Real-time Measurement of Epithelial Barrier Permeability in Human Intestinal Organoids
Summary
This protocol describes the measurement of epithelial barrier permeability in real-time following pharmacologic treatment in human intestinal organoids using fluorescent microscopy and live cell microscopy.
Abstract
Advances in 3D culture of intestinal tissues obtained through biopsy or generated from pluripotent stem cells via directed differentiation, have resulted in sophisticated in vitro models of the intestinal mucosa. Leveraging these emerging model systems will require adaptation of tools and techniques developed for 2D culture systems and animals. Here, we describe a technique for measuring epithelial barrier permeability in human intestinal organoids in real-time. This is accomplished by microinjection of fluorescently-labeled dextran and imaging on an inverted microscope fitted with epifluorescent filters. Real-time measurement of the barrier permeability in intestinal organoids facilitates the generation of high-resolution temporal data in human intestinal epithelial tissue, although this technique can also be applied to fixed timepoint imaging approaches. This protocol is readily adaptable for the measurement of epithelial barrier permeability following exposure to pharmacologic agents, bacterial products or toxins, or live microorganisms. With minor modifications, this protocol can also serve as a general primer on microinjection of intestinal organoids and users may choose to supplement this protocol with additional or alternative downstream applications following microinjection.
Introduction
The intestinal epithelium forms a selective barrier that mediates the directional transport of nutrients, H2O, ions, and waste products while minimizing nonspecific diffusion-mediated exchange of other particles between the lumen and the mesenchymal tissue or blood supply1,2. The nonspecific permeability of the intestinal epithelial barrier has long been considered a key functional parameter in both health and disease3,4,5,6, that reflects the rate of diffusion of small molecules across the epithelium via the paracellular space. Measurement of epithelial barrier permeability can be conducted in animal models7 and in human patients8 through the ingestion of lactulose, which has no specific transporter in the gastrointestinal tract, and the subsequent collection and measurement of lactulose concentrations in peripheral blood. Alternate ingested markers of barrier function such as fluorescently labeled carbohydrates are also available9,10. This approach has been adapted for intestinal epithelial cell cultures grown on Transwell supports11, a simplified approach that allows for greater experimental control but has also been criticized as a poor predictor of in vivo permeability due to the absence of differentiated epithelial subtypes and tissue structure12. Using chambers represent yet another approach and allow for the measurement of epithelial barrier function in whole intestinal mucosa ex vivo13. Application of this technique is frequently limited by tissue availability and condition13,14. Thus new methods which balance reproducibility and throughput with physiologic relevance are necessary.
Recent developments in in vitro organogenesis have led to the adoption of 3D tissue culture model systems as a sophisticated platform for recapitulating the dynamics of complex tissues15,16,17,18,19,20,21,22,23. In particular, human pluripotent stem cell (hPSC) derived human intestinal organoids (HIOs)19,24 have emerged as a reproducible and experimentally tractable model system for the study of host-microbial interactions and epithelial barrier dynamics25,26,27,28. Similarly, human tissue-derived organoids (also known as enteroids) can be derived from a simple biopsy procedure and can be used as a tractable system to study human physiology and disease15,29,30. Microinjection of human intestinal organoids allows for the delivery of experimental compounds25 or live microbes25,31,32,33 to the apical epithelial surface of the organoid lumen. Leslie and Huang et al.25 recently adapted this technique to measure barrier permeability in HIOs microinjected with fluorescein isothiocyanate (FITC) labeled dextran following exposure to bacterial toxins.
This protocol is intended as a guide for the measurement of epithelial barrier permeability in hPSC-derived HIOs and tissue-derived HIOs using fluorescent microscopy. With minor modifications, it can also serve as a general primer on microinjection of HIOs with experimental compounds. Users may supplement this protocol with additional or alternative downstream applications after microinjection.
Protocol
Normal, de-identified human adult intestinal tissue was obtained from deceased organ donors through the Gift of Life, Michigan. Human ES cell line H9 (NIH registry #0062) was obtained from the WiCell Research Institute. All human tissue used in this work was obtained from non-living donors, was de-identified and was conducted with approval from the University of Michigan IRB (protocol # HUM00093465 and HUM00105750).
1. Microinjector Setup
- Obtain the materials listed in Materials Table.
- Place the micromanipulator, dissecting scope, and light source in the biosafety cabinet. Clean the microinjection setup with 70% ethanol or other disinfectant prior to experimental use. Avoid prolonged exposure to disinfectants containing bleach, which may corrode the microinjection equipment.
NOTE: An example of a complete microinjector assembly is shown in Figure 1. - Position the dissecting scope so that the eye piece can be used through the access port in the shield for the biosafety cabinet.
- Alternately, use a clean bench with positive air flow system in place of a biosafety cabinet with microscope access points.
- Assemble the micromanipulator on the right side of the microscope according to the manufacturer's instructions and adjust so that the controls can be easily reached and adjusted. The micromanipulator should be mounted on an iron plate or otherwise stabilized.
NOTE: Left-handed users may want to place the micromanipulator to the left of the dissecting scope. - Mount the micropipette holder to the micromanipulator arm and connect the analog tubing by inserting it into the micropipette holder.
- Fill the 10 mL glass syringe with approximately 5 – 7 mL of sterile mineral oil.
- Connect the 10 mL glass syringe filled with mineral oil to the open end of the analog tubing using the Luer lock mechanism. Place the glass syringe to the left of the dissecting scope, opposite from the micromanipulator.
- Gently depress the syringe, pushing mineral oil through the tubing. Flush approximately 10 drops of mineral oil from the tip of the micropipette holder and collect in a Petri dish or similar vessel and discard.
