Demonstration of Self-Assembled Cell Sheet Culture and Manual Generation of a 3D Tendon/Ligament-Like Organoid by using Human Dermal Fibroblasts
Summary
Herein, we demonstrate a three-step organoid model (two-dimensional [2D] expansion, 2D stimulation, three-dimensional [3D] maturation) offering a promising tool for tendon fundamental research and a potential scaffold-free method for tendon tissue engineering.
Abstract
Tendons and ligaments (T/L) are strong hierarchically organized structures uniting the musculoskeletal system. These tissues have a strictly arranged collagen type I-rich extracellular matrix (ECM) and T/L-lineage cells mainly positioned in parallel rows. After injury, T/L require a long time for rehabilitation with high failure risk and often unsatisfactory repair outcomes. Despite recent advancements in T/L biology research, one of the remaining challenges is that the T/L field still lacks a standardized differentiation protocol that is able to recapitulate T/L formation process in vitro. For example, bone and fat differentiation of mesenchymal precursor cells require just standard two-dimensional (2D) cell culture and the addition of specific stimulation media. For differentiation to cartilage, three-dimensional (3D) pellet culture and supplementation of TGFß is necessary. However, cell differentiation to tendon needs a very orderly 3D culture model, which ideally should also be subjectable to dynamic mechanical stimulation. We have established a 3-step (expansion, stimulation, and maturation) organoid model to form a 3D rod-like structure out of a self-assembled cell sheet, which delivers a natural microenvironment with its own ECM, autocrine, and paracrine factors. These rod-like organoids have a multi-layered cellular architecture within rich ECM and can be handled quite easily for exposure to static mechanical strain. Here, we demonstrated the 3-step protocol by using commercially available dermal fibroblasts. We could show that this cell type forms robust and ECM-abundant organoids. The described procedure can be further optimized in terms of culture media and optimized toward dynamic axial mechanical stimulation. In the same way, alternative cell sources can be tested for their potential to form T/L organoids and thus undergo T/L differentiation. In sum, the established 3D T/L organoid approach can be used as a model for tendon basic research and even for scaffold-free T/L engineering.
Introduction
Tendons and ligaments (T/L) are vital components of the musculoskeletal system that provide essential support and stability to the body. Despite their critical role, these connective tissues are prone to degeneration and injury, causing pain and impairment of mobility1. Moreover, their limited blood supply and slow healing capacity can lead to chronic injuries, whereas factors such as aging, repetitive motion, and improper rehabilitation further increase the risk of degeneration and injury2. Conventional treatments, such as rest, physical therapy, and surgical interventions, are unable to fully restore the T/L structure and function. Over the past few years, researchers have strived to better understand the intricate nature of T/L in order to seek effective treatments for T/L disorders3,4,5. T/L are distinguished by a hierarchically organized, extracellular matrix (ECM)-dominated structure, composed primarily of type I collagen fibers and proteoglycans, a feature that is difficult to be replicated in vitro6. Traditional two-dimensional (2D) cell culture models fail to capture the characteristic three-dimensional (3D) organization of T/L tissues, limiting their translational potential as well as hindering the innovative progress in the field of T/L regeneration.
Recently, the development of 3D organoid models has offered new possibilities for advancing basic research and scaffolds-free tissue engineering of various tissue types7,8,9,10,11,12,13. For example, to investigate myotendinous junction, Larkin et al. 2006 developed 3D skeletal muscle constructs together with self-organized tendon segments derived from rat tail tendon10. Moreover, Schiele et al. 2013, by using micromachined fibronectin-coated growth channels, directed the self-assembly of human dermal fibroblasts to form cellular fibers without the assistance of 3D scaffold, an approach that can capture key traits of embryonic tendon development11. In the study by Florida et al. 2016, bone marrow stromal cells were first expanded into bone and ligament lineages, next used to generate self-assembled monolayer cell sheets, which were then implemented to create a multiphasic bone-ligament-bone construct mimicking the native anterior cruciate ligament, a model aiming at improved understanding of ligament regeneration12. To elucidate tendon mechanotransduction processes, Mubyana et al. 2018 utilized a scaffold-free methodology by which single tendon fibers were created and subjected to mechanical loading protocol13. Organoids are self-organized 3D structures that mimic the native architecture, microenvironment, and functionality of tissues. 3D organoid cultures provide a more physiologically relevant model for studying tissue and organ biology as well as pathophysiology. Such models can also be used to induce tissue-specific differentiation of different stem/progenitor cell types14,15. Hence, implementing 3D organoid models in the field of T/L biology and tissue engineering becomes a very attractive approach9,16. Alternative cell sources can be implemented for the organoid assembly and stimulated toward tenogenic differentiation. One relevant cell type used for demonstration in this study is dermal fibroblasts7,17,18. These cells are easily accessible through a skin biopsy procedure, which is less invasive compared to bone marrow puncture or liposuction and can be multiplied fairly quickly to large numbers due to their good proliferative capacity. In contrast, more specialized cell types, such as T/L-resident fibroblasts, are more challenging to isolate and expand. Therefore, dermal fibroblasts were also used as a starting point for cell reprogramming technologies towards induced pluripotent embryonic stem cells19. Subjecting dermal fibroblasts to specific 3D culture conditions and signaling cues, such as transforming growth factor-beta 3 (TGFß3), which has been reported to act as a key regulator of various cellular processes, including the formation and maintenance of T/L, can potentiate their in vitro tenogenic differentiation leading to the expression of tendon-specific genes and the deposition of T/L-typical ECM20,21.
