM/F12 medium from the plate and seed the cells into a well of 6-well plate at a density of 1 x 105 cells in 2 mL of mTesR.
- Incubate in a 5% CO2 incubator and maintain the cells by replacing with preheated mTesR medium daily. Passage the cells at roughly 70-80% confluence or when the cell colonies begin to make contact.
2. Stem cell passage and differentiation
- For passaging cells, aspirate the medium and incubate the cells with 1 mL per well of enzyme solution (e.g., Accutase for 5 min at 37 °C. Then transfer the cells by pipetting to a 15 mL centrifuge tube containing 4 mL of DMEM/F12 prewarmed to 37 °C.
- Centrifuge the cells at 200 x g at RT for 2 min.
- Aspirate the medium and gently resuspend the cell pellet in 1 mL of mTesR medium.
- Prepare 24-well plates by coating with 250 µL of 1x hESC-qualified Matrigel medium in advance. Seed hESCs as described above at a density of 1 – 1.5 x 105 cells per well in 500 µL of mTesR medium.
- Allow cells to incubate for 24 h at 37 °C in a CO2 incubator.
3. DE formation
- Prepare stock solutions of 100 µg/mL Activin A with DPBS containing 0.2% BSA and 3 mM CHIR99021 with DMSO.
- Prepare Stage I differentiation basic medium by supplementing RPMI medium with 1x B27.
- Add Activin A to a final concentration of 100 ng/mL and CHIR99021 to a final concentration of 3 µM in an appropriate volume of preheated Stage I differentiation basic medium.
- Aspirate mTesR medium from the cells and replace with Stage I differentiation media containing added differentiation factors (e.g., 0.5 mL per well of a 24-well plate).
- Allow cells to incubate for 3 days at 37 °C in a CO2 incubator, replacing Stage I media daily with freshly added differentiation factors (Days 0-2).
NOTE: After 3 days of differentiation, differentiated cells should express markers of DE cells, such as FOXA2, SOX17, GATA4, CRCR4, and FOXA1 (minimum 80% of SOX17 needed for proceeding).
4. Differentiation of hepatic progenitor cells
- Prepare Stage II differentiation basic media by dissolving knockout serum replacement (KOSR) to a final concentration of 20% in Knock Out DMEM.
- Prepare Stage II differentiation medium containing 1% DMSO, 1x GlutaMAX, 1x non-essential amino acid (NEAA) solution, 100 µM 2-mercaptoethanol and 1x penicillin/streptomycin (P/S) to the appropriate volume of KOSR/knockout DMEM.
- On Day 3 of differentiation, aspirate the medium and replace with Stage II medium (e.g., 0.5 mL per well of a 24 well plate). Then incubate for 24 h.
- On Day 4 of differentiation, aspirate the medium and replace with Stage II medium (e.g., 1 mL per well of a 24 well plate).
- Change the medium every other day (replace medium on Day 6).
NOTE: After 8 days of differentiation, differentiated cells should express appropriate markers of hepatic progenitor cells, such as HNF4α, AFP, TBX3, TTR, ALB, NTCP, CEBPA (minimum 80% of HNF4α needed for proceeding).
5. Hepatocyte differentiation
- Prepare 10 µg/mL hepatocyte growth factor (HGF), 20 µg/mL Oncostatin (OSM) stock solutions with DBPS (containing 0.2% BSA) and filter with a 0.22 µm filter membrane.
- Prepare Stage III differentiation basic medium (HepatoZYME-SFM (HZM) medium supplemented with 10 µM hydrocortisone-21-hemisuccinate and 1x P/S).
- Add HGF to a final concentration of 10 ng/mL and OSM to a final concentration of 20 ng/mL to the appropriate volume of preheated Stage III differentiation medium.
- On Day 8 of differentiation, aspirate Stage II medium from cells and replace with Stage III medium (e.g., 1 mL per well of a 24 well plate).
- Allow cells to incubate for 10 days at 37 °C in a CO2 incubator, changing Stage III media every other day with freshly added differentiation factors (Days 8-18).
NOTE: After 18 days of differentiation, differentiated cells should express characteristic markers of hepatocytes, such as AAT, ALB, TTR, HNF4α, NTCP, ASGR1, CYP3A4. The percentage of Albumin-positive cells usually is more than 90%.
6. RNA isolation, cDNA synthesis and RT-qPCR analysis
- Aspirate medium from cells and add the proper amount of RNAiso Plus reagent, (e.g., 0.3 mL per well of a 24-well plate). Collect the supernatant in a 1.5 mL tube and let stand at RT for 5 min.