NOTE: This step removes all air from the tubing and should be performed before each microinjection session.
2. Preparation for microinjection
- 24 h prior to microinjection:
- Prepare FITC dextran solution by re-suspending FITC dextran at a concentration of 2 mg/mL in sterile PBS or saline. Prepare a total volume of >250 µL.
NOTE: Higher or lower concentrations of FITC-dextran may be used, ranging from approximately 0.1 – 10 mg/mL. Adjust the concentration of FITC-dextran to suit downstream the imaging application. - Setup organoid cultures on 4- or 8-well glass chamber slides or other culture vessel suitable for live microscopy, with up to 4 – 6 HIOs per well, embedded in 50 µL cell matrix solution and cultured in EGF, Noggin and R-Spondin (ENR) growth factor media 19,24. Take care to space the organoids evenly (approximately 5 mm apart) to avoid capturing multiple HIOs in a single microscopic field during real-time imaging analysis. For a complete guide to HIO culture and maintenance see McCraken et al.24.
NOTE: This protocol was developed using stem-cell derived HIOs and the representative results (see below) demonstrate the use of this tissue culture model. However, the same protocol is easily adapted to tissue-derived intestinal epithelial organoids15,29. Tissue-derived HIOs are typically smaller than PSC-derived HIOs and lack the supporting mesenchymal basolateral cell structure15,29,30. Microinjection of tissue-derived HIOs may require a greater degree of technical ability and experience. The degree to which epithelial barrier permeability data obtained using tissue-derived HIOs may correlate with hPSC-derived HIOs is unknown. - Incubate HIOs in a standard cell culture incubator at 42 °C and 5% CO2 prior to microinjection.
- Prepare FITC dextran solution by re-suspending FITC dextran at a concentration of 2 mg/mL in sterile PBS or saline. Prepare a total volume of >250 µL.
- 30 minutes prior to microinjection, turn on the biosafety cabinet and raise the glass shield to the optimal working height.
- Remove all unnecessary items from the biosafety cabinet. Clutter increases the risk of spills or other accidents when working in confined spaces.
- Spray and thoroughly clean the work surface with 70% ethanol or other disinfectant. Wipe clean using a paper towel.
- Check the level of mineral oil in the glass syringe attached to the microinjector. If there is less than 3 mL of mineral oil remaining, unscrew the syringe and refill inside the biosafety cabinet, being careful to avoid introducing bubbles. Do not fill more than 7 mL.
- Turn on the lamp to illuminate the dissecting scope. Adjust the eyepiece for personal comfort.
- Position the micromanipulator to the right of the dissecting scope. Secure the micromanipulator to an iron plate using a magnetic stand. Switch the magnetic stand to the OFF position to adjust the position of the micromanipulator and secure the stand to the iron plate by setting the magnetic stand to the ON position.
- Microcapillary installation
- Retrieve a single 1 mm glass filament from the storage container provided by the manufacturer.
- Insert the glass filament through the center of the copper heating coil of the micropipette puller.
NOTE: See Figure 2 for a guide to preparing the micropipette puller. - Position the filament so that the copper heating coil is approximately in the middle of the glass filament. This will ensure that the puller generates two usable microinjection needles from each glass filament.
- Secure the glass filament using the clamps by tightening the top clamp first, making sure that the glass filament is secured within the notched grove to prevent breakage.
- Extend the puller arm to its maximum vertical position before tightening the bottom clamp.
NOTE: This step is essential. Failure to fully extend the puller arm will result in irregular separation of the two sections of the glass filament. - Check the settings on the heating and pulling mechanism. Select Heat #1 with the toggle (990) and set PULL at 059 (Figure 2).
- Turn on the instrument using the On/Off toggle.
- Close the protective plexiglass shield and press the PULL button on the bottom right face of the puller. The copper coil will begin to heat and will glow bright orange. As the temperature rises, the glass filament will begin to stretch and eventually separate. Upon separation of the two ends of the glass filament, the instrument will power down.
NOTE: The copper coil will remain extremely hot for several minutes. The pulled glass filament will be referred to as a 'microcapillary' from this point forward in the protocol. - Being careful to avoid touching the copper coil, remove a one of the glass microcapillaries. The microcapillary should have a very fine point. Handle the microcapillary carefully with latex or nitrile gloves and immediately proceed to the biosafety cabinet containing the microinjector setup.
NOTE: The glass microcapillary is extremely sharp and very fragile. Handle with care. - Loosen the end cap of the micromanipulator arm. Insert the blunt end of the pulled microcapillary into the open end of the micropipette holder and push it inward until it stops.
- Secure the blunt end of the microcapillary to the micromanipulator arm by re-tightening the end cap.
NOTE: When installing the microcapillary on the microinjection system take care to avoid touching the sharpened end. This will reduce the chance of contamination and preserve the fine point for microinjection. Failure to correctly secure the microcapillary can result in instability or breakage of the glass filament during microinjection. - Open the tip of the glass microcapillary. During preparation of the pulled glass microcapillary, the tip of the point will likely melt in such a way as to seal the pointed end of the microcapillary. To remove this blockage:
- Position the micromanipulator such that the microcapillary is pointed down at the glass stage of the dissecting scope without touching this surface.
- Find a sterile plastic surface (the underside of a culture plate lid works perfectly) and place it on the microscope stage directly underneath the microcapillary needle on the stage of the dissecting scope.
- Center the tip of the microcapillary under the viewing area and ensure that it is visible when looking through the eyepiece of the microscope under 1.5X magnification.
- Using the micromanipulator controls, slowly advance the micromanipulator arm and microcapillary towards the sterile plastic culture lid until the tip barely contacts the plastic surface and the tip of the microcapillary breaks free.
