Fabrication and Characterization of Colorectal Cancer Organoids from SW1222 Cell Line in Ultrashort Self-Assembling Peptide Matrix
Summary
This protocol aims to evaluate biofunctional self-assembling peptides for cell adhesion, organoid morphology, and gene expression by immunostaining. We will use a colorectal cancer cell line to provide a cost-effective way of obtaining organoids for intensive testing.
Abstract
Ultrashort self-assembling peptides (SAPs) can spontaneously form nanofibers that resemble the extracellular matrix. These fibers allow the formation of hydrogels that are biocompatible, biodegradable, and non-immunogenic. We have previously proven that SAPs, when biofunctionalized with protein-derived motifs, can mimic the extracellular matrix characteristics that support colorectal organoid formation. These biofunctional peptide hydrogels retain the original parent peptide’s mechanical properties, tunability, and printability while incorporating cues that allow cell-matrix interactions to increase cell adhesion. This paper presents the protocols needed to evaluate and characterize the effects of various biofunctional peptide hydrogels on cell adhesion and lumen formation using an adenocarcinoma cancer cell line able to form colorectal cancer organoids cost-effectively. These protocols will help evaluate biofunctional peptide hydrogel effects on cell adhesion and luminal formation using immunostaining and fluorescence image analysis. The cell line used in this study has been previously utilized for generating organoids in animal-derived matrices.
Introduction
In recent years, self-assembling peptides (SAPs) have emerged as promising biomaterials for tissue engineering applications. SAPs possess unique properties, including spontaneous formation of nanofibers, biocompatibility, biodegradability, and non-immunogenicity, making them attractive candidates for scaffold development1. SAPs have been previously used together with various types of cells, and notably, reported ultrashort SAPs have facilitated the encapsulation of stem cells while upholding their pluripotency over prolonged periods encompassing more than 30 passages and with minimal incidence of chromosomal aberrations2,3,4. Hence, adapting SAPs for their use in organoid culture constituted a rational subsequent step.
Organoids are complex three-dimensional structures that arise from single pluripotent cells. These structures give rise to various cell types, which then self-organize to replicate the developmental processes of embryonic and tissue growth in vitro5. This innovative framework has evolved into a formidable tool for in vitro investigations, efficiently conserving the genetic, phenotypic, and behavioral attributes characteristic of in vivo organs6. Nevertheless, a principal impediment in the bench-to-bedside transition of organoid-based findings is the need for reproducibility7. This variation in results is primarily ascribed to the difficulty in fully differentiating human pluripotent stem cells into specialized cell lineages, compounded by the absence of authentic tissue architecture and the inherent complexity encountered in organoid models. Given the rise in popularity of stem-cell- and organoid-based studies8, there has been an escalated demand for biomaterials with dynamic mechanical properties and integrin-like adhesion sites or scaffolds with controlled degradability7,9. These attributes can be effectively tuned through the strategic utilization of biofunctionalized SAPs.
SAPs are short sequences of amino acids with the inherent ability to spontaneously organize into well-defined structures10. These peptides often contain alternating hydrophobic and hydrophilic residues, which drive their self-assembly through non-covalent interactions, including hydrogen bonding, electrostatic interactions, and hydrophobic effects11,12. The self-assembly process of these peptides is primarily driven by the need to minimize the system's free energy. When in an aqueous environment, the hydrophobic residues tend to cluster together to minimize their exposure to water, while the hydrophilic residues interact with the surrounding water molecules. This phenomenon leads to the formation of various nanostructures. In this case, ultrashort self-assembling peptides form nanofibers with characteristics that depend on the peptide sequence and environmental conditions2,12,13,14,15,16. These peptides adopt a β-turn conformation, where individual peptide strands align parallel or antiparallel to each other, stabilized by hydrogen bonds (Figure 1). The presence of positive ions accelerates the self-assembling processes that lead to nanofiber formation.
Peptide nanofibers have been identified as possessing an innate ability to entrap significant volumes of water. This characteristic facilitates the generation of a hydrogel, which subsequently manifests as a biocompatible and biodegradable three-dimensional space conducive to cellular proliferation and growth. These self-assembling peptide nanofibers intricately emulate the topographical features inherent to the natural extracellular matrix (ECM) environment1. This resemblance endows cells with an environment that imitates their physiological habitat, further promoting optimal cellular activities17. Furthermore, the versatility inherent to these peptides permits a considerable degree of tunability4. Such adaptability enables changes to the matrix's properties, such as stiffness, gelation kinetics, and porosity. These adaptations are achieved through modifications to the peptide sequence. Consequently, self-assembling peptides (SAPs) have emerged as pivotal components in contemporary biomaterial science, extensively used as scaffolds for tissue regeneration and cellular culture13,17.
One of the critical advantages of self-assembling peptides is their ability to be easily synthesized and modified at the molecular level. This advantage allows for the incorporation of specific functional groups or bioactive motifs into the peptide sequence, enabling the design of peptides with tailored properties and functionalities18,19,20 (Figure 1B). For example, biofunctionalized SAPs can be designed to mimic the ECM and promote differentiation using the RGD peptide19,21. Peptide Ben-IKVAV has also been reported to significantly increase the expression of neuronal-specific markers due to its ligand-specific motiety22. These peptides can also be engineered to display bioactive molecules, such as integrin-binding peptides, to enhance cell survival23. Finally, other biofunctionalized SAPs have been developed to promote angiogenesis by including the IKVAV and YIGSR motifs in their structures24.
Biofunctionalized SAPs reported in an earlier publication show that self-assembling can be further modified by including the naïve peptide from which the biofunctionalized SAP was derived25. These SAP mixtures can also provide a wide array of SAP formulations that vary in their biochemical activity and physicochemical properties. For instance, the amphiphilic peptide IIFK is an ultrashort SAP that can withstand biofunctionalization with various motifs, exemplified by the incorporation of the IKVAV motif (Figure 1B). Both peptides can form nanofibers, both alone and combined. Each formulation results in hydrogels with varying physical properties (Figure 2)
However, because of the extensive array of potential permutations and alternatives obtained from the biofunctionalization of SAPs, there is a need to provide a fast and cost-effective option for organoid testing. One candidate for such intensive and cost-effective organoid evaluation is the SW1222 cell line, derived from human colorectal adenocarcinoma26,27. SW1222 cells possess characteristics that enable their aggregation into 3D structures resembling organoids, making them an ideal model for studying tissue development and regenerative medicine applications. SW1222 cells have been identified as capable of generating organoids owing to their intrinsic overexpression of the LGR5 gene27. The creation of organoids from individual LGR5+ stem cells has been convincingly exhibited before, as well as the propensity of SW1222 cells to achieve morphological characteristics of a colorectal cancer organoid28.
In this methods paper, we present detailed protocols for evaluating and characterizing the effects of various biofunctional peptides on cell adhesion and lumen formation using SW1222 cells (Figure 3). By providing step-by-step procedures and imaging analysis methods, we hope to offer valuable insights into the biofabrication of organoids and the evaluation of simplistic SAP matrices for organoid culture.
Protocol
1. Buffer and solution preparation
NOTE: All stated concentrations are final concentrations.
- Add fetal bovine serum (FBS) and penicillin-streptomycin at a final concentration of 10% and 1%, respectively, to prepare complete Iscove's Modified Dulbecco's Medium (IMDM). Store in the dark at 4 °C for up to 1 month.
- Mix MgCl2 (3 mM), sucrose (300 mM), and Triton X-100 (0.5%) in phosphate buffer saline (PBS) to prepare the permeabilization buffer. Store this solution at 4 °C for up to 2 months.