- Add 60 µL of chloroform reagent and leave it at RT for 5 min. Then centrifuge at 12,000 x g for 15 min.
- Collect the 125 µL supernatant and transfer it to another centrifuge tube. Add 160 µL of isopropanol, mix well, and stand at RT for 10 min.
- Centrifuge at 12,000 x g for 10 min.
- Remove the supernatant, add 75% ethanol to the precipitated, and centrifuge at 7,500 x g for 5 min.
- After drying at RT, add 40 µL of DEPC-treated water to dissolve. For qPCR, reverse transcribe 2 µg of total RNA with a reverse transcription kit according to the manufacturer's protocol.
- Perform RT-qPCR using a commercial kit according to the manufacturer's protocol.
- Normalize gene expression relative to non-induced stem cells and determine fold variation following normalization to GAPDH.
7. Immunofluorescence Validation of Differentiation
- Prepare 1x PBST washing buffer by adding 0.1% Tween-20 to PBS.
- Aspirate medium from adherent cells and wash briefly with PBS at RT on a shaker.
- Aspirate PBS and incubate cells with 500 µL of ice-cold methanol per well of a 24-well plate to fix the cells at -20 °C overnight.
- Remove the methanol and wash the cells 3 times with PBS on a shaker for 10 min each time at RT.
- Block cells with 500 µL of 5% BSA prepared in PBS for 1-2 h at RT on a shaker.
- Dilute primary antibody at 1: 200 in the same solution used for blocking.
- Incubate the fixed cells in 300 µL primary antibody solution overnight at 4 °C on a shaker.
- After overnight incubation, wash cells 3 times with PBST for 10 min each time at RT on a shaker.
- Prepare secondary antibody solution in blocking solution at 1:1000.
- Incubate in 300 µL of secondary antibody solution protected from light for 1 h at RT on a shaker.
- Aspirate secondary antibody solution and wash cells 3 times with PBST for 10 min each time at RT on a shaker.
- Incubate cells with 300 µL of 1 µg/mL DAPI for 3-5 min, and then wash twice with PBST.
- After aspirating PBST, add an appropriate amount of PBS for taking pictures under a fluorescence microscope.
8. Western blot analysis
- Prepare 1x TBST washing buffer by adding 0.1% Tween-20 to Tris-buffered saline (TBS).
- Prepare SDS-PAGE electrophoresis buffer and western transfer buffer by using a commercial kit according to the manufacturer's protocol.
- Resolve total cell protein (40 µg) on a 10% sodium dodecyl sulfate polyacrylamide gel and run in SDS-PAGE electrophoresis buffer at 15 mA constant current for about 2 h.
- Transfer the gel to a polyvinylidene difluoride (PVDF) membrane at 300 mA constant current for 2 h at low temperature.
NOTE: The running time and transfer time may vary depending on the equipment used and the type and percentage of the gel. - Block the membrane with 3% BSA prepared in TBST for 1 h at RT on a shaker.
- Incubate in primary antibody solution (1:1000 dilution) overnight at 4 °C on a shaker.
- After overnight incubation, wash blotting membrane 3 times with TBST for 15 min per time at RT on a shaker.
- Incubate in secondary antibody solution (1:2000 dilution) for 2 h at RT on a shaker.
- Discard secondary antibody solution and wash the membrane 3 times with TBST, for 15 min each time, at RT on a shaker.
- Visualize immunoreactive bands using a chemiluminescence reagent followed by autoradiography. Use β-actin as the loading control.
9. Indocyanine green uptake
- Prepare 200 mg/mL indocyanine green stock solution in DMSO.
- Prepare a working solution by supplementing Stage III differentiation medium with indocyanine green solution to a final concentration of 1 mg/mL.
- Incubate in a 37 °C, 5% CO2 incubator for 1-2 h.
- Aspirate medium and wash the cells 3 times with PBS.
- Photograph under a microscope.
10. Periodic Acid-Schiff (PAS) staining and Hematoxylin-Eosin (H&E) staining
- Aspirate medium from adherent cells and briefly wash twice with PBS.
- For cell fixation, aspirate PBS and incubate cells with ice-cold methanol at -20 °C overnight.
- Visualize glycogen storage by PAS staining and observe binucleated cells by H & E staining using a kit following the manufacturer's instructions.
- Take images with a fluorescence microscope.
11. Assay for CYP Activity
- Measure CYP450 activity using commercially available cell-based assays (P450-Glo Assays) according to the manufacturer's instructions.
- Use three independent repeats for testing. Data are represented as the mean ± SD.