NOTE: Aim for the smallest possible break that allows for fluid flow from the glass filament in order to minimize the damage to the HIO during microinjection. - Back the microcapillary away from the sterile surface to minimize the chance of accidental breakage before proceeding.
- Check the microcapillary for flow by depressing the glass syringe, pushing mineral oil through the tubing. If the end of the microcapillary has been opened you will see a small droplet of mineral oil emerge from the tip of the microcapillary after a few seconds. If this does not occur, repeat the previous step and re-test.
3. Sterile Microinjection
NOTE: Once the microcapillary has been prepared, installed, and tested, begin microinjecting HIOs. Figure 3 illustrates a tissue-derived HIO that has been successfully injected with FITC-dextran.
- Fill the microcapillary with the injection material. Submerge the tip of the glass microcapillary in the FITC dextran injection suspension, being careful to avoid breaking the tip of the microcapillary against the sides or bottom of the tube. Once the tip of the microcapillary is submerged in the solution, pull back on the mineral oil syringe to draw approximately 10 µL of the FITC dextran suspension into the microcapillary.
NOTE: It is recommended to use 1.5 mL or 0.5 mL tubes for the injection solutions/cultures since these will be the most easily accessible to the installed microcapillary. Alternatively, injection solutions can be drawn from a sterile Petri dish or sterile parafilm, which may reduce the chance of sharps injury by limiting the need to hold a tube in close proximity to the microcapillary. If the suspension is highly viscous or if the opening of the microcapillary is exceptionally small, it may take several second to fill the microcapillary. Do not draw microinjection suspensions into the plastic microinjection tubing as this may contaminate the entire microinjection system. - Stop filling the microcapillary when the microinjection suspension fills 90% of the length of the glass microcapillary. Depress the syringe slightly before withdrawing the microcapillary from the microinjection solution to ensure that the tip of the microcapillary does not contain pockets of air.
NOTE: If the examination of the microcapillary reveals pockets of air, empty and re-fill. - Remove the HIO culture plate(s) from the cell culture incubator and transfer to the biosafety cabinet with the microinjector.
- Remove the lid from the HIO culture plate within the biosafety cabinet and center the first well on the microscope stage so that it is clearly visible through the eyepiece of the scope at the lowest magnification setting.
- Turn the micromanipulator arm such that the microcapillary is pointed down into the HIO culture well at an angle >45° relative to the microscope stage. This is most easily accomplished by manually turning the micromanipulator arm assembly on its horizontal axis at the point where the horizontal micromanipulator arm support meets the vertical stand, since the fine controls (black dials) have a limited range of motion. Position the tip of the microcapillary above the first culture well at approximately 1 cm above the surface of the media (Figure 3A). A representative image demonstrating microinjection of tissue-derived intestinal epithelial organoids is shown in Figure 3B.
- Check that both the tip of the glass filament and the HIO(s) are visible through the eyepiece. Re-position if necessary.
- Advance the microcapillary slowly by turning the Z-axis control knob clockwise.
NOTE: Judging the position of the microcapillary tip, particularly the depth, requires practice. As the tip breaches the surface of the media, notice a slight visual distortion of the microcapillary tip. Proceed with care from this point to avoid breaking the microcapillary against the bottom of the culture plate or damaging the HIOs. - Pierce the HIO with the microcapillary tip. The outer surface of the HIO will depress slightly as the microcapillary begins to apply pressure and will pop back into shape as the tip penetrates the lumen. It may be difficult to identify the tip of the microcapillary within the HIO lumen. Adjust the lighting or magnification settings of the dissecting scope if there is difficulty seeing the microcapillary.
- Remove hands from the micromanipulator controls when the microcapillary is correctly positioned with the tip of the microcapillary near the center of the HIO.
- Depress the mineral oil syringe slightly to push the microinjection solution out of the microcapillary and into the HIO lumen. The HIO may expand slightly to accommodate the volume of the injection.
NOTE: Take care to avoid over-filling the HIO as this will cause the organoid to burst. As a rule of thumb, any visible expansion of the HIO volume means it is time to stop. The mean volume of the mature HIO lumen is approximately 1 µL, but may vary significantly. Automated syringe pumps may be used in place of a manual syringe to allow for fine control of the injected volume. - Withdraw the microcapillary from the HIO using the Z-axis control and position above the surface of the media.
- Move to the next HIO target with the microcapillary positioned above the media to avoid accidental damage to the HIOs during maneuvering and inject in a similar manner (see steps 3.6 – 3.11).
- Refill the microcapillary using the approach described above.
NOTE: If the tip breaks at any point during the microinjection, do not attempt to continue injections. Change the tip according to the instructions below.
- Refill the microcapillary using the approach described above.
- Change the microcapillary between treatments or in the event of breakage. Loosen the clamp on the end of the micromanipulator arm and remove the microcapillary from the micropipette holder.
Caution: Do not handle the microcapillary near the sharp end. The fine point of the microcapillary is extremely sharp and will easily puncture gloves and skin. Use caution and be aware of all movements, handling the microcapillary using the manipulator only and never placing hands between the point of the microcapillary and the HIO cultures. Seek medical treatment immediately in the event of a needle stick from a microcapillary containing infectious agents or toxins.
NOTE: In some cases, it may be difficult to visually confirm successful HIO microinjection. The visibility of clear microinjection suspensions can be enhanced by use of addition of higher concentrations of FITC-dextran.
