Manipulation of Ploidy in Caenorhabditis elegans
Summary
This method allows for the generation of tetraploid and triploid Caenorhabditis nematodes from any diploid strain. Polyploid strains generated by this method have been used to study chromosome interactions in meiotic prophase, and this method is useful for examining important basic questions in cell, developmental, evolutionary, and cancer biology.
Abstract
Mechanisms that involve whole genome polyploidy play important roles in development and evolution; also, an abnormal generation of tetraploid cells has been associated with both the progression of cancer and the development of drug resistance. Until now, it has not been feasible to easily manipulate the ploidy of a multicellular animal without generating mostly sterile progeny. Presented here is a simple and rapid protocol for generating tetraploid Caenorhabditis elegans animals from any diploid strain. This method allows the user to create a bias in chromosome segregation during meiosis, ultimately increasing ploidy in C. elegans. This strategy relies on the transient reduction of expression of the rec-8 gene to generate diploid gametes. A rec-8 mutant produces diploid gametes that can potentially produce tetraploids upon fertilization. This tractable scheme has been used to generate tetraploid strains carrying mutations and chromosome rearrangements to gain insight into chromosomal dynamics and interactions during pairing and synapsis in meiosis. This method is efficient for generating stable tetraploid strains without genetic markers, can be applied to any diploid strain, and can be used to derive triploid C. elegans. This straightforward method is useful for investigating other fundamental biological questions relevant to genome instability, gene dosage, biological scaling, extracellular signaling, adaptation to stress, development of resistance to drugs, and mechanisms of speciation.
Introduction
Whole genome polyploidy exists throughout nature and is often a necessary step in adaptation, speciation, organogenesis, wound healing, and biological scaling; it is also known to promote both cancer and resistance to drugs1,2,3,4,5,6,7,8,9,10,11,12. Agriculture and fishery industries generate polyploid plants, fish, and shellfish by chemical treatment (e.g., colchicine and orzalin) to obtain faster growth rates and bulkier crops and livestock13,14,15. Experimental and inefficient production of tetraploids exists for the mouse and zebrafish model systems16,17. However, most or all polyploid multicellular animals generated are embryonically lethal or sterile and thus not ideal for laboratory studies on the effects of polyploidy in a multicellular organism. Consequently, any understanding of whole genome polyploidization in multicellular organisms has been limited to closely related species from evolutionary recent polyploidization events18,19,20. A path to advance queries of the biological role or consequences of polyploidization is the use of the C. elegans model system. Importantly, C. elegans is a tractable genetic system that normally exists as a diploid, contains only five autosomes (A) and one sex chromosome (X) per genome, is transparent allowing for in vivo observation of biological processes, and has a short life cycle of 3 – 4 days (from egg to sexually mature adult). C. elegans has been shown to be able to reproduce as a tetraploid, which is the most common type of whole genome polyploidy in nature. Triploid animals can be generated by crossing tetraploids with diploids, but their ploidy is not stable, and the strains become diploid within a couple of generations.
In the last few decades only a handful of viable and fertile C. elegans tetraploids strains were isolated in the laboratory, using a strategy that is laborious and generates only limited types of strains21,22,23. This strategy generates C. elegans tetraploids by heat-shock treatments, which presumably affects chromosome segregation in the gametes, followed by a screening for putative polyploid animals using genetic markers. These tetraploids were extremely useful in the enquiry of how this nematode determines whether to become male or hermaphrodite. Later studies used the available strains to investigate the growth, gene dose, and regulation of synapsis during meiosis in this nematode21,24,25,26. Unfortunately, these studies were limited by the difficulty in generating new tetraploid strains and the background genetic markers these strains contained. Shown here is a simple and rapid protocol to generate stable tetraploids, which has been used to generate strains to study regulation of synapsis during meiosis27.
As in nature, polyploidization can arise by the formation of diploid instead of haploid gametes. Our finding that a meiosis-specific cohesin component mutant rec-8 generates diploid sperm and oocytes indicated that knocking down the rec-8 gene would result in the production of tetraploid progeny (Figure 1)27,28,29. Generating tetraploid strains simply involves knocking down rec-8 by RNA interference (RNAi) for two generations in order to phenocopy the rec-8 mutant diploid gametes. Putative polyploids can be easily identified by their longer than normal body size. Polyploids are confirmed as full or partial tetraploids by counting the number of chromosomes per nucleus once stable lines are established.
The strategy described here enables the generation of stable tetraploid Caenorhabditis nematode strains from any initial diploid genetic background or karyotype without the use of genetic markers. Since this protocol is more efficient, versatile, and simple than the scheme previously used, it will expand the tools needed to query the roles of polyploidization in fundamental processes of development, genome stability, and evolution in multicellular organisms. The only foreseeable limitation in the use of this protocol is in genetic backgrounds resistant to RNAi.