Representative Results
The schematic diagram of HLC induction from hESCs and representative bright-field images of each differentiation stage are shown in Figure 1. In Stage I, Activin A and CHIR99021 were added for 3 days to induce stem cells to form endoderm cells. In Stage II, the endoderm cells differentiated into hepatic progenitor cells after being treated with differentiation medium for 5 days. In Stage III, early hepatocytes had matured and differentiated into HLCs after 10 days in HGF and OSM (Figure 1A). In the final stage of differentiation, cells showed a typical hepatocyte phenotype (The cells are polygon and distributed evenly and regularly) (Figure 1B).
To further confirm hepatic differentiation, RT-qPCR, immunofluorescence staining and western blotting were used to detect markers of endoderm cells, hepatic progenitor cells, and mature hepatocytes (Figure 2). The differentiated cells showed high expression levels of differentiation-related genes and proteins in each stage, such as endoderm markers SOX17 (89.7± 0.8%) at Day 3, hepatic progenitor marker HNF4α (81.3±2.9%) at Day 8, AFP (86.6±0.3%) at Day 14, and mature hepatocyte marker ALB (94.5±1.1%) at Day 18. These results indicate that hepatocyte-like cells are generated by this protocol.
In order to detect whether HLCs have hepatocyte functions, we detected ICG uptake, glycogen storage, and CYP activity of HLCs, as well as performed H&E staining. As can be seen the HLCs exhibited green staining (Figure 3A) and extensive cytoplasmic periodic acid-Schiff staining (pink to purple) was observed (Figure 3B), which is consistent with glycogen storage. In addition, we can see the representative morphology of a typical binucleated hepatocyte (Figure 3C). Finally, the enzymatic activity of CYP3A4, the most important metabolic CYP in the liver, was confirmed by enzymatic activity assay (Figure 3D).
| Name of Reagent | Company | Catalog Number | Stock Concentration | Final Concentration |
| 2-Mercaptoethanol | Sigma | M7522 | 50 mM | 100 µM |
| 488 labeled goat against mouse IgG | ZSGB-BIO | ZF-0512 | – | IF: 1:1000 |
| 488 labeled goat against Rabbit IgG | ZSGB-BIO | ZF-0516 | – | IF: 1:1000 |
| Accutase | Stem Cell Technologies | 7920 | 1 x | 1 x |
| Activin A | peproTech | 120-14E | 100 µg/ml | 100 ng/ml |
| Anti – Albumin (ALB) | Sigma-Aldrich | A6684 | – | IF: 1:200; WB:1:1000 |
| Anti – Human Oct4 | Abcam | Ab19587 | – | IF: 1:200 |
| Anti – α-Fetoprotein (AFP) | Sigma-Aldrich | A8452 | – | IF: 1:200; WB:1:1000 |
| Anti -SOX17 | Abcam | ab224637 | – | IF: 1:200 |
| Anti-Hepatocyte Nuclear Factor 4 alpha (HNF4α) | Sigma-Aldrich | SAB1412164 | – | IF: 1:200 |
| B-27 Supplement | Gibco | 17504-044 | 50 x | 1 x |
| BSA | Beyotime | ST023-200g | – | 5.00% |
| CHIR99021 | Sigma-Aldrich | SML1046 | 3 mM | 3 µM |
| DAPI | Beyotime | C1006 | – | – |
| DEPC-water | Beyotime | R0021 | – | – |
| DM3189 | MCE | HY-12071 | 500 µM | 250 nM |
| DMEM/F12 | Gibco | 11320-033 | 1 x | 1 x |
| DMSO | Sigma-Aldrich | D5879 | 1 x | 1 x |
| DPBS | Gibco | 14190-144 | 1 x | 1 x |
| GlutaMAX | Gibco | 35050-061 | 100 x | 1 x |
| H&E staining kit | Beyotime | C0105S | – | – |
| Hepatocyte growth factor (HGF) | peproTech | 100-39 | 10 µg/ml | 10 ng/ml |
| HepatoZYME-SFM (HZM) | Gibco | 17705-021 | – | – |
| Hydrocortisone-21-hemisuccinate | Sigma-Aldrich | H4881 | 1 mM | 10 µM |