- Combine bovine serum albumin (BSA, 1%), glycine (0.3 M), and Tween-20 (0.1%) in PBS to prepare a blocking buffer. Store this solution at 4 °C for up to 2 months.
- Dilute 10x to 2x PBS with ultrapure water under sterile conditions. Store this solution at room temperature for over 6 months.
- Sterilize 3 mg of the peptide powder under ultraviolet light for 30 min. Add 500 µL of ultrapure water to dissolve using a vortex mixer.
NOTE: The final concentration must be twice the target concentration, i.e., 6 mg/mL, as the 1x concentration will be 3 mg/mL.
2. Colorectal cancer organoid fabrication from SW1222 in peptide
- Thaw an aliquot overnight at 4 °C and keep it in ice until needed to prepare the basement matrix control.
- Place a 10 µL droplet of 2x peptide solution in the center of each well in a 24-well plate. Allow the plate to incubate at room temperature for 10 min. Alternatively, deposit 8-10 droplets per well in a 6-well plate.
NOTE: Samples meant to be used in confocal and electron microscopy are more accessible to manipulate if seeded on top of a sterile coverslip. - Add 10 µL of 2x PBS. Mix the PBS solution gently using the tip. Allow the gels to stabilize for at least 20 min, ensuring the droplets remain almost intact.
NOTE: The goal of PBS addition and gentle mixing is to maintain the integrity of the droplet and avoid spreading the hydrogel into a layer. - Detach SW1222 cells using 0.125% trypsin-EDTA solution (5 mL). Incubate the cells with trypsin at 37 °C for 5-10 min until they detach from the flask. Abstain from tapping the flask to avoid cell clumping.
- Add 5 mL of complete medium to inactivate the trypsin. Filter the cell suspension using a 30 µm cell strainer. Alternatively, use a syringe with a 25 G needle to disrupt the aggregates.
- Prepare a cell suspension of 0.4 × 106 cells/mL and inject 2 µL of the cell suspension in each peptide droplet using a 10 µL micropipette.
- Incubate the droplets for 30 min at 37 °C, 5% CO2. Add 0.5 mL of supplemented medium to each well after incubation.
- While waiting, prepare the basement membrane matrix control by diluting the main stock (8-12 mg/mL, depending on the lot) to a final concentration of 3 mg/mL using the supplemented medium. Add cells to this solution (800 cells per 25 µL).
NOTE: Basement membrane manipulation must be done on ice at all times. Tips must be rinsed in ice-cold PBS before touching the hydrogel. Otherwise, polymerization will occur inside the tips. - Plate 20 µL droplets of the basement membrane solution in each well. Allow the droplets to gel by incubating for 20 min at 37 °C. After the material is gelled, add 0.5 mL of complete medium.
- Change the medium every 3-4 days. Observe the cells organizing and showing signals of lumen formation as early as day 4.
- Image cells in brightfield on day 1, day 4, and day 7 to evaluate organoid growth.
3. Viability and proliferation
- Prepare three black-well plates for the viability assay and three black-well plates for the proliferation assay. Add 25 µL of 2x peptide solution per well and gelate using the same volume of 2x PBS. Seed the cells as described above.
- Prepare three wells for each formulation of peptide hydrogel to be used as blanks in the proliferation plate. Do not seed cells on these.
- Remove the medium from the viability well plate and wash with 1x PBS for 3 x 5 min.
- Dilute Calcein-AM and ethidium homodimer to 8 µM from the viability/cytotoxicity Kit for mammalian cells (see Table of Materials) into the same light-protected conical tube. To prepare 2 mL of this solution to 2 mL of PBS in a light-protected conical tube, add 4 µL of the Calcein-AM stock solution and 8 µL of the Ethidium homodimer-1 stock solution.
- Add the calcein/ethidium homodimer solution prepared in step 3.4 to the 96-well plate. Add 100 µL per well. Protect from light and incubate at room temperature for 30 min.
- Wash for 3 x 5 min with 1x PBS. Image a minimum of three fields (center, top, bottom) when using a 4x objective or a minimum of five fields (center, top left, top right, bottom left, bottom right) when using a 10x and 20x objective under a fluorescence microscope.
- Remove the medium from the proliferation well plate to ensure a final volume of 90 µL.
- Add 10 µL of Alamar Blue solution (see Table of Materials) to reach a final concentration of 10%.
- Protect from light and incubate for 2-4 h at 37 °C, 5% CO2
- Use a well plate reader to measure fluorescence (Ex/Em = 530-560/590 nm/nm) or absorbance (570 nm). Average the values obtained for the blanks and subtract them from the values from each well that contains cells.
- Calculate proliferation values by averaging the signal of each condition and subtracting the average of their respective blanks. Create graphs by generating an indexed box plot of relative absorbance/fluorescence signal versus time.
4. Adherence assay of cells to matrix
- Prepare 100 µL of peptide hydrogels in two independent 96-well plates.
- Seed 20,000 cells in each well and add 100 µL of medium. Incubate the plates for 90 min at 37 °C, 5% CO2.
- After incubation, take only one plate and carefully resuspend the medium using a P200 micropipette for 20-30 s. Be careful not to disrupt the hydrogel by keeping the tip from the gel. Alternatively, if the hydrogel mechanical properties permit it (G' > 8,000 Pa), tap the four sides of the well plate using a moving vortex.
NOTE: Resuspending the media (or tapping the well plate with the vortex) will remove both unattached cells and cells with tenuous adhesion to the surface of the matrix. The cellular attachment will occur within 90 min, although the strength of this attachment will vary depending on the specific peptide hydrogels and their adhesive and binding properties. Colorectal organoid growth is known to be favored in matrices with stiffness values of under 2,000 Pa. Hence, because the hydrogels presented in this protocol are soft and brittle, we make a point of using gentle mechanical stress. - Remove 100 µL of medium from both well plates and add wash once with 1x PBS.
- Prepare a 1 µg/mL DAPI solution. Add 100 µL of the DAPI solution to all wells and incubate for 5 min at room temperature.
- Image a representative number of fields under a fluorescence microscope. Ensure all images are obtained using the same objective to keep the pixel/distance ratio constant.
NOTE: Image nine fields for a 10x objective or 15 fields with a 20x objective. - Load the fluorescence images on image analysis software (e.g., ImageJ) and analyze the particles on all images.
- From the Image menu, select Type and click on 8-bit.
- From the Image menu, select Adjust and click on Brightness/Contrast. Click on the Auto button | Apply.
- From the Image menu, select Adjust and click on Threshold. Click on the Auto button and close the window.
- From the Analyze menu, click on Analyze Particles. Tick the option to summarize and click OK.
- On the Summary window, hover over the File menu and click Save to save the summary values.
- Export the results for all images to standard statistics software.
- Calculate the percentage of area covered by cells before and after stress. Make an indexed box plot of the percentage of area against each condition.
5. Immunostaining of organoids in peptide hydrogel
- Seed 800 cells in 20 µL droplets of peptide hydrogel for 4 days, as described in steps 2.1-2.10.
- On day 4, remove the medium and wash each well 2x using 1x PBS.
- Add 400 µL of a 4% formaldehyde solution. Incubate the well plate for 30 min at room temperature.
- Remove the formaldehyde solution with a P1000 and wash once with 1x PBS.
- Add 1 mL of permeabilization buffer and incubate for 5 min at room temperature.
- Remove the permeabilization buffer with a P1000 and wash once with 1x PBS.
- Add 0.5 mL of blocking buffer and incubate for 30 min at room temperature.
- Remove the blocking buffer with a P1000 and wash with 1x PBS.