Here, we describe and demonstrate a previously established and implemented 3-step (2D expansion, 2D stimulation, and 3D maturation) organoid protocol using commercially available normal adult human dermal fibroblasts (NHDFs) as a cell source, offering a valuable model for studying in vitro tenogenesis7. Despite the fact that this model is not equivalent to in vivo T/L tissue, it still provides a more physiologically relevant system that can be used for investigating cellular differentiation mechanisms, mimicking T/L pathophysiology in vitro, and establishing T/L personalized medicine and drug screening platforms. Moreover, in the future, studies can evaluate whether the 3D organoids are suitable for scaffold-free T/L engineering by further optimization as well as utilizable for the development of scaled-up mechanically robust constructs that closely resemble the dimensions and structural and biophysical properties of native T/L tissues.
Protocol
NOTE: All the steps must be conducted using aseptic techniques.
1. Culture and pre-expansion of NHDFs
- Rapidly thaw the cryo-vial containing adult cryopreserved normal human dermal fibroblasts (NHDFs, 1 x 106 cells) at 37 °C until they are almost defrosting.
- Slowly add 1 mL of prewarmed fibroblast growth medium 2 (ready-to-use kit including basal medium, 2% fetal calf serum (FCS), basic fibroblast growth factor (bFGF) and insulin) supplemented with 1% penicillin/streptomycin (pen/strep) (NHDF medium), prewarmed at 37 °C to the cells.
- Transfer the cells to a 15 mL centrifuge tube. Centrifuge the cells at 300 x g for 5 min at room temperature (RT).
- Remove the supernatant. Resuspend the cell pellet in 12 mL of prewarmed NHDF medium.
- Transfer the cells to a T-75 flask and shake briefly in a cross-wise manner. Place the T-75 flask at 37 °C in a 5% CO2 humidified incubator.
- Change the medium every 2 days. Observe the NHDFs under a microscope until they reach 70% – 80% confluency.
2. 2D expansion
- Take out the T-75 flask of the incubator and remove the NHDF medium. Wash the cells with prewarmed phosphate buffer saline (PBS).
- Add 3 mL of 0.05% trypsin-EDTA to the cells. Incubate the cells at 37 °C in a 5% CO2 humidified incubator until the cells detach from the flask (approximately 3 min).
- Add 6 mL of prewarmed NHDF medium to neutralize the trypsin action. Transfer the cells to a 15 mL centrifuge tube.
- Centrifuge the cells at 300 x g for 5 min at RT and remove the supernatant.
- Resuspend the cell pellet in Dulbecco's Modified Eagles Medium (DMEM) low glucose supplemented with 10% fetal bovine serum (FBS), 1x MEM amino acids, 1% pen/strep, prewarmed at 37 °C.
- Plate the NHDFs in a 10 cm adherent cell culture dish at a density of 8 x 103 NHDFs/cm2 (in total, 4.4 x 105 NHDFs per 10 cm dish). Gently rock in a cross-wise manner.
- Place the 10 cm cell culture dish at 37 °C in a 5% CO2 humidified incubator. Change the medium every 2 days
- Monitor the cells under a microscope until reaching 100% confluency (ca. 5 days).
3. 2D stimulation
- Take out the 10 cm cell culture dishes containing the NHDFs (from steps 2.6 and 2.7) from the incubator. Remove the culture medium and wash the cells with prewarmed PBS.
- Add 10 mL of DMEM high glucose medium supplemented with 10% FBS, 50 µg/mL ascorbic acid, and 1% pen/strep, prewarmed at 37 °C.
- Place the Petri dish at 37 °C in a 5% CO2 humidified atmosphere. Change the medium every 2 days.
- Monitor the NHDFs under a microscope for 14 days.
4. 3D maturation
- Take out the 10 cm cell culture dish from the incubator and remove the DMEM high glucose medium.
- Gently and quickly detach the formed cell sheet from the dish with a cell scraper. While detaching, simultaneously roll the cell sheet into a 3D rod-like organoid.