| Indocyanine | Sangon Biotech | A606326 | 200 mg/mL | 1 mg/mL |
| Knock Out DMEM | Gibco | 10829-018 | 1 x | 1 x |
| Knock Out SR Multi-Species | Gibco | A31815-02 | – | 20% |
| Matrigel hESC-qualified | Corning | 354277 | 60 x | 1 x |
| MEM NeAA | Gibco | 11140-050 | 100 x | 1 x |
| mTesR 5X Supplement | Stem Cell Technologies | 85852 | 5 x | 1 x |
| mTesR Basal Medium | Stem Cell Technologies | 85851 | 1 x | 1 x |
| Oncostatin (OSM) | peproTech | 300-10 | 20 µg/ml | 20 ng/ml |
| P450 – CYP3A4 (Luciferin – PFBE) | Promega | V8901 | – | – |
| PAS staining kit | Solarbio | G1281 | – | – |
| Pen/Strep | Gibco | 15140-122 | 100 x | 1 x |
| Peroxidase-Conjugated Goat anti-Mouse IgG | ZSGB-BIO | ZB-2305 | – | WB: 1:2000 |
| Primary Antibody Dilution Buffer for Western Blot | Beyotime | P0256 | – | – |
| ReverTraAce qPCR RT Kit | TOYOBO | FSQ-101 | – | – |
| RNAiso Plus | TaKaRa | 9109 | – | – |
| RPMI 1640 | Gibco | 11875093 | 1 x | 1 x |
| Skim milk | Sangon Biotech | A600669 | – | 5.00% |
| SYBR Green Master Mix | Thermo Fisher Scientific | A25742 | – | – |
| Torin2 | MCE | HY-13002 | 15 µM | 15 nM |
| Tween | Sigma-Aldrich | WXBB7485V | – | 0.10% |
Table 1: Components of cell culture and differentiation base media.
| Name of Gene | abbreviations | primers sequences(F) | primers sequences (R) |
| POU Class 5 Homeobox 1 | OCT4 | AGCGAACCAG TATCGAGAAC |
TTACAGAACC ACACTCGGAC |
| Forkhead Box A2 | FOXA2 | GCATTCCCAATC TTGACACGGTGA |
GCCCTTGCAGC CAGAATACACATT |
| SRY-Box Transcription Factor 17 | SOX17 | TATTTTGTCTGC CACTTGAACAGT |
TTGGGACACAT TCAAAGCTAGTTA |
| Frizzled Class Receptor 8 | FZD8 | ATCGGCTACAA CTACACCTACA |
GTACATGCTGC ACAGGAAGAA |
| C-X-C Motif Chemokine Receptor 4 | CXCR4 | CACCGCATCT GGAGAACCA |
GCCCATTTCCT CGGTGTAGTT |
| Forkhead Box A1 | FOXA1 | AAGGCATACGA ACAGGCACTG |
TACACACCTTG GTAGTACGCC |
| Msh Homeobox 1 | MSX1 | CGCCAAGGCA AAGAGACTAC |
GCCATCTTCAG CTTCTCCAG |
| Mesogenin 1 | MSGN1 | GCTGGAATCCTA TTCTTCTTCTCC |
TGGAAAGCTAACA TATTGTAGTCCAC |
| Caudal Type Homeobox 1 | CDX1 | AAGGAGTTTCAT TACAGCCGTTAC |
TGCTGTTTCTTCT TGTTCACTTTG |
| Even-Skipped Homeobox 1 | EVX1 | GCTGTCTCTCTG AACAAAATGCT |
CATCTCTCACTCT CTCCTCCAAA |
| Mesoderm Posterior BHLH Transcription Factor 2 | MESP2 | AGCTTGGGTG CCTCCTTATT |
TGCTTCCCTGAA AGACATCA |
| NK2 Homeobox 5 | NKX2.5 | CAAGTGTGCG TCTGCCTTT |
CAGCTCTTTCTT TTCGGCTCTA |
| ISL LIM Homeobox 1 | ISL1 | AGATTATATCAGGT TGTACGGGATCA |
ACACAGCGGA AACACTCGAT |
| Hepatocyte Nuclear Factor 4 Alpha | HNF4α | ACATGGACATG GCCGACTAC |
CGTTGAGGTTG GTGCCTTCT |
| Alpha Fetoprotein | AFP | ACTGAATCCAG AACACTGCA |
TGCAGTCAATG CATCTTTCA |
| T-Box Transcription Factor 3 | TBX3 | TTATGTCCCAG CGAGGGTGA |
ACGTGGTGGTG GAGATCTTG |
| Transthyretin | TTR | AGAAAGGCTG CTGATGAC |
GTGCCTTCCA GTAAGATTTG |
| Albumin | ALB | CCCCAAGTGT CAACTCCA |
GTTCAGGACCA CGGATAG |
| Sodium taurocholate cotransporting polypeptide | NTCP | CATAGGGATCGT CCTCAAATCCA |
GCCACACTGCAC AAGAGAATG |
| CCAAT Enhancer Binding Protein Alpha | CEBPA | AGGAGGATGAA GCCAAGCAGCT |
AGTGCGCGATC TGGAACTGCAG |
| Serpin Family A Member 1 | AAT | AAATGAACTCA CCCACGAT |
ACCTTAGTGATG CCCAGT |
| Asialoglycoprotein Receptor 1 | ASGR1 | CAGACCCTGAGA CCCTGAGCAA |
TCCTGCAGCTGG GAGTCTTTTCT |
| Cytochrome P450 Family 3 Subfamily A Member 4 | CYP3A4 | TTGTAATCACTG TTGGCGTGGG |
AATGGGCAAAGT CACAGTGGA |
Table 2: Primer sequences for q-PCR analysis.