- Dilute the primary antibody in the blocking buffer according to the manufacturer's instructions (see Table of Materials). Add 200 µL to two wells.
- Incubate overnight at 4 °C.
- Remove the staining solution with a P200 and wash once with 1x PBS.
- Dilute the secondary antibody in blocking buffer, according to the manufacturer's instructions (see Table of Materials). Add the phalloidin conjugate at a ratio of 1:40 in the secondary antibody solution.
- Protect from light and incubate at room temperature for no more than 3 h. Remove the staining solution with a P200 and wash once with 1x PBS.
- Prepare a DAPI solution of 1 µg/mL in the blocking buffer. Add 300 µL of the DAPI solution to each well.
- Protect from light and incubate for 5 min at room temperature.
- Remove the staining solution with a P200 and wash once with 1x PBS.
- Protect from light and store at 4 °C for up to 7 days.
6. Imaging and image analysis of organoids in matrix
- Image the stained samples using an epifluorescence microscope at 10x. Use only the phalloidin and DAPI channels.
- Open the Cellopose software and press ctrl + L to load an image. As the software will have access to all the files in the location of the selected image, be sure to save all fluorescence images in the exact location without having any subfolders.
- Type the average diameter of the colonies in the Segmentation panel and click ENTER.
- Select a custom model for colon organoids and click on the Run Model button.
NOTE: Here, we suggest using two from-scratch models in Cellpose2.029,30 - Press ctrl + s to save the masks as a .npy file.
- Use the keyboard arrows to move through the images of the folder and run the model for colon organoids on all images in the folder.
- Make a copy of the folder and repeat steps 6.2-6.6 using a model for the colon organoid lumen.
- Open ImageJ and click on Plugin | Macros | Run… to run the imagej_roi_converter.py file available from the Cellpose Github site and wait for a window to appear.
- Select the first file from the colon organoid folder and click Open.
- Select the seg.npy file corresponding to the first file and click Open.
NOTE: ImageJ will convert the masks into regions of interest and overlap them with the corresponding fluorescence images. - Click Select All in the ROI Manager window and click Measure.
- Click File | Save as… on the Results window and save the results.
NOTE: This will result in a .csv file containing all the shape properties of each colony in the field. - Repeat steps 6.8-6.12 for the same image in the folder for colon organoid lumen.
- Load OriginPro and press ctrl + 3 to open the Import Wizard. Click on the three dots button and select both .csv files corresponding to organoid and lumen shape properties for the same image.
- Copy and paste the results from the lumen shape properties in one column next to the results of the colony to ensure that each lumen measurement is paired with its respective colony.
- In a new column, divide the areas of the lumen and the colony to yield the relative lumen area of its containing colony.
- Select the column corresponding to the circularity of each colony and the lumen area. Click on Plot | Basic 2D | Scatter.
- Right-click on the plot window and select Plot details. Click on the Centroid(PRO) tab and tick the options reading Show Centroid Point for Subset, Connect to Data Points, and Show Ellipse.
Representative Results
First, we evaluated the cells grown in a 24-well plate for 7 days using brightfield imaging. We identified small clusters of cells assembling into organoids during the week, as seen in Figure 4. A controlled scan method can follow the mobility of the cells and the organoids between different days. In general, we looked at the evolution of the morphology of the cells during the whole week. SW1222-derived organoids should have a round morphology and a light appearance. A darker appearance indicates an undesired higher cell density, as seen in some of the colonies cultured in Peptide P, day 7 (Figure 4).
Figure 5 shows the results of the cell viability (Figure 5A) and proliferation assays (Figure 5B), which point towards the amount of dead and live cells and the metabolic activity, respectively. Here, we used a calcein and ethidium homodimer-based viability assay and noticed that these cells showed a small population of dead cells by day 7 when cultured in Matrigel. This phenomenon was also present in all peptide-based hydrogels but not in the 2D culture. Proliferation results showed a total increase in metabolic activity between day 1 and day 7. Moreover, we found slight decreases in activity between day 4 and day 7 for several conditions. This observation correlates to the increase in the dead cell population seen in the viability assay (Figure 5A).
For a preliminary evaluation of the interaction between the cells and the biofunctionalization of the peptide hydrogels, we performed an adhesion assay based on mechanical stress to induce cell detachment. As can be observed in Figure 6, the use of different concentrations of the biofunctional peptide P2 significantly affected the number of cells retained before and after the detachment process. Here, we used a peptide that contains only the biofunctional motif of peptide P2 as a positive control. We also used the non-biofunctional peptide P as a negative control. As can be observed in the graph, cells seeded in peptide P had a relatively low initial adhesion as well as a low retention value. Moreover, the change in the ratio of the biofunctional peptide P2 was correlated to the amount of cells retained after the stress. Hence, using this assay, we can conclude that the P2 peptide affected cell adhesion that may or may not be driven by the biofunctional motif but was correlated with the amount of biofunctional peptide present in the mixture.
Then, we investigated the effect of the matrices in the fabricated organoids. We evaluated the circularity of the colony and the relative size of the lumen with respect to the size of the colony by performing shape analysis of cytoskeleton-stained samples cultured for 4 days. The data points had a particular distribution along the x- and y-axes, and each condition had a centroid with a particular value for circularity and relative lumen area. Figure 7 shows that the cells cultured in 2D had a very high variation in colony circularity. In contrast, the cells cultured in Matrigel had a more extensive distribution for the relative size of the lumen.
Interestingly, the centroid values for all peptide-containing hydrogels had a very similar position. However, the distance between data points showed different distribution profiles. For instance, the P3 peptide had a smaller distribution among the three peptide hydrogels, while the data points from the non-biofunctional peptide P appeared to have a larger distribution. These data suggest that both hydrogels with P1 and P3 have the potential to be optimized for organoid culture.
Finally, we evaluated the expression of specific markers using immunocytochemistry on a sample stained for the apical markers ZO-1 and ezrin. These markers allowed for the evaluation of the polarization of the cells within each organoid. We also evaluated pan cadherin markers to visualize cell-cell junctions and a cytoskeleton-specific marker (phalloidin) to visualize the lumen. Figure 8A shows the organoids found in non-biofunctional peptide P, biofunctionalized peptide P1, and Matrigel on day 4 and day 7. ZO-1 and ezrin markers should be localized within the apical and basolateral membranes of the organoid; proper cell polarization will allow us to observe the expression of both molecules by day 4 near the lumen area, otherwise invisible. Indeed, as seen in Figure 8, ZO-1 and ezrin markers were located in the apical membranes of the organoids cultured in Matrigel. Interestingly, ezrin markers were barely visible in the non-biofunctional peptide P. From this, we infer that the inefficient polarization of the cells resulted in a low expression of these molecules in peptide P.
Moreover, the dispersion of the nuclei tended to be irregular in the peptide hydrogels, whereas the organoids embedded in Matrigel formed a single, asymmetrical layer of nuclei. This description matches the ideal morphology for a colorectal organoid; however, although the orientation of the nuclei was not completely asymmetrical in the peptide hydrogels, denser colonies with multilayers of nuclei were found mainly in the non-biofunctional peptide P. In contrast, cell-cell junctions must be localized perpendicular to the apical and basolateral membranes (Figure 8B). This marker allowed us to observe the edges of all cells located within the colony. When grown in Matrigel, cells had an almost perfectly radial distribution of pan-cadherin (cell-cell junctions) by day 7. In contrast, non-polarized colonies, such as the ones in peptide P, presented a disordered arrangement of cells, and expression of cell-cell junctions near the basal and apical membrane.