4. Pharmacological Treatment of HIOs
- To test compounds delivered to the apical epithelium, resuspend in sterile PBS containing 2 mg/mL FITC-dextran and microinject into the HIO lumen as indicated above (Section 3). For the representative experiment, resuspend Clostridium difficile toxin TcdA at 12.8 ng/µL in PBS containing FITC-dextran.
- To test compounds delivered to the basolateral compartment, replace the external culture media with new media containing 2 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid (EGTA)(positive control), PBS vehicle alone (negative control), or other experimental compound after microinjection of FITC-dextran.
NOTE: Dosing and timing of the application of pharmacologic compounds, toxins, or other agents may vary according to the experimental question.
5. Live Imaging of Microinjected Organoids
- Begin live imaging immediately following microinjection. Transfer the culture plates to a fluorescent microscope equipped with a humidified chamber maintained at 37 °C and 21% O2 and 5% CO2 with an automated live cell imaging system. Close the environmental chamber and start the imaging process.
- Launch the image analysis software (e.g., softWoRx) from the desktop. Click "File" and select "Acquire (Resolve 3D)" to initialize the imaging and microscope control software.
- Set excitation/emission to FITC (475 nm excitation/523 nm emission), set the transmission power to 5%, and the lens to 4X. Set the exposure time to 0.025 ms (Figure 4A).
NOTE: Exposure time should be varied to suit the strength of the fluorescent signal. In general, the FITC fluorescence signal will only decrease. Therefore, to ensure maximum sensitivity, the initial exposure times should be adjusted such that the recorded fluorescent signal is just below the saturation point of the camera and imaging software. - Find the HIOs at 4X magnification using the digital controller attached to the microscope. Center the HIO within the viewing Field (Figure 4A).
- Click "View" on the toolbar and select "Point list" to reveal a new window. Click "Mark point" to store the current position of the stage in the point list (Figure 4A).
- Repeat steps 5.4 and 5.5 until the positions of all HIOs have been recorded. Using a notepad or digital record, make note of the unique ID number assigned to each programmed microscopy position and the images captured at that position. The image files will be tagged with this ID number and it will be important to associate image ID numbers with specific HIOs and treatments after data collection is complete.
- To setup automated image collection, click "File" and then "Experiment" in the toolbar. Name the experiment with the date or another unique name.
- Set the parameters for image collection by going through each of the option tabs as shown in Figure 4B.
- Un-check "Z Sectioning" under the tab "Sectioning".
- Under the tab "Channels", enter the parameters from the top left "Resolve 3D" window. To save time, the first row will automatically populate when the box on the left in checked.
- Under the tab 'Time-lapse", check the box labeled "Time lapse" and enter the time interval between images in the row labeled "Time-lapse" and the total duration of the experiment in the row labeled "Total Time". The software will automatically calculate the number of Time points.
- Under the tab "Points", Check the box labeled "Visit Point list". You may also manually enter the specific point numbers to be included in the experiment by typing in the text box.
- Do not change the default options under the tabs "Panels" or "Actions".
- Click on the "Run" tab. Enter the date of the experiment in the box labeled "image file name" and set the data save location under "Settings".
- Click on the green arrow to run the experiment macro. A time lapse counter will track the experimental progress.
- At the end of the planned time course, export and save all image files as 24-bit RGB TIFF images (Figure 4C).
- On the toolbar, select Process followed by Task builder.
- In the Task builder menu, add files by navigating to the data folder specified in 5.9. Highlight all files ending in ".dv" by typing "*.dv" in the prompt. Select all (Ctl+A) and click "Add".
- Click on + under the section labeled "Task." Select "Export as…" and click on the "Options" tab.
- Set "Export Format" to "TIFF Images" and check the box below to add a "Custom Output Folder". This is the location where the TIFF images will be exported. Select 24-bit RGB under "TIFF output type" and click "Done".
- Click "Submit to Queue" to run the Export Macro.
- Copy the TIFF images exported in 5.11 to an external driver or server if you will be performing the Post-imaging analysis (Section 6) on a different computer.
NOTE: The HIO tissue and media may be stored for histology34, PCR35), Western blot36, or other downstream analysis.
6. Post-imaging analysis
- Ensure that ImageJ37 is installed and working properly on the computer to be used for analysis.
NOTE: Fiji37 is an alternate distribution of the ImageJ core program that will work equally well for this analysis. - Start the batch analysis by selecting "Process" then "Batch" and finally "Macro" from the ImageJ menu.
- Set the Input to the directory containing the TIFF images collected during the experimental time course. Open the "thesholdmeasure.ijm" ImageJ macro file or directly copy/paste the macro code into the window. Click "Process" to begin processing the files.
NOTE: The minimum threshold value x (setThreshold(x,255);) can be set to any number 0 – 255 and should be adjusted so as to eliminate background fluorescence. Values <100 are recommended. To empirically determine the appropriate threshold value, run the imaging macro on a single image representing an organoid with no fluorescent signal. The mean intensity of this image can serve as a guide for setting the threshold appropriately. For the representative analysis presented below, the threshold was set to "42". - ImageJ will produce a large table containing the area of all pixels within the threshold intensity range, and the mean, median, minimum, and maximum (limit) intensity value for the area within the threshold intensity limits. Save this as a CSV or XLS.
- Change in intensity over time can be computed in a spreadsheet39 by manipulating the data table to perform the calculation below or can be automated using a suitable programming language. An example analysis script written in R 40 is provided with this manuscript.