- Pick up and place the NHDF organoids in a 10 cm non-adherent (for cells) Petri dish. This dish type is used to avoid cell outmigration from the organoid to the plastic.
- At this time point or a day after (day 0 or day 1 of 3D maturation), collect some of the organoids for further analyses such as wet weight, histological evaluation (day 1 organoids are preferable as they become more compact), RNA and protein isolation.
- Fix the edges of the organoid with metal pins as follows:
- Hold one side of the organoid carefully with a tweezer, and with another tweezer, gently apply manual stretching of approximately 10% axial elongation. To estimate the 10% axial elongation, use a millimeter paper or ruler.
- Next, pick up one metal pin with the tweezer and manually press down through the organoid edge into the plastic dish to fix it. Repeat this procedure with the second pin onto the second edge of the organoid.
- Add 10 mL of DMEM high glucose medium supplemented with 10% FBS, 1x MEM amino acids, 50 µg/mL ascorbic acid, 10 ng/mL TGFß3, and 1% pen/strep, prewarmed at 37 °C.
- Place the 10 cm dish at 37 °C in a 5% CO2 humidified atmosphere. Change the medium every 2 days.
- Monitor the organoids regularly for 14 days.
NOTE: Pins may loosen up, hence re-refix using the same technique as described in step 4.5. - After 14 days, remove the DMEM high glucose medium from the organoids. Wash the organoids with prewarmed PBS.
- Measure organoids for wet weight at day 0 and day 14 using a precision scale.
- Place an empty 10 cm dish on the scale and tare the weight to zero to ensure that only the weight of the organoids is measured. Remove the culture medium fully from the 10 cm dish and measure the organoid wet weight one by one.
NOTE: In this study, at day 0, three organoids per donor, and at day 14, two organoids per donor were weighed.
- Place an empty 10 cm dish on the scale and tare the weight to zero to ensure that only the weight of the organoids is measured. Remove the culture medium fully from the 10 cm dish and measure the organoid wet weight one by one.
- According to the study purposes, implement some of the NHDF organoids in further histological analyses or immediately freeze them in liquid nitrogen using sterile DNA/RNA-free tubes for subsequent DNA, RNA, and protein isolation and then store them at -80 °C.
- For histological investigation, fix each organoid in 5 mL of precooled (4 °C) 4% paraformaldehyde in PBS (adjusted to neutral pH) for 45 min on ice, followed by PBS washing at RT (2 x 5 min each).
- Cryoprotect the fixed organoids using a sucrose gradient by placing the organoids in glass jars for histology. Incubate the organoids in 10% sucrose in PBS solution for 2 h at 4 °C, following 20% sucrose/PBS for 2 h at 4 °C, and finally, 30% sucrose/PBS overnight incubation at 4 °C. Change the sucrose solutions by first pipetting out and then filling up with the new solution.
- Embed the organoids in cryo-medium.
- Place a copper plate onto dry ice in a Styrofoam box and cool down for 10 min.
- Next, place a plastic cryomold, prefilled with cryo-medium, on the copper plate and gently press the organoid to the bottom of the cryomold using tweezers. Wait until the cryomedium is fully frozen.
- For long organoids, first cut in half with a scalpel and put the two halves parallel to each other in the cryomold. Repeat the procedure for each organoid intended to undergo histological analysis.
- Store the samples at -20 °C till cryosectioning.
- Using a cryotome, cut the organoids longitudinally in 10 µm thick sections and subject to histological staining such as hematoxylin and eosin (H&E) using standard protocol8.
Representative Results
The 3D T/L organoid model was previously established and demonstrated here by implementing commercially purchased NHDF (n=3, 3 organoids per donor, NHDF were used at passages 5-8). The model workflow is summarized in Figure 1. Figure 2 shows representative phase-contrast images of NHDF culture during the pre-expansion in T-75 flasks (Figure 2A) as well as at the beginning and after 5 days of culture in the 2D expansion step in 10 cm cell culture dishes (Figure 2B). Figure 2C (representative phase-contrast images) demonstrates the confluence of NHDFs during 2D stimulation after 14 days of culture in the 10 cm cell culture dishes. Subsequently, the cell sheets were manually detached and simultaneously rolled into 3D rod-like organoids (at day 0, organoid length is approximately 6 cm, width approximately 0.4 cm), which were cultivated under 10 % static tension for 14 days (Figure 3, day 0 and day 14, representative macroscopic images). By day 14 of the 3D maturation process, the organoids displayed a glistening white appearance, contracted, and appeared denser than the initial organoids. The wet weight of the organoids was measured at the time of formation (day 0) and again on day 14 of the 3D maturation step (Figure 4). A significant reduction in the wet weight was observed, indicating contraction and ECM structural reorganization within the organoids during the time span of this protocol step. In addition to weight change, due to the contraction, the organoid length also reduced (at day 14, the organoid length is approximately 3.5 cm, and the width is approximately 0.3 cm). In this process, the metal pins loosened up and had to be re-fixed; hence, during the 14 days of 3D maturation, we recommend monitoring the organoids frequently. To evaluate the morphology of the organoids, longitudinal sections were obtained from NHDF organoids collected on day 1 and day 14 of the 3D maturation step and subjected to H&E staining (Figure 5). On day 1, the organoids displayed disconnected layers, primarily composed of cells surrounded by low amounts of ECM. Furthermore, the cells within the organoid layers were not aligned along the longitudinal organoid axis used as direction for the applied static tensile load. The NHDF nuclei exhibited a round shape with varying sizes and shapes. H&E analysis of day 14 organoids revealed a noticeable increase in the eosin signal, indicating ECM deposition during the 3D maturation process. At this time point, the organoids exhibited fused layers, with areas containing aligned cells. Cells were frequently in groups; however, they were also organized in cell rows in some locations. The majority of the nuclei had similar sizes and occasionally were elongated.