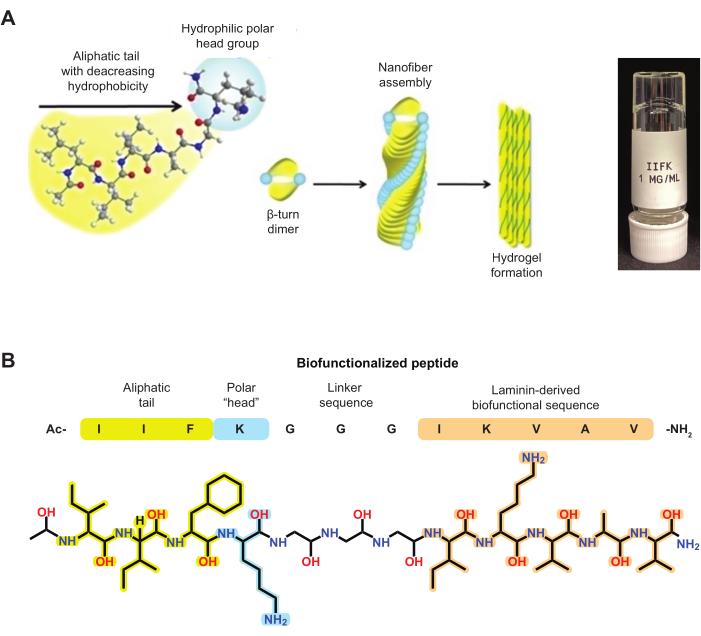
Figure 1: Ultrashort self-assembling peptides forming nanofibers. (A) Depiction of the self-assembling process for aliphatic ultrashort peptides. (B) Chemical structure of ultrashort SAP IIFK biofunctionalized with the IKVAV ligand. This figure was modified from Loo et al.4. Please click here to view a larger version of this figure.
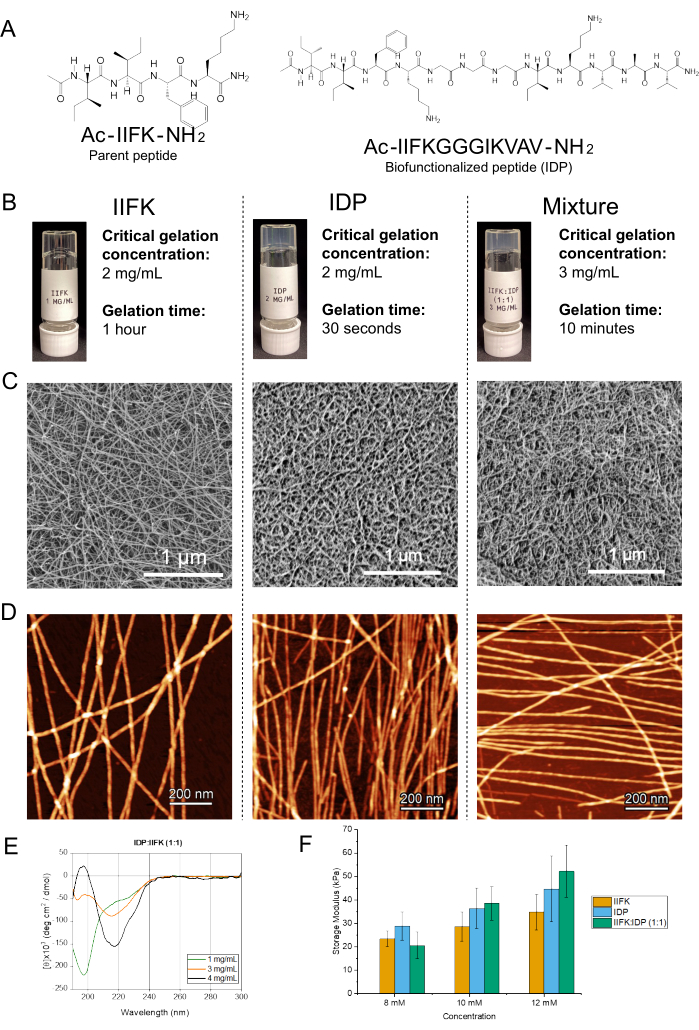
Figure 2: Structure and properties of peptides. (A) The chemical structures of the parent peptide IIFK and the biofunctional peptide P2 are illustrated. (B) The inverted vial test results reveal critical gelation concentration and gelation time differences. P2 forms a translucent gel with a higher CGC than the parent peptide IIFK. Notably, the IIFK:P2 (1:1) mixture exhibits an even higher CGC and longer gelation time than either peptide alone. (C) Scanning electron micrographs provide visual insights into the structural characteristics of IIFK, P2, and the IIFK:P2 (1:1) mixture. (D) The atomic force microscopy height topography images depict the gelated peptides at a concentration of 8 mM, offering a detailed view of their surface features. (E) Circular dichroism spectroscopy reveals the conformational changes of the IIFK:P2 mixture at various concentrations, shedding light on their secondary structure alterations. (F) Rheological analyses measure the stiffness of the gelated peptides at different concentrations, providing valuable insights into their mechanical properties. This figure is reproduced from Perez-Pedroza et al.25. Scale bars = 1 µm (C), 200 nm (D). Abbreviation: CGC = critical gelation concentration. Please click here to view a larger version of this figure.
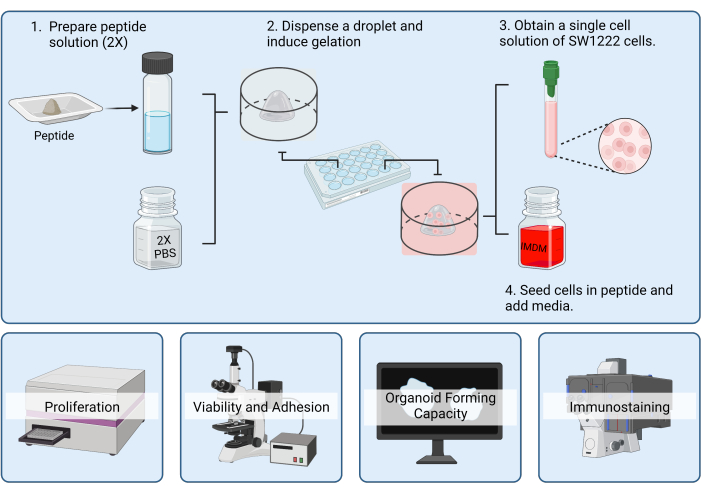
Figure 3: Diagram depicting peptide preparation and cell seeding for organoid fabrication and characterization by viability, adhesion, organoid forming capacity, and immunostaining in peptide hydrogels. Please click here to view a larger version of this figure.
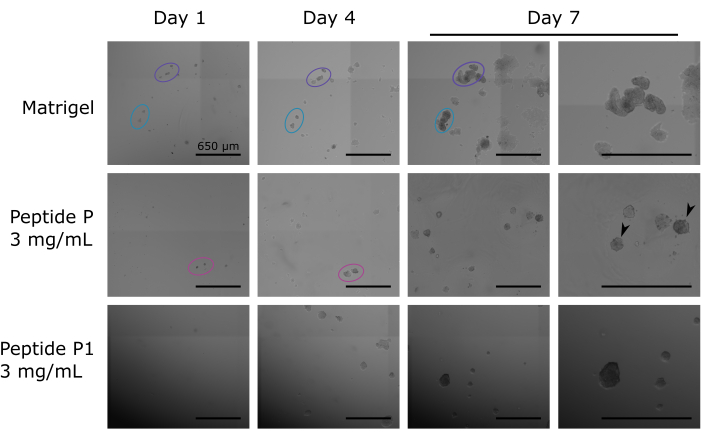
Figure 4: Brightfield microscopy images captured from SW1222 cells cultivated within peptide hydrogels labeled as P and P1 compared with Matrigel control. Interestingly, organoids cultured in the peptide hydrogels manifest a predominantly spherical morphology by the seventh day. In contrast, those situated within the Matrigel seem to be exhibiting a lowered cell density, which can be told by their bright body, compared to the darkened body of the organoids culture in SAP (black arrowheads). Nevertheless, they exhibit an apparent clustering between adjacent colonies. Furthermore, it becomes evident that the migratory capacity of cells within the Matrigel environment is comparatively diminished. Scale bars = 650 µm. Please click here to view a larger version of this figure.