Protocol
1. Setting Up rec-8 RNAi (Day 1 – 3 in Figure 2)
This protocol is modified from Kamath and Ahringer30.
- For induction of rec-8 dsRNA expression in Escherichia coli, prepare Nematode Growth Medium (NGM) agar plates31 supplemented with a final concentration of 1 mM isopropyl-β-D-2-thiogalactopyranoside (IPTG) and 100 µg/mL ampicillin. Store in the dark at 4 °C until use, for up to 4 weeks.
- Streak HT115 bacteria carrying the rec-8 (W02A2.6) clone from the Ahringer laboratory library30 rec-8 clone onto Luria Broth (LB) plates supplemented with 100 µg/mL ampicillin and 50 µg/mL tetracycline. Grow overnight in a shaker at 37 °C.
- On Day 1, inoculate single colonies from the freshly streaked single colonies of the HT115 E. coli bacteria carrying the rec-8 RNAi clone into 4 mL LB containing 100 µg/mL ampicillin and 50 µg/mL tetracycline. Grow the bacteria culture overnight in a shaker or a roller drum at 37 °C.
- The next morning (Day 2), induce production of double stranded (ds) RNA for rec-8 W02A2.6 in the HT115 bacterial culture by adding a final concentration of 1 mM IPTG and shaking for 40 min at 37 °C.
- After induction, seed NGM/IPTG plates with 100 – 200 µL of HT115 rec-8 bacteria and store plates at room temperature in the dark overnight (Day 3).
- The next morning (Day 4), add the desired nematode strain to the induced HT115 rec-8 bacteria plates as described in step 2.1 below.
2. Generating and Isolating Tetraploids (Day 4 – 16 in Figure 2)
- On Day 4, place 3 – 4 young L4 (fourth larval) stage hermaphrodites of the desired diploid strain on the NMG/IPTG plates seeded with HT115 rec-8 bacteria that had been induced the day before (step 1.5, above). Grow nematodes at 15 °C in the dark.
- Repeat steps 1.3 and 1.4 starting 3 days after step 2.1, which will be three days before the first progeny (F1s) from that step become L4s. The timing of this step will depend on how fast a nematode strain grows at 15 °C. Some diploid mutant strains are slow growers, and this timing will need fine tuning in order to isolate tetraploids.
- On Day 8 (after 4 days of rec-8 RNAi feeding treatment), transfer 20 (2 hermaphrodites/Petri dish) of the F1 (first filial generation) L4 hermaphrodites grown in the HT115 rec-8 bacteria onto freshly induced HT115 rec-8 RNAi and allow them to self-fertilize.
- Alternatively, transfer 20 of the F1 L4 hermaphrodites grown in the HT115 rec-8 bacteria onto freshly induced HT115 rec-8 RNAi bacteria together with untreated males of the same strain (2 hermaphrodites with 4 – 6 males/plate).
- On Day 13, start screening for F2 (second filial generation) progeny that appear long (Lon) or overall larger than the wild type; then transfer individual Lon animals onto regular OP50 or HB101 strains of E. coli bacteria. Continue screening F3 (third filial) progeny; however, F3 progeny from the same plate cannot be considered independent strains if they become established, because they could be siblings from the same already established F2 mother.
NOTE: Putative tetraploids are easily identified because they are clearly longer than diploids. Wild type hermaphrodites are two thirds shorter than tetraploids. Because it is longer, a tetraploid body makes an extra turn as it moves forward by generating sinusoidal body-bending waves from head to tail (Figure 3A). - Allow Lon worms to self-fertilize and propagate by picking Lon progeny until only Lon progeny are sired in the absence of rec-8 RNAi treatment. This may take two to three additional generations. Lon worms often are sterile and do not yield progeny.
3. Verifying Tetraploid Strains (Movie 1 and Figure 3A, B)
NOTE: Tetraploid strains can be validated by counting the number of chromosomes in their unfertilized oocytes. Fluorescent microscopy can be used to screen for the number of chromosome pairs in unfertilized diploid oocytes (prior to the meiotic divisions), if the strain has a fluorescent marker for chromosomes. In the absence of fluorescent chromosome markers, tetraploid nematodes can be screened by fixing the nematodes and staining with a DNA dye; see below for a protocol for ethanol fixing and whole-animal 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) staining.
- Whole animal DAPI staining
NOTE: Tetraploid strains carrying 12 connected chromosome pairs can be validated by DAPI staining these animals and counting the number of DAPI bodies in its unfertilized oocytes.- Place 5 to 10 µL of M9 buffer31 on a microscope slide, then transfer 6 – 10 nematodes to the drop.