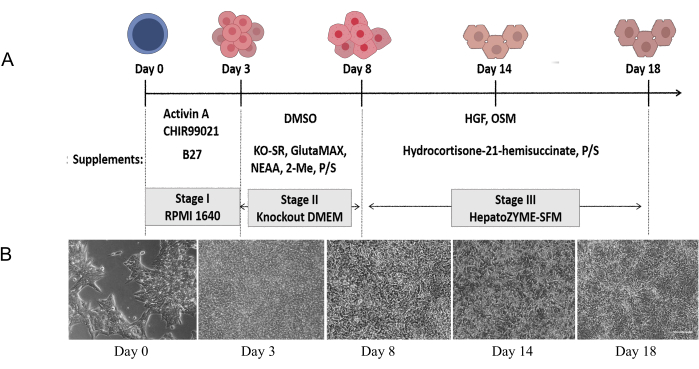
Figure 1: Schematic diagram of protocol for the specification of DE and HLCs. (A) Schematic presentation of the 3-stage differentiation strategy used in this study. (B) Morphology of the differentiated cells at Days 0, 3, 8, 14 and 18. Scale bar = 100 µm. Please click here to view a larger version of this figure.
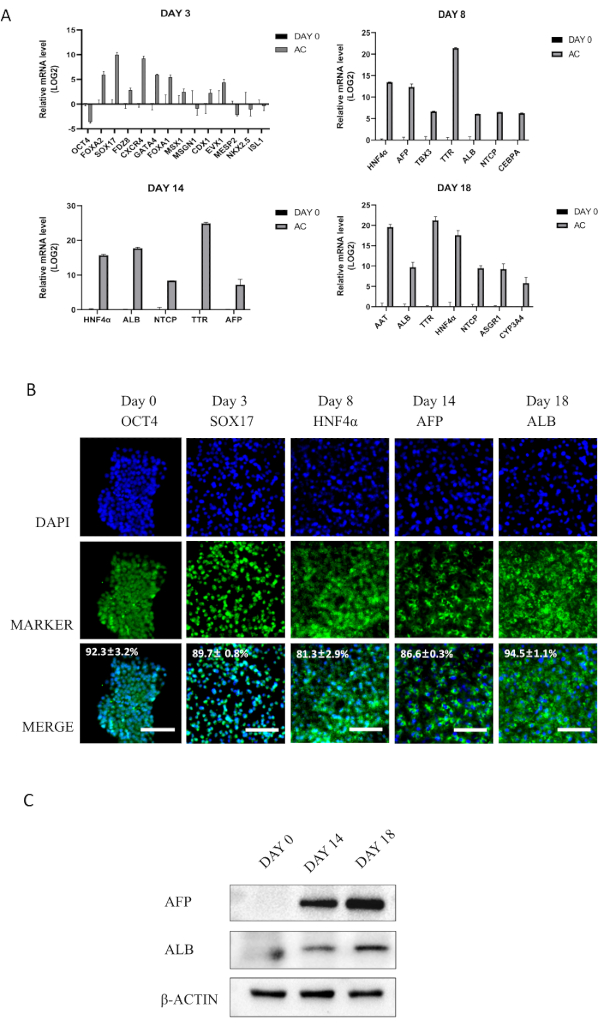
Figure 2: Stage-specific marker expression during hESC differentiation into HLCs. (A) mRNA expression of stage-specific genes. (B-C) Protein expression of stage-specific genes. Please click here to view a larger version of this figure.