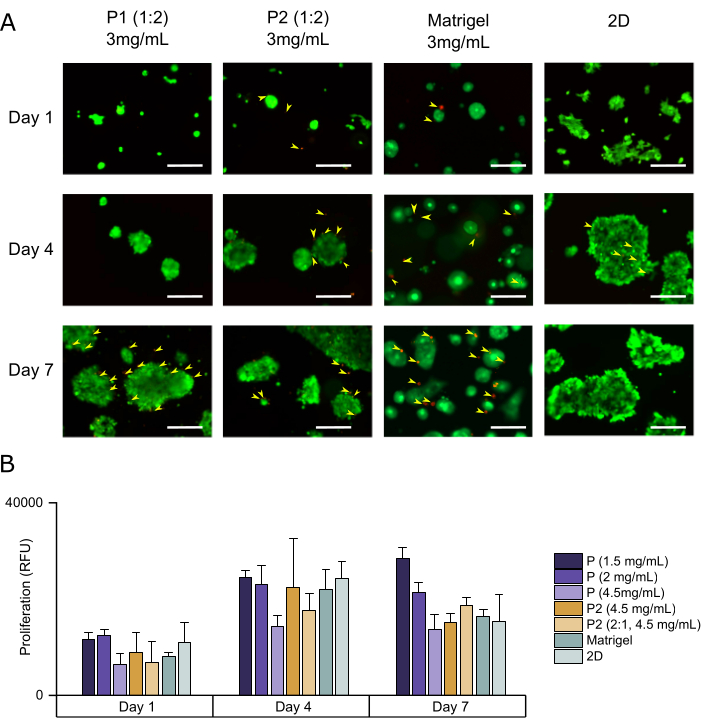
Figure 5: Viability and proliferation assays of SW1222 cells in biofunctionalized SAP at various concentrations. (A) Viability assessment of cells cultured in biofunctional peptide P1 and P2, compared to Matrigel and 2D. (B) Proliferation assessment of cell culture in non-biofunctional peptide P1 and biofunctional peptide P2, compared to Matrigel and 2D controls. Here, we see a behavior similar to that of the cells growing in Matrigel for the live/dead assay. A slight increase in dead cells (yellow arrowheads) is found in several conditions, mainly Matrigel. For proliferation, the trends vary depending on the type and concentration of the peptide, but generally, there is a positive trend. Scale bars = 250 µm. This figure was adapted from Perez-Pedroza et al.25. Please click here to view a larger version of this figure.
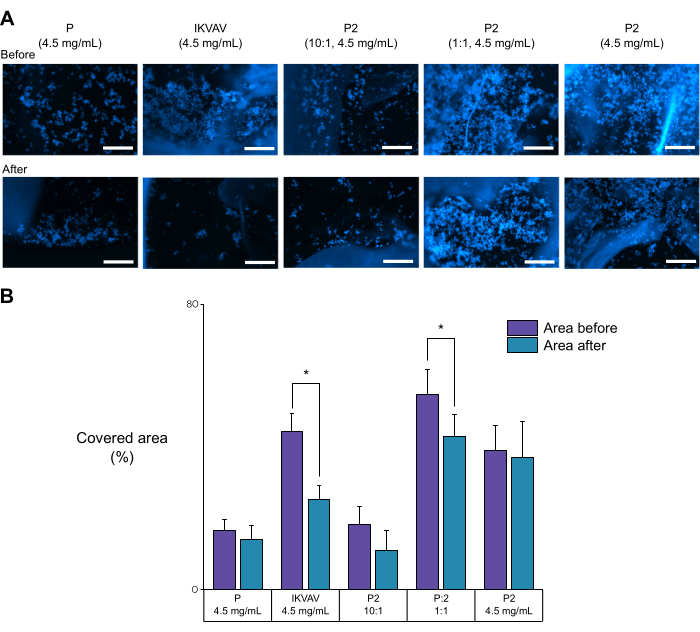
Figure 6: Effects of the peptide on the number of cells detached. (A) Fluorescence images of cells stained with DAPI before and after mechanical stress. Here, we use peptide P, modified peptide P2, and the IKVAV motif. (B) Change in cell adhesion was determined by measuring the covered area in ImageJ. The comparative analysis revealed that the biofunctionalized peptide P2 had a heightened retention capacity compared to the non-biofunctional peptide P. Scale bars = 100 µm. This figure was adapted from Perez-Pedroza, et al.25. Please click here to view a larger version of this figure.
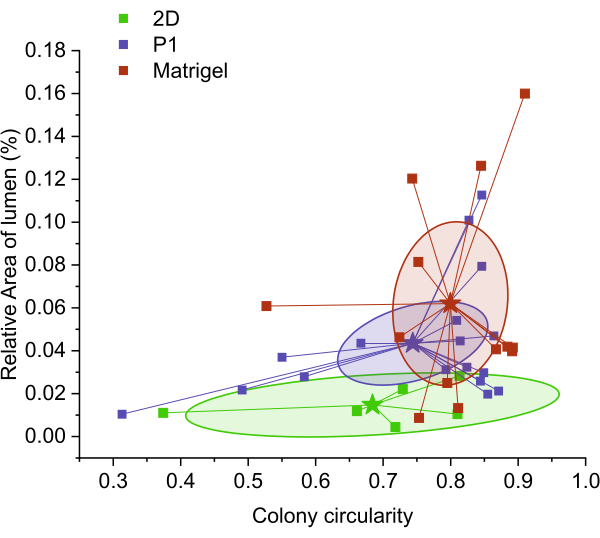
Figure 7: Evaluation of organoid forming capacity by centroid calculation of relative area of the lumen versus circularity. The centroid and dispersion of the data varied among treatments. As expected, the worst-performing condition was that of the 2D measurements, which expanded in an extensive range of circularity values and had the lowest relative area of the lumen. Matrigel had not only the highest circularity but also the biggest lumen. The peptide conditions varied slightly in both values, but the P3 condition held the most negligible variation of the data. Please click here to view a larger version of this figure.