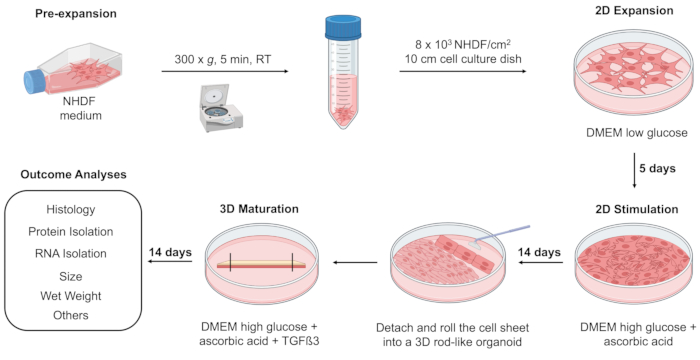
Figure 1: Cartoon of 3D T/L organoid model composed of 3 steps: 2D expansion, 2D stimulation, and 3D maturation. Initially, NHDFs were pre-expanded in a T-75 flask until they reached 70%-80% confluence. Subsequently, NHDFs were cultivated using Dulbecco's Modified Eagles Medium (DMEM) low glucose medium in a 10 cm cell culture dish for 5 days (2D expansion). The cells were then stimulated with DMEM high glucose medium supplemented with ascorbic acid for 14 days of culture (2D stimulation). Following, NHDF were detached from the 10 cm adherent dish, rolled into a 3D rod-like organoid, transferred, and fixed in a 10 cm non-adherent cell culture dish filled with DMEM high glucose medium supplemented with ascorbic acid and Transforming Growth Factor beta 3 (TGFß3) for 14 days (3D maturation). The formed 3D organoids may undergo an assessment of wet weight, histological evaluation, RNA and protein isolation, and other analyses (e.g., transmission electron microscopy, biomechanical measurements) at different time points such as days 0, 1, and 14. The cartoon was generated in BioRender software. Please click here to view a larger version of this figure.
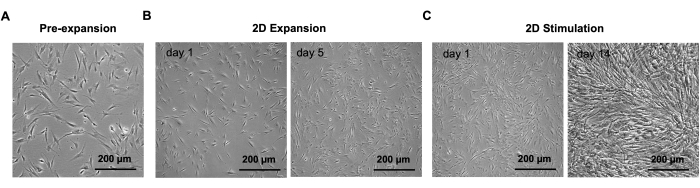
Figure 2: Representative phase-contrast images of NHDFs during pre-expansion, 2D expansion, and 2D stimulation steps. (A) NHDFs during pre-expansion in T-75 flask. (B) NHDFs at days 1 and 5 of the 2D expansion step in a 10 cm cell culture dish. (C) NHDFs at days 1 and 14 of 2D stimulation step in a 10 cm cell culture dish. Scale bars: 200 µm. Images were taken with an inverted microscope equipped with a high-resolution camera. Please click here to view a larger version of this figure.
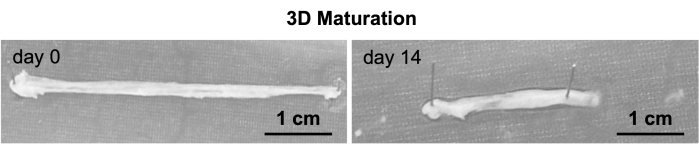
Figure 3: Gross morphology of NHDF 3D organoids. Representative macroscopic images of NHDF organoids at days 0 and 14 of the 3D maturation step. Scale bar: 1 cm. Images were taken with a mobile phone camera. Please click here to view a larger version of this figure.