NOTE: For each HIO, relative fluorescence intensity may be quantified as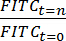 . Elimination time 41 (t1/2) of FITC-dextran in the HIO lumen was calculated as follows:
. Elimination time 41 (t1/2) of FITC-dextran in the HIO lumen was calculated as follows:
- First, calculate the "area under the curve" (AUC) from the curve describing the relative fluorescence intensity over time (t) as:
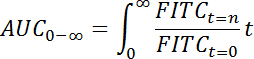
- Then, calculate the "clearance" (
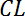 ) rate with the volume of distribution (Vd) defined as 1 for the normalized fluorescence at t = 0:
) rate with the volume of distribution (Vd) defined as 1 for the normalized fluorescence at t = 0:
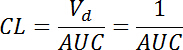
- Next, define the "elimination rate constant" (
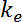 ) as:
) as:
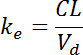
- And finally, calculate the "elimination time" (t1/2) as:
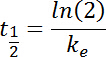
The reduced equation is thus:
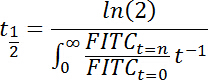
- First, calculate the "area under the curve" (AUC) from the curve describing the relative fluorescence intensity over time (t) as:
Representative Results
HIOs were differentiated from human pluripotent stem cells and cultured in cell matrix solution as previously described19,24. After 4 weeks in culture, the HIOs had expanded sufficiently to allow for microinjection. HIOs were microinjected with 4 kDa FITC-conjugated dextran suspended in PBS or PBS containing Clostridium difficile toxin TcdA. C. difficile is an opportunistic gastrointestinal pathogen that exhibits toxin-mediated epithelial toxicity in HIOs25. As a positive control, EGTA was added to the HIO culture media in a subset of HIOs injected with PBS and FITC-dextran. EGTA is a calcium chelator that causes rapid de-polymerization of the actin cytoskeleton42. FITC fluorescence was monitored in real time on a live imaging microscope within a controlled environmental chamber and images were captured in 10-minute intervals.
Post-hoc analysis of imaging data revealed substantial differences in the retention of FITC fluorescence (Figure 5). HIOs injected with PBS retained nearly all of the fluorescent signal present at t = 0, however HIOs that were also injected with TcdA of treated with EGTA exhibited a substantial decrease in fluorescent intensity by 8 hours post-microinjection (Figure 5A). Imaging data were quantified for all HIOs at all time points to generate a high-resolution dataset representing the relative change in fluorescent intensity over time in each experimental condition (Figure 5B). Differences in epithelial permeability were evaluated by calculating the mean elimination time (t1/2 ) of FITC for each treatment group (Table 1) and comparing differences in t t1/2 between groups using the Student's t-test. Control-treated HIOs retained the majority of FITC fluorescent signal for more than 16 hours (t1/2 = 17 ± 0.3 h). Treatment with EGTA significantly reduced FITC-dextran elimination time relative to control HIOs (t1/2 = 2.6 ± 0.2 h; P = 1.3 x 10-8). Consistent with previously published results25, microinjection of TcdA significantly increased epithelial barrier permeability relative to control treatment (t1/2 = 7.6 ± 0.6 h; P = 5.4 x 10-4). Thus, both external (EGTA) and microinjected (TcdA) compounds are capable of inducing significant alterations in epithelial barrier permeability in HIOs. These results suggest that the effects of a wide range of pharmacologic agents, metabolites, bacterial products, cytokines, growth factors, and other compounds on epithelial barrier function may be evaluated using this approach.
| Treatment | Half-life (h) | SEM (h) | Lower 95% CI (h) | Upper 95% CI (h) | n |
| Control | 17.11 | 0.3 | 16.45 | 17.78 | 11 |
| EGTA | 2.58 | 0.19 | 1.78 | 3.38 | 3 |
| TcdA | 7.56 | 0.63 | 5.93 | 9.18 | 6 |
Table 1: Mean elimination time (t1/2) for FITC-dextran in HIOs treated with EGTA or TcdA. Units are hours post-microinjection.

Figure 1: Basic layout of a microinjector and micromanipulator for HIO microinjection. This image shows the complete complement of equipment used for performing microinjection of HIOs. Key components are labeled and ordering information can be found in the Materials table. Please click here to view a larger version of this figure.
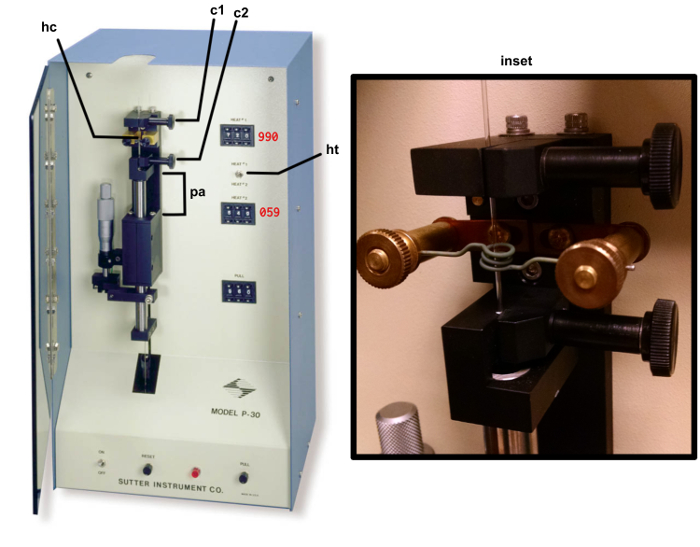
Figure 2: Micropipette puller. The copper heating coil hc, top clamp c1, bottom clamp (c2), puller arm (pa), heat selection toggle (ht) are identified by the arrows. The correct setting for HEAT 1 and PULL are indicated in red text. The inset shows a correctly mounted glass filament ready for heating. Please click here to view a larger version of this figure.