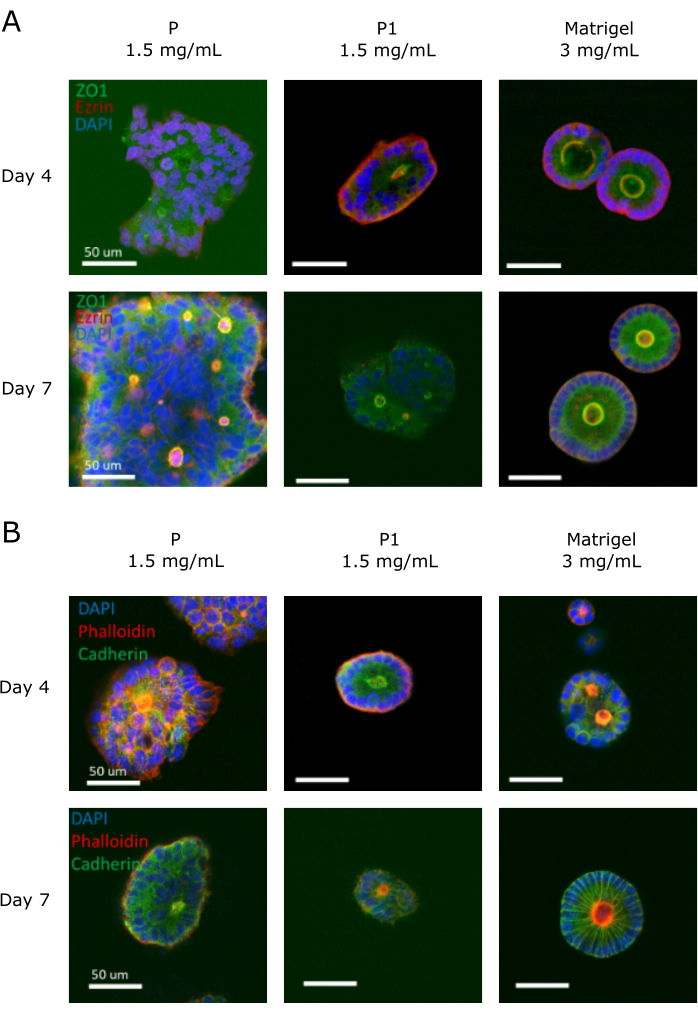
Figure 8: Confocal microscopy images of organoids at day 4 and day 7 for various markers. (A) ZO-1 (green), ezrin (red), and DAPI (blue) markers are used to find the apical distribution of the cells within the organoids. As seen in the Matrigel control, these molecules are typically localized in the apical and basolateral membranes. Non-biofunctional peptide P shows expression of these molecules only by day 7. (B) Cell-cell junction markers stained with pan-cadherin (green) antibodies should be perpendicular to the basolateral and apical membranes of the organoids. Cell nuclei (blue) and cytoskeleton (red) are used as counterstaining. The non-biofunctional peptide P induced cell-cell junction expression in the basolateral membrane, contrary to the other two treatments (P3 and Matrigel). Scale bars = 100 µm. Please click here to view a larger version of this figure.
Discussion
The vast potential of SAPs in biomedical research is underscored by their malleability and adaptability through biofunctionalization. With such an extensive array of permutations, the challenge arises in efficiently testing and determining the most promising configurations for specific applications, especially in organoid research. A fast, cost-effective solution is paramount. The SW1222 cell line, derived from human colorectal adenocarcinoma, emerges as a pivotal candidate for such intensive evaluation. Here, we presented a method for the evaluation and characterization of organoids grown in a peptide-based matrix. This protocol promises an integrative, cost-effective strategy that focuses on monitoring and analysis of organoids using viability and proliferation assessments, adhesion assays, shape analysis, and immunocytochemistry. The selection of varying peptide concentrations in our cell culture experiments is underpinned by a carefully considered rationale that aligns with specific experimental objectives and contributes significantly to advancing our comprehension of organoid culture within the context of peptide matrices. Our research journey has evolved over time, necessitating a diverse approach to explore different formulations. First, the utilization of a range of peptide concentrations, from 1.5 mg/mL to 4.5 mg/mL, was motivated by the desire to investigate a spectrum of potential outcomes. By employing lower concentrations, we aimed to assess the minimal peptide concentration required to support organoid growth and development effectively. This information is vital for understanding the feasibility of cost-effective, minimal-resource applications. Conversely, higher concentrations were chosen to examine the upper limits of peptide concentration and its potential impact on organoid behavior, addressing questions regarding saturation effects and potential drawbacks of excessive peptide concentrations.
Second, specific experimental goals guided our choices. For instance, when investigating cell adhesion, peptide concentrations were employed to diminish the fracture of the hydrogel under stress. Conversely, in studies focusing on long-term viability and tissue maturation, lower concentrations were utilized to achieve the lowest rheological values possible while still providing a gel. Moreover, proliferation values were chosen to show the various profiles obtained through time on the organoid culture when employing similar formulations. The significance of this diverse approach lies in its capacity to yield a comprehensive dataset that encompasses a broad spectrum of conditions and outcomes. By examining the effects of different peptide concentrations on organoid culture, we can better understand the nuanced interplay between matrix properties and organoid behavior.
The initial evaluation involves the monitoring of the progression of SW1222 cells over a week in a 24-well plate using brightfield imaging. The general analysis of the morphology and the cell density is a routine technique for organoid lines9 that is being applied to the cell line. The non-invasive nature of this protocol allows for uninterrupted observation of the cell culture, enabling us to capture crucial data on morphological changes and cell density for each particular colony. This initial analysis can provide valuable insights into the suitability of the ultrashort self-assembling peptide matrix. These observations lay the foundation for further in-depth analyses, demonstrating the suitability of the peptide matrix as an effective scaffold for supporting the development and growth of colorectal cancer organoids. Moreover, interesting observations may arise from monitoring the growth and changes in each colony. For instance, in Figure 4, we can observe that colonies growing in Matrigel may collide with each other, but it appears that a boundary is consistently maintained. However, cells cultured in Peptides reorganize the peptide in such a way that by day 7, the cell locations are not precisely the same as they were before. This is something to take into account if this protocol was exported to an automated system that would tag and follow up on the growth of the organoids.
Subsequent scrutiny involves assessing cell viability and proliferation using a viability assay based on calcein and ethidium homodimer. The viability assay will provide qualitative information on the population of living cells compared to the dead cells. The proliferation assay will provide quantitative information regarding the metabolic state of the cells to assess cell division31. These are standard assays widely used in cell culture and provide a general overview of the cells' proliferative status. While SAP matrices have been extensively demonstrated to be biocompatible, organoids are known to exhibit reduced viability due to suboptimal culture conditions, including the matrix in which they are embedded. Furthermore, the mechanical properties of the peptide matrices can influence cell viability. If the matrix is too stiff or rigid, it can subject the cells to mechanical stress, potentially leading to cell damage. Conversely, if the matrix is too soft or unstable, it may not provide sufficient support for cell adhesion and survival. Additionally, the bioactive motifs or functional groups present in the peptides can interact with specific cell receptors or signaling pathways, potentially resulting in cell death. Therefore, it is crucial to monitor cell viability and proliferation continuously.
In earlier studies25, Peptide 1 and Peptide 2 showed similar levels of cell death compared to Matrigel (Figure 5). However, fluctuations were observed in the proliferation profiles of cells in the parent peptide with varying concentrations. Notably, the rate of proliferation from day 4 to day 7 remained constant rather than increasing, suggesting that the cells may be entering a less active stage. Furthermore, the changes in cell viability observed between day 4 and day 7 also remained relatively consistent, hinting at a potential plateau in growth and reproduction.
To proceed with the evaluation and characterization, we employed an adhesion assay adapted for 3D culture, which relies on mechanical stress to assess cell detachment effects32. The method to induce mechanical stress should be chosen based on the mechanical properties of the hydrogel. The method's effectiveness is inherently tied to the chosen model system, and the insights obtained are contingent on how well it represents the biological context. Variability in adhesion and organoid characteristics can result from numerous physical and chemical interactions, which may be influenced by factors such as the hydrogel composition and the adhesive properties of the nanofibers. These factors have the potential to affect the generalizability of our findings33. However, if the reader were to adapt this protocol for stiffer or highly ductile hydrogels, consider using a more robust stressing method.