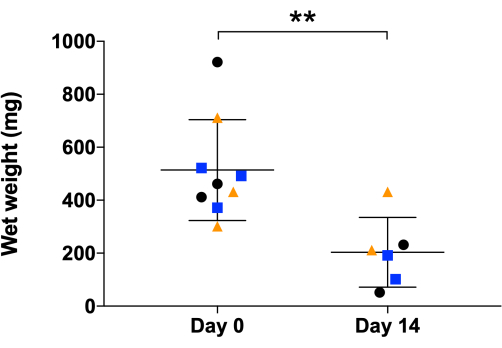
Figure 4: Wet weight analysis of NHDF 3D organoids. Organoids' wet weight measurement was at day 0 and day 14, the end of the 3D maturation process. The graph shows mean values and standard deviations (NHDF n = 3, 3 organoids/donor at day 0 and 2 organoids/donor at day 14 due to 1 organoid/donor taken for histology at day 1), each dot represents individual organoids while each colored shape represents a different individual donor. Data is presented as mean ± standard deviation, and an unpaired parametric t-test was used. Statistical differences: **p ≤0.01. Please click here to view a larger version of this figure.
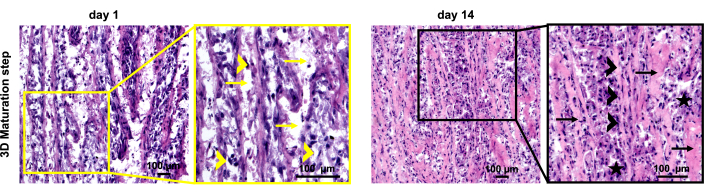
Figure 5: Histological analysis of NHDF 3D organoids. Representative images of hematoxylin and eosin (H&E) stained sections of NHDF organoids during the 3D maturation process at days 1 and 14. Right image – low magnification images, left image – magnified view with indicators as follows: yellow arrows – disconnected layers; yellow arrowheads – nuclei with varying sizes and shapes; black arrows – fused layers and ECM deposition; black arrowheads – cell rows, nuclei with similar size and elongation; black stars -cells in groups. Scale bars: 100 µm. Please click here to view a larger version of this figure.
Discussion
The results demonstrated in this study provide valuable insights into the establishment and characterization of the NHDF 3D organoid model for studying T/L tissues. The 3-step protocol led to the formation of 3D rod-like organoids that exhibit typical features of T/L niche. This model was previously reported in Kroner-Weigl et al. 20237 and demonstrated in great detail here.
The phase-contrast images presented in Figure 2 showed the progression of the NHDF confluence and sheet formation during the expansion and stimulation steps. The 3D maturation process was performed by manually rolling the cell sheets into 3D rod-like organoids, which were then cultured under static tension for 14 days. The organoids displayed a glistening white appearance and increased density by day 14, suggesting contraction and structural reorganization within the organoids. This indicated that the maturation period is critical for achieving a more organized and T/L tissue-mimicking structure.
Furthermore, weight analysis revealed a significant decrease in the wet weight of organoids between days 0 and 14, yet again indicating significant contraction and ECM reorganization within the organoids during the maturation process. Importantly, wet weight measurements may vary between experiments due to handling and residual medium within the organoid or in the dishes. Moreover, the organoid might contain more medium at the beginning because, as discussed below, at day 0, the layers formed by the cell sheet rolling are segregated and not fused. With time, the layers joined and became more compact, and thus, the medium might be extruded.
Morphological evaluation by H&E staining provided additional insights into the organoid structural characteristics. On day 1, the organoids exhibited disconnected layers, primarily composed of cells with round nuclei surrounded by thin peri-cellular ECM. The lack of cell alignment parallel to the axis of the tensile load suggested that further maturation is needed to achieve a more organized cellular and ECM architecture of the organoids. Despite the contraction and organoid weight and dimension reduction between days 1 and 14, the H&E staining showed an increase in eosin-positive areas, suggesting augmented ECM deposition. Moreover, there were no longer visible layers, and occasionally, the cells had elongated nuclei and were positioned in parallel cell rows. This pointed out that the cells within the organoids undergo self-organization by changing their own arrangement as well as ECM deposition, structure, and alignment, thus mimicking the behavior of the native T/L tissue formation.
Overall, these findings highlight the potential of the NHDF organoid model as a valuable tool for studying T/L biology, in vitro tenogenesis, and even for the further development of scaffold-free T/L engineering. However, there are three critical steps for the successful handling of the 3D organoid model: 1) The formation of a tight cell sheet during 2D stimulation stage of the protocol. When using primary cells, this predominantly depends on the donor cell properties; therefore, it is important to assess multiple donors in parallel; 2) The manual rolling of the cell sheet. This requires rolling quickly, but at the same time gently, the cell sheet using a cell scraper. Therefore, it is advisable to plan initial organoids to first train this procedure; and 3) The stable fixation of the pins at the organoid edges. This fixation ensures the organoid axial elongation and prevents deformities as well as collapsing into lump-like structure. Moreover, during the 3D maturation stage, since the organoids experience remodeling of the ECM, the pins can suddenly detach, therefore, it is important to frequently monitor each organoid and to re-fix the pins.