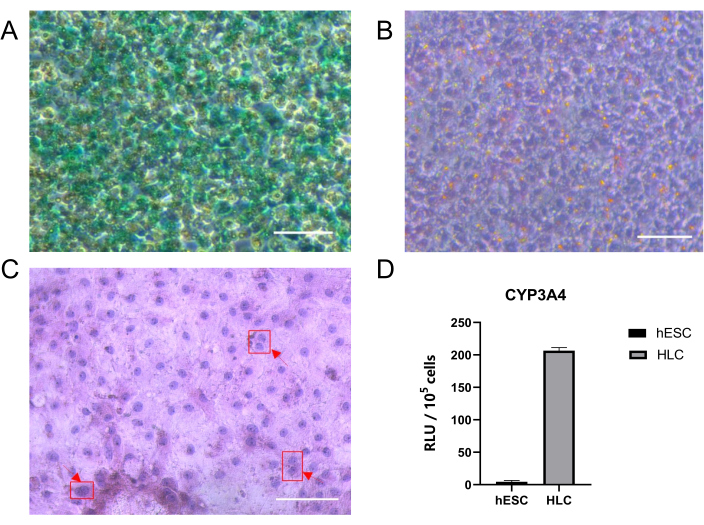
Figure 3: Functions of hESC-derived HLCs. (A) HLCs treated for 1 h with 1 mg/mL indocyanine green demonstrate uptake as assessed by phase microscopy. (B) Representative PAS staining images indicating glycogen storage. (C) Representative HE staining images showing binucleates cells. (D) Basal level activity of major cytochrome P450 enzymes. All values are presented as mean ± SD. Scale bar = 100 µm. Please click here to view a larger version of this figure.
Discussion
Here, we present a stepwise method that induces HLCs from hESCs in three stages. In the first stage, Activin A and CHIR99021 were used to differentiate hESCs into DE. In the second stage, KO-DMEM and DMSO were used to differentiate DE into hepatic progenitor cells. In the third stage, HZM plus HGF, OSM and hydrocortisone 21-hemisuccinate sodium salt were used to continue to differentiate hepatic progenitor cells into HLCs.
The following critical steps need to be taken into consideration while using the protocol. The initial differentiation density of cells and the accurate timing of medium changes are important factors for successful differentiation. When we begin to differentiate, we must ensure that the initial seeding density is appropriate, at a confluence of about 30-40%, when cells are just beginning to come into contact with each other, and that the intercellular spaces are evenly arranged in a network under the field of vision. During differentiation, the corresponding differentiation medium must be changed precisely at the indicated time, especially in the first stage and the key moment of each replacement stage, to ensure that cells differentiate in the mode and stage imitating the process of embryonic development.
The first stage of differentiation of hESCs into DE is particularly important. There are usually a large number of cells that detach and float up during Stage I of differentiation, which is a critical stage for the cell fate, but at this time the cells still retain the ability to proliferate. Therefore, the confluence of cells can reach about 90-100% at the end of Stage I. In addition, it should be noted that at the beginning of Stage III of differentiation (i.e., Day 8), the cytoplasm of the cells begins to condense, resulting in the cells gradually acquiring a slender morphology. However at Day 10, the cytoplasm gradually recovers, and at Day 14, the cells begin to show the obvious polyhedral morphology of hepatocytes.
In this study, the HLCs obtained are actually closer to fetal hepatocytes as AFP is higher expressed than ALB at day 18. Although the generated HLCs exert hepatocyte characteristics and functions, there is still a gap between HLCs and primary human hepatocytes, indicating our differentiated cells are still not functionally mature. Therefore, future work will need to focus on further optimizing the differentiation method.
At present, growth factors and small molecular compounds such as Activin A9,10,14, Wnt3a15, CHIR16, Torin217 and IDE118,19 are commonly used to induce DE differentiation in various differentiation systems. The Song group used Activin A to differentiate stem cells into DE, and used FGF4 (Fibroblast Growth Factor 4), BMP2(Bone morphogenetic protein 2), HGF, KGF (Keratinocyte growth factor), OSM and Dex for hepatic specification and HLCs maturation20. The Sullivan group induced DE by Activin A and Wnt 3a, and induced HLCs by β-ME (2-mercaptoethanol), DMSO, Insulin, HGF, OSM21. The Siller team used CHIR99021 for DE differentiation, DMSO, Dihexa (Hepatocyte growth factor receptor agonist N-hexanoic-Tyr), and Dex for HLC differentiation to obtain HLC with liver function characteristics16. However, some of these methods use many recombinant growth factors to generate DE with high cost and some of them use only small molecules to generate DE with lower efficiency. Activin A is critical factor for DE generation. We have tried to replace Activin A with other small molecules but usually get a lower efficiency. Therefore, we adopt the method of adding Activin A and CHIR99021 as inducing factors of DE, and detect differentiation-related genes in each stage by q-PCR, immunofluorescence and western blotting.