We utilized this technique to assess the overall adhesiveness provided to the SAP by the biofunctional motif (Figure 6). We observed two phenomena. First, the peptides exhibited varying initial adhesive properties. Specifically, the IKVAV motif and hydrogels containing a high concentration of the biofunctionalized peptide were found to adhere cells across more than 40% of their surface area. Second, after the application of induced stress, some peptides experienced a significant loss of adhered cells. Notably, the retention capacity was relatively high in peptides that contained high concentrations of the non-biofunctionalized peptide. Thus, the IKVAV ligand and the SAP offer different types of adherence of the cells to the matrix. This method allows for the finding of patterns and elucidation of the adherence mechanism for the material. Further techniques could be used, such as adhesiveness measurement using atomic force microscopy, which can be used to probe the adhesive forces between a probe tip and a sample surface at the nanoscale level, providing valuable information about the adhesion properties of materials and biological samples.
The capacity of various peptide materials to induce organoid formation was evaluated by means of colony circularity and the relative lumen area of the organoids grown in such peptide hydrogels. Circularity refers to the roundness or shape of an object. In the context of organoids, circularity can be used to assess the morphology and symmetry of the structure. Several studies have mentioned the circular morphologies of organoids. For example, breast cancer organoids tend to have strict circular morphologies with cord-like basolateral peripheries34. This suggests that circularity can be used as a parameter to evaluate the shape of organoids. Lumen size refers to the central cavity or space within an organoid. It is an essential feature as it reflects the differentiation and maturation of the organoid. During the differentiation process, organoids undergo a decrease in lumen size and an increase in the thickness of the epithelial layer35. This indicates that lumen size can be used as an indicator of the developmental stage of the organoid. Assessing circularity and lumen size can provide valuable information about the structural and functional characteristics of organoids. Circular organoids with well-defined lumens are more representative of the native tissue architecture and can better mimic organ function36.
These properties can be evaluated by obtaining fluorescence images of the cytoskeleton and the nuclei from the previously stained samples. The images were segmented using Cellpose, and shape measurements of the resultant masks were obtained using ImageJ. Shape analysis of cytoskeleton-stained samples after 4 days of culture further revealed organoid characteristics. Similar methods, primarily focused on brightfield imaging, have been employed to determine the responses of the organoids to their environment based on shape analysis and determination of characteristics of the organoid by image analysis36,37,38. Finally, we used immunocytochemistry to evaluate cell polarity with various polarity markers. Notably, the use of a peptide hydrogel will reduce the handling of the sample, as this does not require the removal of the matrix and retrieval of the organoids39.
The importance of this method relies on the versatility of biofunctional SAP hydrogels compared to other animal-derived matrices to develop methods that allow the translation of peptide hydrogels into matrices for organoid culture. Comparing the ultrashort self-assembling peptide matrix with traditional matrices such as Matrigel or collagen reveals numerous advantages. First, the peptide matrix offers superior reproducibility as the peptide sequence can be precisely controlled and synthesized, ensuring consistent properties and performance across different batches40. Additionally, it exhibits high stability in terms of mechanical properties and resistance to degradation, maintaining structural integrity over extended periods and providing a stable microenvironment for organoid growth41. Furthermore, the ultrashort peptide matrix enables precise control and fine-tuning of the microenvironment. The peptide sequence can be modified to incorporate specific bioactive motifs or functional groups, presenting desired cues for cell differentiation42.
In summary, the method's significance lies in its potential to inform the rational design of peptide-based hydrogels for fostering organoid culture. Through systematically evaluating cell behaviors, this protocol facilitates the identification of promising peptide compositions for optimizing organoid growth. Such insights are poised to extend to broader research areas, contributing to the advancement of organoid-based studies and regenerative medicine approaches.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was financially supported by King Abdullah University of Science and Technology. The authors acknowledge KAUST's Seed Fund Grant and KAUST's Innovation Fund awarded by KAUST's Innovation and Economic Development. The authors would like to acknowledge KAUST's Bioscience and Imaging Core Labs for supporting the biological characterization and microscopy analyses.
Materials
| 1x PBS | Gibco | 14190144 | |
| 6-well plate, tissue culture treated | Corning | 07-200-83 | |
| 10x PBS, no calcium, no magnesium | Gibco | 70011044 | |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Scientific | 28906 | |
| 24-well plate, tissue culture treated | Corning | 09-761-146 | |
| 96-well black plate, tissue culture treated | Corning | 07-200-565 | |
| alamarBlue Cell Viability Reagent | Invitrogen | DAL1025 | |
| Anti-Ezrin antibody, rabbit monoclonal | Abcam | ab40839 | Secondary used: anti-rabbit Dylight 633 |
| Anti-pan Cadherin antibody, rabbit polyclonal | Abcam | ab16505 | Secondary used: anti-rabbit Alexa 488 |
| Anti-ZO1 tight junction antibody, goat polyclonal | Abcam | ab190085 | Secondary used: anti-goat Alexa 488 |
| BSA | Sigma-Aldrich | A9418 | |
| Cellpose 2.0 | NA | NA | Obtained from https://github.com/MouseLand/cellpose |
| Confocal Laser Scanning Microscope with Airyscan | ZEISS | LSM 880 | |
| DAPI | Invitrogen | D1306 | |
| Donkey anti-Goat IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Invitrogen | A-11055 | |
| Glycine | Cytiva | GE17-1323-01 | |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 | Invitrogen | A-11008 | |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, DyLight 633 | Invitrogen | 35562 | |
| Heat Inactivated Fetal Bovine Serum (HI FBS) | Gibco | 16140071 | |
| ImageJ 1.54f | NIH | NA | |
| IMDM | Gibco | 12440079 | |
| LIVE/DEAD Viability/Cytotoxicity Kit, for mammalian cells | Invitrogen | L3224 | |
| Magnesium Chloride, hexahydrate (MgCl2 6 H2O) | Sigma-Aldrich | M2393 | |
| Matrigel for Organoid Culture, phenol-red free | Corning | 356255 | Refered in the manuscript as Matrigel or basement membrane matrix. |
| Microscope, brightfield | |||
| Microscope, EVOS | Thermo Scientific | EVOS M7000 | |
| OriginPro 2023 (64-bit) 10.0.0.154 | OriginLab Corp | NA | |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 | |
| Peptide P (Ac,Ile,Ile,Phe,Lys,NH2) | Lab-made | NA | Can be custom-made by peptide manufacturers such as Bachem. |
| Peptide P1 (Ac, Ile, Ile, Phe, Lys, Gly, Gly, Gly, Arg, Gly, Asp, Ser, NH2) | Lab-made | NA | Can be custom-made by peptide manufacturers such as Bachem. |
| Peptide P2 (Ac, Ile,Ile,Phe,Lys,Gly,Gly,Gly,Ile,Lys,Val,Ala,Val,NH2) | Lab-made | NA | Can be custom-made by peptide manufacturers such as Bachem. |
| Rhodamine Phalloidin | Invitrogen | R415 | |
| Round Cover Slip, 10 mm diameter | VWR | 631-0170 | |
| Scanning Electron Microscope | Thermo Fisher – FEI | TENEO VS | |
| Sterile 30 μm strainer | Sysmex | 04-004-2326 | |
| sucrose | Sigma-Aldrich | S1888 | |
| SW1222 cell line | ECACC | 12022910 | |
| Triton x-100 | Thermo Scientific | 85111 | |
| Trypsin-EDTA 0.25% | Gibco | 25200056 | |
| Tween 20 | Sigma-Aldrich | P1379 | |
| UltraPure water | Invitrogen | 10977015 |
References
- Susapto, H. H., et al. Ultrashort peptide bioinks support automated printing of large-scale constructs assuring long-term survival of printed tissue constructs. Nano Letters. 21 (7), 2719-2729 (2021).
- Loo, Y., et al. A chemically well-defined, self-assembling 3D substrate for long-term culture of human pluripotent stem cells. ACS Applied Bio Materials. 2 (4), 1406-1412 (2019).