The current model has not yet reached the level of in vivo mimicry to T/L tissue and harbors a number of limitations. For example, it requires a large number of cells per organoid as well as 35 days for the procedure to be completed, while most differentiation protocols, such as 3D chondrogenic pellet model, are set to 21 days22. The washing steps in the protocol procedure should preferably be carried out with physiologically-balanced wash buffer, for example, Hanks Balanced Salt Solution (HBSS) or Earles Balanced Salt Solution (EBSS), rather than PBS which might have metabolic consequences for primary human cells cultivated in vitro. Regarding the culture medium used in the 2D stimulation and 3D maturation stages in the organoid, it has been chosen DMEM plus 10% FBS formulation because such is the foundation of widely accepted differentiation protocols as well as with the intention to standardize and compare between different cell types for their organoidogenic properties23. Reproduction of the 3-step organoid protocol across different laboratory settings might be influenced by cell type used, cell quality and passage, as well as donor-dependent cell properties. Variations in the equipment, especially in the quality of the 10 cm cell culture dishes24 used during the 2D stimulation stage, could potentially impact the robustness of the cell sheet formation. Moreover, due to ECM contraction over the 3D maturation, the axial fixation of the organoids via pins might loosen up; therefore, as mentioned above, frequent monitoring is recommended. Testing and selecting different pins in terms of diameter and strength can also reduce the risk of pin detachment25. However, using pins for manual stretching is not robust, and it is prone to handling variability; hence, it is very important to develop in follow-up research a clamping mechanism for holding the organoid edges, which can result not only in standardization of this step, but also can permit dynamic stretching of the organoids. In general, it will be of interest to investigate two ways of organoid model modification, one downscaling – to reduce the number of cells per organoid and procedure time, thus making it more user-friendly, especially for in vitro studies; and the second upscaling – to increase the dimensions of the organoids in order to test their behavior in large animal models as potential tendon replacement strategy.
The model demonstrated here can provide a platform for investigating the mechanisms underlying tendon development, pathophysiology, and perhaps repair and regeneration if the organoids are combined with different cell types and subjected to micro-ruptures. It is important to mention that the study of Kroner-Weigl et al. 20237 identified several limitations regarding the NHDF cell type, namely, the NHDFs formed organoids are larger and more cellular than those derived from T/L cells, making it challenging to control cell proliferation and behavior. With regards to cell apoptosis, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-based assay showed no evidence of dead cells throughout the organoid at day 147, suggesting appropriate diffusion of nutrients, waste, and oxygen during the 3D maturation step. Interestingly, despite the morphological differences, pilot screening by quantitative PCR of a number of T/L-related genes (including different collagen types, Scleraxis (SCX), Mohawk (MKX), etc.) suggested comparable gene expression between NHDF and T/L organoids7, a finding that should be further investigated as well as should be validated at the protein level. Hence, for this cell type, further optimization is required in order to support T/L differentiation and can be based on adjusting the medium used in 2D stimulation and 3D maturation steps (e.g., serum concentration, growth factors, vitamins), extending the time of maturation or even subjecting the organoids to dynamic mechanical stretch. Evolving the protocol could further improve the organoid compositional, structural, and functional properties, thus enabling the model to better mimic the complex in vivo T/L tissue niche. Moreover, alternative cell sources can be investigated for their capacity to form T/L organoids and whether they can undergo T/L differentiation. Yan et al. 20209, employed the 3D organoid model by using young/healthy and aged/degenerative tendon stem/progenitor cells (Y-TSPCs and A-TSPCs) to investigate cellular behavior in 3D as well as whether tendon-forming capacity changes with T/L aging. They reported that A-TSPC 3D organoids exhibit poor tissue morphology, and the cells within have a lower proliferation rate but higher apoptosis paralleled with elevated levels of senescence-related markers9. Hence, the T/L 3D organoid model is a promising platform for studying molecular and cellular mechanisms involved in tendon aging and further can be used to test novel pharmacological therapies for tendon repair and regeneration. In a different approach, Chu et al. 2021 implemented dental follicle cells (DFCs) in the 3D organoid model in order to assess their ligamentogenic differentiation. This cell type formed very well-organized organoids, thus suggesting DFCs as an attractive cell source to differentiate in periodontal ligament (PDL) tissue, which could, in turn, develop a promising therapeutic strategy for periodontitis8.
In the future, the model could become even more complex by including different cell types (e.g., myofibroblasts, macrophages), which may offer novel insights into cell-specific behaviors and cellular crosstalk within organoids and contribute to a more comprehensive understanding of T/L biology, physiology and pathophysiology8,9,16,26.