In recent years, the establishment of an experimental system for directed HLC induction from hESCs is an important basis for the modeling of hepatocyte development and screening drugs for liver-related diseases. At the same time, it can also provide an effective source of cells for hepatocyte transplantation, liver tissue engineering, bioartificial livers and other research. It has been reported that HLCs derived from stem cells have been used to conduct various studies on viral infection as a drug screening model to predict hepatotoxic drug-induced responses and to study the innate immune pathway and signaling pathways of cells10,22,23,24,25. Generally, this method introduces in detail an efficient hepatocyte-like cell differentiation technique from hESCs that can successfully differentiate hESCs into mature functional hepatocytes. The results provide a platform for the development of HLC-based applications.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81870432 and 81570567 to X.L.Z.), (No. 81571994 to P.N.S.); the Natural Science Foundation of Guangdong Province, China (No. 2020A1515010054 to P.N.S.), The Li Ka Shing Shantou University Foundation (No. L1111 2008 to P.N.S.). We would like to thank Prof. Stanley Lin from Shantou University Medical College for useful advice.
Materials
| 2-Mercaptoethanol | Sigma | M7522 | For hepatic progenitor differentiation |
| 488 labeled goat against mouse IgG | ZSGB-BIO | ZF-0512 | For IF,second antibody |
| 488 labeled goat against Rabbit IgG | ZSGB-BIO | ZF-0516 | For IF,second antibody |
| Accutase | Stem Cell Technologies | 7920 | For cell passage |
| Activin A | peproTech | 120-14E | For definitive endoderm formation |
| Anti – Albumin (ALB) | Sigma-Aldrich | A6684 | For IF and WB, primary antibody |
| Anti – Human Oct4 | Abcam | Ab19587 | For IF, primary antibody |
| Anti – α-Fetoprotein (AFP) | Sigma-Aldrich | A8452 | For IF and WB, primary antibody |
| Anti -SOX17 | Abcam | ab224637 | For IF, primary antibody |
| Anti-Hepatocyte Nuclear Factor 4 alpha (HNF4α) | Sigma-Aldrich | SAB1412164 | For IF, primary antibody |
| B-27 Supplement | Gibco | 17504-044 | For definitive endoderm formation |
| BSA | Beyotime | ST023-200g | For cell blocking |
| CHIR99021 | Sigma-Aldrich | SML1046 | For definitive endoderm formation |
| DAPI | Beyotime | C1006 | For nuclear staining |
| DEPC-water | Beyotime | R0021 | For RNA dissolution |
| DM3189 | MCE | HY-12071 | For definitive endoderm formation |
| DMEM/F12 | Gibco | 11320-033 | For cell culture |
| DMSO | Sigma-Aldrich | D5879 | For hepatic progenitor differentiation |
| DPBS | Gibco | 14190-144 | For cell culture |
| GlutaMAX | Gibco | 35050-061 | For hepatic progenitor differentiation |
| H&E staining kit | Beyotime | C0105S | For H&E staining |
| Hepatocyte growth factor (HGF) | peproTech | 100-39 | For hepatocyte differentiation |
| HepatoZYME-SFM (HZM) | Gibco | 17705-021 | For hepatocyte differentiation |
| Hydrocortisone-21-hemisuccinate | Sigma-Aldrich | H4881 | For hepatocyte differentiation |
| Indocyanine | Sangon Biotech | A606326 | For Indocyanine staining |
| Knock Out DMEM | Gibco | 10829-018 | For hepatic progenitor differentiation |
| Knock Out SR Multi-Species | Gibco | A31815-02 | For hepatic progenitor differentiation |
| Matrigel hESC-qualified | Corning | 354277 | For cell culture |
| MEM NeAA | Gibco | 11140-050 | For hepatic progenitor differentiation |
| mTesR 5X Supplement | Stem Cell Technologies | 85852 | For cell culture |
| mTesR Basal Medium | Stem Cell Technologies | 85851 | For cell culture |
| Oncostatin (OSM) | peproTech | 300-10 | For hepatocyte differentiation |
| P450 – CYP3A4 (Luciferin – PFBE) | Promega | V8901 | For CYP450 activity |
| PAS staining kit | Solarbio | G1281 | For PAS staining |
| Pen/Strep | Gibco | 15140-122 | For cell differentiation |
| Peroxidase-Conjugated Goat anti-Mouse IgG | ZSGB-BIO | ZB-2305 | For WB,second antibody |
| Primary Antibody Dilution Buffer for Western Blot | Beyotime | P0256 | For primary antibody dilution |
| ReverTraAce qPCR RT Kit | TOYOBO | FSQ-101 | For cDNA Synthesis |
| RNAiso Plus | TaKaRa | 9109 | For RNA Isolation |
| RPMI 1640 | Gibco | 11875093 | For definitive endoderm formation |
| Skim milk | Sangon Biotech | A600669 | For second antibody preparation |
| SYBR Green Master Mix | Thermo Fisher Scientific | A25742 | For RT-PCR Analysis |
| Torin2 | MCE | HY-13002 | For definitive endoderm formation |
| Tween | Sigma-Aldrich | WXBB7485V | For washing buffer preparation |
Referencias
- Hou, Y., Hu, S., Li, X., He, W., Wu, G. Amino Acid Metabolism in the Liver: Nutritional and Physiological Significance. Advances in Experimental Medicine and Biology. 1265, 21-37 (2020).