- Loo, Y., et al. Self-assembled proteins and peptides as scaffolds for tissue regeneration. Advanced Healthcare Materials. 4 (16), 2557-2586 (2015).
- Loo, Y., et al. Peptide bioink: self-assembling nanofibrous scaffolds for three-dimensional organotypic cultures. Nano Letters. 15 (10), 6919-6925 (2015).
- Ho, B. X., Pek, N. M. Q., Soh, B. -. S. Disease modeling using 3D organoids derived from human induced pluripotent stem cells. International Journal of Molecular Sciences. 19 (4), 936 (2018).
- Li, Y., Tang, P., Cai, S., Peng, J., Hua, G. Organoid based personalized medicine: from bench to bedside. Cell Regeneration. 9 (1), 21 (2020).
- Kim, S., et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nature Communications. 13 (1), 1692 (2022).
- Kaushik, G., Ponnusamy, M. P., Batra, S. K. Concise review: current status of three-dimensional organoids as preclinical models. Stem Cells. 36 (9), 1329-1340 (2018).
- Gjorevski, N., et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 539 (7630), 560-564 (2016).
- Hauser, C. A., et al. Natural tri- to hexapeptides self-assemble in water to amyloid beta-type fiber aggregates by unexpected alpha-helical intermediate structures. Proceedings of the National Academy of Sciences of the United States of America. 108 (4), 1361-1366 (2011).
- Lakshmanan, A., Zhang, S., Hauser, C. A. E. Short self-assembling peptides as building blocks for modern nanodevices. Trends in Biotechnology. 30 (3), 155-165 (2012).
- Rauf, S., et al. Self-assembling tetrameric peptides allow in situ 3D bioprinting under physiological conditions. Journal of Materials Chemistry B. 9 (4), 1069-1081 (2021).
- Bilalis, P., et al. Dipeptide-based photoreactive instant glue for environmental and biomedical applications. ACS Applied Materials & Interfaces. 15 (40), 46710-46720 (2023).
- Abdelrahman, S., et al. The impact of mechanical cues on the metabolomic and transcriptomic profiles of human dermal fibroblasts cultured in ultrashort self-assembling peptide 3D scaffolds. ACS Nano. 17 (15), 14508-14531 (2023).
- Moretti, M., et al. Selectively positioned catechol moiety supports ultrashort self-assembling peptide hydrogel adhesion for coral restoration. Langmuir. 39 (49), 17903-17920 (2023).
- Alhattab, D. M., et al. Fabrication of a three-dimensional bone marrow niche-like acute myeloid Leukemia disease model by an automated and controlled process using a robotic multicellular bioprinting system. Biomaterials Research. 27 (1), 111 (2023).
- Abdelrahman, S., et al. The impact of mechanical cues on the metabolomic and transcriptomic profiles of human dermal fibroblasts cultured in ultrashort self-assembling peptide 3D scaffolds. ACS Nano. 17 (15), 14508-14531 (2023).
- Habibi, N., Kamaly, N., Memić, A., Shafiee, H. Self-assembled peptide-based nanostructures: smart nanomaterials toward targeted drug delivery. Nano Today. 11 (1), 41-60 (2016).
- Yan, Y., et al. Enhanced osteogenesis of bone marrow-derived mesenchymal stem cells by a functionalized silk fibroin hydrogel for bone defect repair. Advanced Healthcare Materials. 8 (3), 1801043 (2018).
- Kisiday, J., et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proceedings of the National Academy of Sciences of the United States of America. 99 (15), 9996-10001 (2002).
- Hogrebe, N. J., et al. Independent control of matrix adhesiveness and stiffness within a 3D self-assembling peptide hydrogel. Acta Biomaterialia. 70, 110-119 (2018).
- Wu, F. J., Lin, H. C. Synthesis, self-assembly, and cell responses of aromatic IKVAV peptide amphiphiles. Molecules. 27 (13), 4115 (2022).
- Cringoli, M. C., et al. Bioadhesive supramolecular hydrogel from unprotected, short d,l-peptides with Phe-Phe and Leu-Asp-Val motifs. Chemical Communications. 56 (20), 3015-3018 (2020).
- Pulat, G. O., Gökmen, O., Çevik, Z. B. Y., Karaman, O. Role of functionalized self-assembled peptide hydrogels in in vitro vasculogenesis. Soft Matter. 17 (27), 6616-6626 (2021).
- Pérez-Pedroza, R., et al. Fabrication of lumen-forming colorectal cancer organoids using a newly designed laminin-derived bioink. International Journal of Bioprinting. 9 (1), 633 (2022).
- Ashley, N., Yeung, T. M., Bodmer, W. F. Stem cell differentiation and lumen formation in colorectal cancer cell lines and primary tumors. Cancer Research. 73 (18), 5798-5809 (2013).
- Yeung, T. M., Gandhi, S. C., Wilding, J. L., Muschel, R., Bodmer, W. F. Cancer stem cells from colorectal cancer-derived cell lines. Proceedings of the National Academy of Sciences of the United States of America. 107 (8), 3722-3727 (2010).
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262-265 (2009).
- Pachitariu, M., Stringer, C. Cellpose 2.0: how to train your own model. Nature Methods. 19 (12), 1634-1641 (2022).
- Stringer, C., Wang, T., Michaelos, M., Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nature Methods. 18 (1), 100-106 (2021).
- Adan, A., Kiraz, Y., Baran, Y. Cell proliferation and cytotoxicity assays. Current Pharmaceutical Biotechnology. 17 (14), 1213-1221 (2016).
- Humphries, M. J. Cell-substrate adhesion assays. Current Protocols in Cell Biology. , (2001).
- Munevar, S., Wang, Y. -. l., Dembo, M. Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Molecular Biology of the Cell. 12 (12), 3947-3954 (2001).
- Djomehri, S., Burman, B., González, M. E., Takayama, S., Kleer, C. G. A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer. Journal of Cell Communication and Signaling. 13 (1), 129-143 (2018).
- Puschhof, J., et al. Intestinal organoid cocultures with microbes. Nature Protocols. 16 (10), 4633-4649 (2021).
- Matthews, J. M., et al. OrganoID: A versatile deep learning platform for tracking and analysis of single-organoid dynamics. PLOS Computational Biology. 18 (11), 1010584 (2022).
- Herpers, B., et al. Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR × LGR5 bispecific antibody with efficacy in epithelial tumors. Nature Cancer. 3 (4), 418-436 (2022).
- Borten, M. A., Bajikar, S. S., Sasaki, N., Clevers, H., Janes, K. A. Automated brightfield morphometry of 3D organoid populations by OrganoSeg. Scientific Reports. 8 (1), 5319 (2018).
- Bergdorf, K. N., et al. Immunofluorescent staining of cancer spheroids and fine-needle aspiration-derived organoids. STAR Protocols. 2 (2), 100578-100578 (2021).
- Yang, Z., Xu, H., Zhao, X. Designer self-assembling peptide hydrogels to engineer 3D cell microenvironments for cell constructs formation and precise oncology remodeling in ovarian cancer. Advanced Science. 7 (9), 1903718 (2020).
- Yadav, N., et al. Ultrashort peptide-based hydrogel for the healing of critical bone defects in rabbits. ACS Applied Materials & Interfaces. 14 (48), 54111-54126 (2022).
- Alshehri, S., Susapto, H. H., Hauser, C. A. E. Scaffolds from self-assembling tetrapeptides support 3D spreading, osteogenic differentiation, and angiogenesis of mesenchymal stem cells. Biomacromolecules. 22 (5), 2094-2106 (2021).