The 3D organoid models hold several advantages for tissue engineering and regenerative medicine applications. They mimic closely the cellular composition and organization as well as cell-to-cell and cell-to-matrix interactions of human tissues, thereby offering a physiologically relevant platform for studying disease mechanisms and drug responses27. Organoids can be used as a high-throughput screening platform to assess the effectiveness but also potential side effects of different drugs and provide a reliable and low-cost alternative to animal testing. Furthermore, they enable direct comparisons between healthy and diseased conditions, allowing the discovery of key molecular and cellular changes associated with diverse pathologies, identification of biomarkers for early disease detection as well as facilitation of ascended understanding of disease mechanisms, and the development of targeted therapies28,29.
Taken together, we demonstrated in great detail the steps of the previously established 3D T/L organoid model that provides a promising platform for studying T/L tissue biology and in vitro tenogenic processes of various cell types. Further implementation, optimization, and research of this model are strongly encouraged as they may lead to advancing scaffold-free T/L engineering strategies, which, in turn, can contribute to improving T/L therapy.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
D.D. and S.M.-D. acknowledge the BMBF Grant "CellWiTaL: Reproducible cell systems for drug research – transfer layer-free laser printing of highly specific single cells in three-dimensional cellular structures" Proposal Nr. 13N15874. D.D. and V.R.A. acknowledge the EU MSCA-COFUND Grant OSTASKILLS "Holistic training of next-generation Osteoarthritis researches" GA Nr. 101034412. All authors acknowledge Mrs. Beate Geyer for technical assistance.
Materials
| Ascorbic acid | Sigma-Aldrich, Taufkirchen,Germany | A8960 | |
| 10 cm adherent cell culture dish | Sigma-Aldrich, Taufkirchen,Germany | CLS430167 | |
| 10 cm non-adherent petri dish | Sigma-Aldrich, Taufkirchen,Germany | CLS430591 | |
| Cryo-medium | Tissue-Tek, Sakura Finetek, Alphen aan den Rijn, Netherlands | 4583 | |
| Cryomold standard | Tissue-Tek, Sakura Finetek, Alphen aan den Rijn, Netherlands | 4557 | |
| D(+)-Sucrose | AppliChem Avantor VWR International GmbH, Darmstadt, Germany | A2211 | |
| DMEM high glucose medium | Capricorn Scientific, Ebsdorfergrund, Germany | DMEM-HA | |
| DMEM low glucose | Capricorn Scientific, Ebsdorfergrund, Germany | DMEM-LPXA | |
| Fetal bovine serum | Anprotec, Bruckberg, Germany | AC-SM-0027 | |
| Fibroblast growth medium 2 | PromoCell, Heidelberg, Germany | C-23020 | |
| Inverted microscope with high resolution camera | Zeiss | NA | Zeiss Axio Observer with Axiocam 506 |
| MEM amino acids | Capricorn Scientific, Ebsdorfergrund, Germany | NEAA-B | |
| Metal pins | EntoSphinx, Pardubice, Czech Republic | 04.31 | |
| Normal human dermal fibroblasts | PromoCell, Heidelberg, Germany | C-12302 | |
| Paraformaldehyde | AppliChem, Sigma-Aldrich, Taufkirchen, Germany | A3813 | |
| Penicillin/streptomycin | Gibco, Thermo Fisher Scientific, Darmstadt, Germany | 15140122 | |
| Phosphate buffer saline | Sigma-Aldrich, Taufkirchen, Germany | P4417 | |
| TGFß3 | R&D Systems, Wiesbaden, Germany | 8420-B3 | |
| Trypsin-EDTA 0,05% DPBS | Capricorn Scientific, Ebsdorfergrund, Germany | TRY-1B |
Referencias
- Schneider, M., Angele, P., Järvinen, T. A. H., Docheva, D. Rescue plan for Achilles: Therapeutics steering the fate and functions of stem cells in tendon wound healing. Adv Drug Deliv Rev. 129, 352-375 (2018).
- Steinmann, S., Pfeifer, C. G., Brochhausen, C., Docheva, D. Spectrum of tendon pathologies: Triggers, trails and end-state. Int J Mol Sci. 21 (3), 844 (2020).
- Snedeker, J. G., Foolen, J. Tendon injury and repair – A perspective on the basic mechanisms of tendon disease and future clinical therapy. Acta Biomater. 63, 18-36 (2017).
- Lomas, A. J., et al. The past, present and future in scaffold-based tendon treatments. Adv Drug Deliv Rev. 84, 257-277 (2015).
- Gomez-Florit, M., Labrador-Rached, C. J., Domingues, R. M. A., Gomes, M. E. The tendon microenvironment: Engineered in vitro models to study cellular crosstalk. Adv Drug Deliv Rev. 185, 114299 (2022).