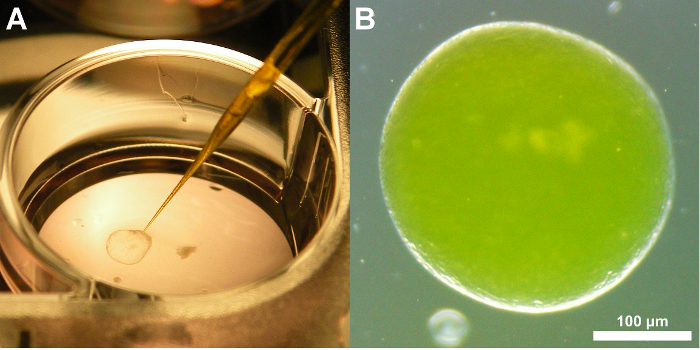
Figure 3: Representative images of a tissue-derived HIO injected with FITC-dextran. (A) Image demonstrating the positioning of the glass microcapillary just prior to insertion into the HIO and microinjection. (B) Brightfield image of a tissue-derived human intestinal organoids after microinjection of FITC-dextran. Note that the fluorescence signal is apparent even without the use of a specific filter set. This coloration aids microinjection precision. 3X magnification Please click here to view a larger version of this figure.
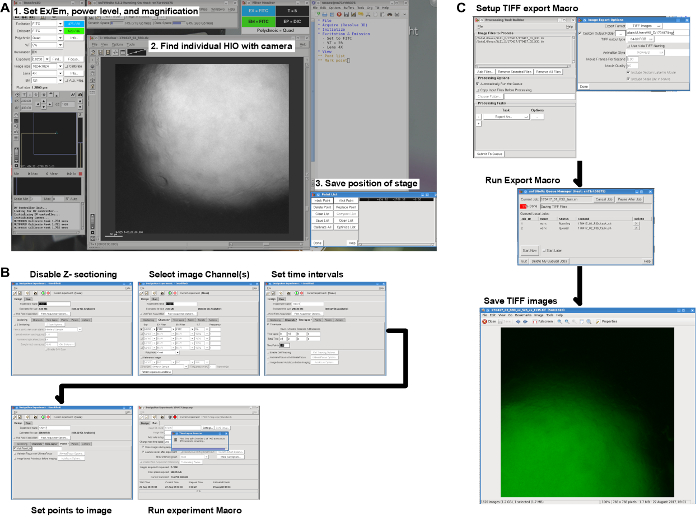
Figure 4: Live imaging workflow. (A) Screenshot illustrating the steps necessary to set the microscope parameters, find the HIOs on the slide, and save the position of the stage for each HIO to be imaged. (B) Pre-imaging settings for capturing the FITC channel at regular intervals at each of the positions specified in A. (C) Post-imaging export of DV format data files as TIFF images with one TIFF image per HIO per time point. Please click here to view a larger version of this figure.
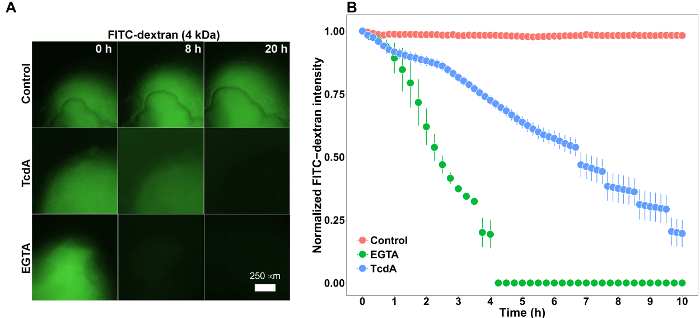
Figure 5: Representative results. (A) Stem-cell derived human intestinal organoids (HIO) microinjected with 2 mg/mL FITC-dextran (4 kDa) imaged for 20 hours. HIOs were also microinjected with PBS (control) or the Clostridium difficile toxin TcdA (12.8 ng/µL) or treated with 2 mM EGTA added to the external culture media. 4X Magnification. (B) Plot of mean normalized FITC intensity over time in HIOs treated with PBS (control), TcdA, or EGTA. Error bars represent S.E.M. and n = 11 HIOs (Control), 3 HIOs (EGTA), 6 HIOs (TcdA). Please click here to view a larger version of this figure.
Discussion
This protocol establishes a general-purpose method for the microinjection of hPSC-derived HIOs and tissue-derived intestinal organoids and the measurement of epithelial barrier permeability in real time. We have also demonstrated our approach to analysis and interpretation of the data generated using these methods. Given the growing adoption of intestinal organoids model systems16,20,21,28 and the long-standing interest in intestinal barrier permeability as a physiologically relevant functional outcome3,4,5,6, we anticipate that others working in this field will be able to apply and build upon these methods.
There are several steps which are critical to the application of this technique. Access to high quality hPSC- or tissue derived HIO tissue should be established prior to extensive experimentation with microinjection. HIO macrostructure may be heterogenous, with variation in both size and shape, although tissue identity and cellular morphology is highly reproducible when utilizing established methodology to generate HIOs24. Spherical HIOs consisting of a single semi-transparent lumen and measuring approximately 1 mm in diameter are ideal for microinjection and measurement of luminal fluorescence in real-time. In some cases, microinjection will fail, resulting in collapse of the HIO or obvious leakage of injected material. Failed HIOs can be removed from the culture well at the user's discretion using a standard micropipette. Consider the objective lenses available on an imaging platform when selecting HIOs for microinjection and imaging. In general, 2 – 4X objective lenses are ideal for capturing the complete HIO fluorescent signal, although a 10X objective may be used if low power lenses are not available or if the available HIOs are <1 mm in diameter. Imaging software must allow for the automated capture of fluorescent images at defined points over time.