- Huang, C., Li, Q., Xu, W., Chen, L. Molecular and cellular mechanisms of liver dysfunction in COVID-19. Discovery Medicine. 30, 107-112 (2020).
- Todorovic Vukotic, N., Dordevic, J., Pejic, S., Dordevic, N., Pajovic, S. B. Antidepressants- and antipsychotics-induced hepatotoxicity. Archives of Toxicology. , (2021).
- Thomson, J. A., et al. Embryonic stem cell lines derived from human blastocysts. Science. 282, 1145-1147 (1998).
- Li, Z., et al. Generation of qualified clinical-grade functional hepatocytes from human embryonic stem cells in chemically defined conditions. Cell Death & Disease. 10, 763 (2019).
- Rassouli, H., et al. Gene Expression Patterns of Royan Human Embryonic Stem Cells Correlate with Their Propensity and Culture Systems. Cell Journal. 21, 290-299 (2019).
- Loh, K. M., et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 14, 237-252 (2014).
- Mukherjee, S., et al. Sox17 and beta-catenin co-occupy Wnt-responsive enhancers to govern the endoderm gene regulatory network. Elife. 9, (2020).
- Ang, L. T., et al. A Roadmap for Human Liver Differentiation from Pluripotent Stem Cells. Cell Reports. 22, 2190-2205 (2018).
- Carpentier, A., et al. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. Journal of Clinical Investigation. 124, 4953-4964 (2014).
- Gomez, G. A., et al. Human neural crest induction by temporal modulation of WNT activation. Biología del desarrollo. 449, 99-106 (2019).
- Lee, J., Choi, S. H., Lee, D. R., Kim, D. S., Kim, D. W. Generation of Isthmic Organizer-Like Cells from Human Embryonic Stem Cells. Molecules and Cells. 41, 110-118 (2018).
- Matsuno, K., et al. Redefining definitive endoderm subtypes by robust induction of human induced pluripotent stem cells. Differentiation. 92, 281-290 (2016).
- Mathapati, S., et al. Small-Molecule-Directed Hepatocyte-Like Cell Differentiation of Human Pluripotent Stem Cells. Current Protocols in Stem Cell Biology. 38, 1-18 (2016).
- Hay, D. C., et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proceedings of the National Academy of Sciences of the United States of America. 105, 12301-12306 (2008).
- Siller, R., Greenhough, S., Naumovska, E., Sullivan, G. J. Small-molecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Reports. 4, 939-952 (2015).
- Yu, J. S., et al. PI3K/mTORC2 regulates TGF-beta/Activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nature Communications. 6, 7212 (2015).
- Borowiak, M., et al. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 4, 348-358 (2009).
- Tahamtani, Y., et al. Treatment of human embryonic stem cells with different combinations of priming and inducing factors toward definitive endoderm. Stem Cells and Development. 22, 1419-1432 (2013).
- Song, Z., et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Research. 19, 1233-1242 (2009).
- Sullivan, G. J., et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 51, 329-335 (2010).
- Xia, Y., et al. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. Journal of Hepatology. 66, 494-503 (2017).
- Li, S., et al. Derivation and applications of human hepatocyte-like cells. World Journal of Stem Cells. 11, 535-547 (2019).
- Ogawa, S., et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 140, 3285-3296 (2013).
- Kim, D. E., et al. Prediction of drug-induced immune-mediated hepatotoxicity using hepatocyte-like cells derived from human embryonic stem cells. Toxicology. 387, 1-9 (2017).