- Docheva, D., Müller, S. A., Majewski, M., Evans, C. H. Biologics for tendon repair. Adv Drug Deliv Rev. 84, 222-239 (2015).
- Kroner-Weigl, N., Chu, J., Rudert, M., Alt, V., Shukunami, C., Docheva, D. Dexamethasone Is not sufficient to facilitate tenogenic differentiation of dermal fibroblasts in a 3D organoid model. Biomedicines. 11 (3), 772 (2023).
- Chu, J., Pieles, O., Pfeifer, C. G., Alt, V., Morsczeck, C., Docheva, D. Dental follicle cell differentiation towards periodontal ligament-like tissue in a self-assembly three-dimensional organoid model. Eur Cell Mater. 42, 20-33 (2021).
- Yan, Z., Yin, H., Brochhausen, C., Pfeifer, C. G., Alt, V., Docheva, D. Aged tendon stem/progenitor cells are less competent to form 3D tendon organoids due to cell autonomous and matrix production deficits. Front Bioeng Biotechnol. 8, 406 (2020).
- Larkin, L. M., Calve, S., Kostrominova, T. Y., Arruda, E. M. Structure and functional evaluation of tendon-skeletal muscle constructs engineered in vitro. Tissue Eng. 12 (11), 3149-3158 (2006).
- Schiele, N. R., Koppes, R. A., Chrisey, D. B., Corr, D. T. Engineering cellular fibers for musculoskeletal soft tissues using directed self-assembly. Tissue Eng Part A. 19 (9-10), 1223-1232 (2013).
- Florida, S. E., et al. In vivo structural and cellular remodeling of engineered bone-ligament-bone constructs used for anterior cruciate ligament reconstruction in sheep. Connect Tissue Res. 57 (6), 526-538 (2016).
- Mubyana, K., Corr, D. T. Cyclic uniaxial tensile strain enhances the mechanical properties of engineered, scaffold-free tendon fibers. Tissue Eng Part A. 24 (23-24), 1808-1817 (2018).
- Brassard, J. A., Lutolf, M. P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 24 (6), 860-876 (2019).
- Kim, J., Koo, B. K., Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 21 (10), 571-584 (2020).
- Hsieh, C. F., et al. In vitro comparison of 2D-cell culture and 3D-cell sheets of scleraxis-programmed bone marrow derived mesenchymal stem cells to primary tendon stem/progenitor cells for tendon repair. Int J Mol Sci. 19 (8), 2272 (2018).
- Chu, J., Lu, M., Pfeifer, C. G., Alt, V., Docheva, D. Rebuilding tendons: A concise review on the potential of dermal fibroblasts. Cells. 9 (9), 2047 (2020).
- Gaspar, D., Spanoudes, K., Holladay, C., Pandit, A., Zeugolis, D. Progress in cell-based therapies for tendon repair. Adv Drug Deliv Rev. 84, 240-256 (2015).
- Sacco, A. M., et al. Diversity of dermal fibroblasts as major determinant of variability in cell reprogramming. J Cell Mol Med. 23 (6), 4256-4268 (2019).
- Wang, W., et al. Induction of predominant tenogenic phenotype in human dermal fibroblasts via synergistic effect of TGF-β and elongated cell shape. Am J Physiol Cell Physiol. 310 (5), C357-C372 (2016).
- Pryce, B. A., Watson, S. S., Murchison, N. D., Staverosky, J. A., Dünker, N., Schweitzer, R. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 136 (8), 1351-1361 (2009).
- Pattappa, G., et al. Physioxia has a beneficial effect on cartilage matrix production in interleukin-1 beta-inhibited mesenchymal stem cell chondrogenesis. Cells. 8 (8), 936 (2019).
- Shafiee, A., et al. Development of physiologically relevant skin organoids from human induced pluripotent stem cells. Small. 2304879, (2023).
- Zeiger, A. S., Hinton, B., Van Vliet, K. J. Why the dish makes a difference: quantitative comparison of polystyrene culture surfaces. Acta Biomater. 9 (7), 7354-7361 (2013).
- Koh, T. M., Feih, S., Mouritz, A. P. Strengthening mechanics of thin and thick composite T-joints reinforced with z-pins. Compos Part A Appl Sci. 43 (8), 1308-1317 (2012).
- Gelberman, R. H., et al. Combined administration of ASCs and BMP-12 promotes an M2 macrophage phenotype and enhances tendon healing. Clin Orthop Relat Res. 475 (9), 2318-2331 (2017).
- Hofer, M., Lutolf, M. P. Engineering organoids. Nat Rev Mater. 6 (5), 402-420 (2021).
- Driehuis, E., et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 116 (52), 26580-26590 (2019).
- Yan, H. H. N., et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 23 (6), 882-897 (2018).