Several modifications of this protocol are possible in order to suit the experimental requirements. For example, the results of barrier function tests may be dependent on the molecular size of the compounds in use43 and it may be appropriate to test dextran preparations of varying molecular weight. In addition, brightfield imaging may be performed in addition to fluorescence imaging as an indicator of the overall structural integrity of the tissue25. When performing microinjection of live bacteria25,28,31,32,33,44, it may be necessary to add penicillin and streptomycin or gentamicin to the HIO culture media prior to or after microinjection. The outside of the microcapillary will become contaminated during filling with the bacterial culture suspension and this may be transferred to the HIO media. Alternately, microinjection can be performed on HIOs suspended in extracellular matrix (e.g., Matrigel) without media, adding the media after the microinjection is completed. This may limit contamination to the extracellular matrix and external face of the HIO. When planning microbial growth assays, it may be necessary to remove antibiotics in the media after 1 – 2 h to avoid slowing or preventing growth of microinjected organisms.
Finally, recognizing that not all researchers will have access to microscopy equipment suited to in vitro imaging, it is important to point out that the procedures outlined in this protocol for collecting fluorescence data can be applied to images taken at fixed timepoints using standard epifluorescent microscopy without automated image capturing or environmental controls. Examples of this approach can be found in the reports by Leslie and Huang et al.25, who examined C. difficile toxin activity in hPSC-derived intestinal organoids, and Karve and Pradan et al.44, who examined epithelial barrier permeability in similar hPSC-derived intestinal organoids microinjected with live E. coli. Manual operation of imaging equipment may result in greater variation and difficulty in normalizing the fluorescent signal. When performing manual imaging of FITC-dextran injected HIOs it is essential to maintain fixed magnification, fluorescent excitation intensity, and exposure times throughout the experiment to avoid distorting the fluorescent intensity measurements.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Drs. Stephanie Spohn and Basel Abuaita for many useful discussions on organoid microinjection. JRS is supported by the Intestinal Stem Cell Consortium (U01DK103141), a collaborative research project funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of Allergy and Infectious Diseases (NIAID). JRS and VBY are supported by the NIAID Novel, Alternative Model Systems for Enteric Diseases (NAMSED) consortium (U19AI116482). DRH is supported the Mechanisms of Microbial Pathogenesis training grant from the National Institute of Allergy and Infectious Disease (NIAID, T32AI007528) and the Clinical and Translational Science award to the Michigan Institute for Clinical and Health Research (UL1TR000433).
The complete data files and data analysis code used in this manuscript are available at https://github.com/hilldr/HIO_microinjection.
Materials
| EGTA 0.5 M sterile (pH 8.0) | Bioworld | 405200081 | |
| Cell matrix solution (Matrigel) | Corning | 354230 | |
| Deltavision RT live cell imaging system | GE Life Sciences | 29065728 | http://www.gelifesciences.com/webapp/wcs/stores/servlet/catalog/en/GELifeSciences/brands/deltavision/ |
| Camera | GE Life Sciences | 29065728 | Included with Deltavision system |
| softWoRx Imaging software | GE Life Sciences | 29065728 | Included with Deltavision system |
| Biosafety cabinet | Labconco | Cell Logic+ | http://www.labconco.com/product/purifier-cell-logic-class-ii-type-a2-biosafety-cabinets-2/4262 |
| 1X PBS | Life Technologies | 10010-023 | |
| Advanced DMEM-F12 | Life Technologies | 12634-010 | Component of ENR media; see McCraken et al. 24 |
| B27 supplement (50X) | Life Technologies | 17504044 | Component of ENR media; see McCraken et al. 24 |
| L-glutamine (100X) | Life Technologies | 25030-081 | Component of ENR media; see McCraken et al. 24 |
| HEPES buffer | Life Technologies | 15630080 | Component of ENR media; see McCraken et al. 24 |
| Manipulator | Narshge | UM-3C | |
| Micromanipulator | Narshge | UM-1PF | |
| Pipette Holder | Narshge | UP-1 | Alternate to Xenoworks pipette holder |
| Magnetic stand | Narshge | GJ-1 | |
| Dissecting scope | Olympus | SX61 | Recommended scope, although other models are likely compatible |
| Olympus IX71 Fluorescent microscope | Olympus | IX71 | Included with Deltavision system |
| CoolSNAP HQ2 | Photometrics | 29065728 | Included with Deltavision system |
| Recombinant C. difficile Toxin A/TcdA Protein | R&D Systems | 8619-GT-020 | |
| EGF | R&D Systems | 236-EG | Component of ENR media; see McCraken et al. 24 |
| R-spondin 1 | R&D Systems | 4645-RS | Component of ENR media; see McCraken et al. 24 |
| Noggin | R&D Systems | 6057-NG | Component of ENR media; see McCraken et al. 24 |
| Mineral oil | Sigma-Aldrich | M8410 | |
| FITC-dextran (4 kDa) | Sigma-Aldrich | 46944 | |
| Micropipette puller | Sutter Instruments | P-30 | |
| Nunc Lab-Tek II Chamber Slides | ThermoFisher Scientific | 154526PK | |
| Glass filaments | WPI | TW100F-4 | |
| Micropipette holder | Xenoworks | BR-MH2 | Preferred device |
| Analog Tubing kit | Xenoworks | BR-AT | |
| 1/16 in clear ferrule | Xenoworks | V001104 | |
| 1-1.2 mm O-ring | Xenoworks | V300450 |
Referencias
- Standring, S., Borley, N. . Gray’s anatomy: The anatomical basis of clinical practice. , (2008).
- Buckley, A., Turner, J. R. Cell biology of tight junction barrier regulation and mucosal disease. Cold Spring Harb Perspect Biol. , (2017).
- Clayburgh, D. R., Shen, L., Turner, J. R. A porous defense: The leaky epithelial barrier in intestinal disease. Lab Invest. 84, 282-291 (2004).
- Turner, J. R. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 9, 799-809 (2009).
- Bischoff, S. C., et al. Intestinal permeability-a new target for disease prevention and therapy. BMC Gastroenterol. 14, 189 (2014).
- Odenwald, M. A., Turner, J. R. The intestinal epithelial barrier: A therapeutic target?. Nat Rev Gastroenterol Hepatol. 14, 9-21 (2017).
- Krugliak, P., Hollander, D., Schlaepfer, C. C., Nguyen, H., Ma, T. Y. Mechanisms and sites of mannitol permeability of small and large intestine in the rat. Dig Dis Sci. 39, 796-801 (1994).
- Johnston, S. D., Smye, M., Watson, R. P. Intestinal permeability tests in coeliac disease. Clin Lab. 47, 143-150 (2001).
- Salles Teixeira, T. F., et al. Intestinal permeability measurements: General aspects and possible pitfalls. Nutr Hosp. 29, 269-281 (2014).
- Wang, L., et al. Methods to determine intestinal permeability and bacterial translocation during liver disease. J Immunol Methods. 421, 44-53 (2015).
- Donato, R. P., et al. Studying permeability in a commonly used epithelial cell line: T84 intestinal epithelial cells. Methods Mol Biol. 763, 115-137 (2011).
- Balimane, P. V., Chong, S. Cell culture-based models for intestinal permeability: A critique. Drug Discov Today. 10, 335-343 (2005).
- Vidyasagar, S., MacGregor, G. Ussing chamber technique to measure intestinal epithelial permeability. Methods Mol Biol. 1422, 49-61 (2016).
- Herrmann, J. R., Turner, J. R. Beyond Ussing’s chambers: contemporary thoughts on integration of transepithelial transport. Am J Physiol Cell Physiol. 310, C423-C431 (2016).
- Sato, T., et al. Single lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459, 262-265 (2009).
- Clevers, H. Modeling development and disease with organoids. Cell. 165, 1586-1597 (2016).
- Drost, J., et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 11, 347-358 (2016).
- Rookmaaker, M. B., Schutgens, F., Verhaar, M. C., Clevers, H. Development and application of human adult stem or progenitor cell organoids. Nat Rev Nephrol. 11, 546-554 (2015).
- Spence, J. R., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 470, 105-109 (2011).
- Aurora, M., Spence, J. R. HPSC-derived lung and intestinal organoids as models of human fetal tissue. Dev Biol. 420, 230-238 (2016).
- Dedhia, P. H., Bertaux-Skeirik, N., Zavros, Y., Spence, J. R. Organoid models of human gastrointestinal development and disease. Gastroenterology. 150, 1098-1112 (2016).
- Dye, B. R., et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 4, (2015).
- Dye, B. R., Miller, A. J., Spence, J. R. How to grow a lung: Applying principles of developmental biology to generate lung lineages from human pluripotent stem cells. Curr Pathobiol Rep. 4, 47-57 (2016).
- McCracken, K. W., Howell, J. C., Wells, J. M., Spence, J. R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 6, 1920-1928 (2011).
- Leslie, J. L., et al. Persistence and toxin production by clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 83, 138-145 (2015).
- Leslie, J. L., Young, V. B. A whole new ball game: Stem cell-derived epithelia in the study of host-microbe interactions. Anaerobe. 37, 25-28 (2016).
- Zachos, N. C., et al. Human enteroids/Colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem. 291, 3759-3766 (2016).
- Hill, D. R., Spence, J. R. Gastrointestinal organoids: Understanding the molecular basis of the host-microbe interface. Cell Mol Gastroenterol Hepatol. 3, 138-149 (2017).
- Miyoshi, H., Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 8, 2471-2482 (2013).
- Sato, T., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and barrett’s epithelium. Gastroenterology. 141, 1762-1772 (2011).
- Engevik, M. A., et al. Loss of nHE3 alters gut microbiota composition and influences bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol. 305, G697-G711 (2013).
- Forbester, J. L., et al. Interaction of salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 83, 2926-2934 (2015).
- Engevik, M. A., et al. Human clostridium difficile infection: Inhibition of nHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol. 308, G497-G509 (2015).
- Bancroft, J., Gamble, M. . Theory and practice of histological techniques. , (2008).
- Kennedy, S., Oswald, N. . PCR troubleshooting and optimization: The essential guide. , (2011).
- Kurien, B., Scofield, R. . Western blotting: Methods and protocols. , (2015).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH image to imageJ: 25 years of image analysis. Nat Methods. 9, 671-675 (2012).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 9, 676-682 (2012).
- Winston, W. . Microsoft excel data analysis and business modeling. , (2016).
- R Core Team. A language and environment for statistical computing. R Foundation for Statistical Computing. , (2017).
- Rosenbaum, S. . Basic pharmacokinetics and pharmacodynamics: An integrated textbook and computer simulations. , (2016).
- Selden, L. A., Estes, J. E., Gershman, L. C. The tightly bound divalent cation regulates actin polymerization. Biochem Biophys Res Commun. 116, 478-485 (1983).
- Vojdani, A. For the assessment of intestinal permeability, size matters. Altern Ther Health Med. 19, 12-24 (2013).
- Karve, S. S., Pradhan, S., Ward, D. V., Weiss, A. A. Intestinal organoids model human responses to infection by commensal and shiga toxin producing escherichia coli. PLoS One. 12, e0178966 (2017).