- Under a dissecting microscope, draw most of the M9 from the drop without removing the nematodes with a lint-free cleaning tissue. Then, immediately add a 10 µL drop of 90% ethanol onto the worms. Allow the worms to dry completely, but for no longer than a couple of seconds.
- As soon as the ethanol evaporates (this can be seen as it happens under the dissecting microscope), add an additional 10 µL of 90% ethanol onto the worms.
- Repeat step 3.1.3 three more times.
- Once the last drop of ethanol has evaporated, add 6 µL of DAPI or Hoechst dye at the recommended final concentration in the mounting media of choice (for example, a 1:1,000 dilution of a 2 ng/µL DAPI stock concentration in M9). For long-term storage of the slides, 0.5% p-phenylenediamine dissolved in 20 mM Tris-HCl, pH 8.8, in 90% glycerol as an antifade solution, instead of M9 only, may be used.
- Cover the worms on the slide with a coverslip and seal the edges of the coverslip with nail polish. Score in a fluorescence microscope (step 3.1.7) at least 10 min after adding the coverslip. Slides without antifade can be stored for a few days at 4 °C before scoring, but the quality of fluorescence starts to decline after a few days.
- Using a fluorescent microscope at 100X magnification, score the number of single DAPI bodies (presumably single chromosome pairs) in the most mature unfertilized oocyte, which is immediately adjacent to the spermatheca and has not yet entered the spermatheca or the uterus.
- To score individual DAPI bodies within an oocyte nucleus, use the microscope's fine focus to move slowly from the top of the oocyte nucleus to the bottom during counting. Then, recount the same nucleus while moving the focus in the opposite direction (i.e., from the bottom to the top of the nucleus) to confirm the counted number of DAPI bodies.
- Score the most mature unfertilized oocyte in each of the two gonads in at least ten hermaphrodites per stable Lon strain. Wild type oocytes have 6 DAPI bodies on average, 5 autosome pairs, and the sex chromosome pair. The presence of 12 DAPI bodies in the oocyte of stable Lon strains indicates that the animals in this strain are either partial (4 sets of Autosomes, 3 X-chromosomes) or complete (4 sets of Autosomes, 4 X-chromosomes) tetraploids.
- Calculate the average number of DAPI bodies observed from multiple (at least 10) oocytes per strain. Some chromosome pairs will be touching or right on top of one another, so the number of DAPI bodies in an oocyte is often smaller than the actual number of chromosome pairs.
- (Optional) Further validate stable Lon strains where the average number of DAPI bodies is 12 to test whether they are full (4A, 4X) or partial (4A, 3X) tetraploids by immunofluorescence staining against chromosome axis proteins such as HTP-332. This staining will distinguish chromosome pairs by showing a cruciform pattern from single unconnected chromosomes present in the partial tetraploid.
Representative Results
Impairment of the rec-8 Meiotic Cohesin Component Function Results in Diploid Gametes:
Imaging of the meiotic divisions of cohesin component mutant rec-8 sperm and oocytes revealed possible mechanisms for generating tetraploid animals (Figure 1)27,29,33. Mechanistically, the meiotic division defects of rec-8 mutant sperm and oocyte are different; however, both male and female diploid gametes are produced by the rec-8 mutant.
In wild type meiosis, homologous chromosomes become temporarily connected to one another by crossover recombination and enter the first meiotic division as a unit or bivalent (Figure 1A)34. During the first division, homologous chromosomes (homologs) segregate away from one another, whereas sister chromatids in each homolog remain together until the second division. Although the pattern of chromosome segregation is the same in female and male gametes, oocyte divisions are asymmetric whereas spermatocyte divisions are symmetric and undergo a specialized cytokinesis. In each division, the oocyte discards half of the division product into a small polar body. Thus, each oocyte precursor gives rise to a single haploid oocyte and two polar bodies (Figure 1B, C)35. Conversely, a single spermatocyte precursor gives rise to four functional spermatids by undergoing two symmetric divisions. The second division culminates with four spermatids budding off from a residual body. This cytokinesis is special in that the pattern and number of spermatids is determined by the centrosomes36.
In the precursor to the rec-8 mutant gametes, crossover recombination between homologous chromosomes does not take place and homologs are not connected as a bivalent at the beginning of the first meiotic division29,34. Sister chromatids of each homolog segregate away from one another in the first division instead of remaining together until the second division, as they do in wild type gametes. Interestingly, the rec-8 mutant phenotype during the second division is different in male and female gametes in that oocytes fail to undergo the cytokinesis whereas spermatocytes undergo a relatively normal cytokinesis (Figure 1B–F)27,28,29. Diploid oocytes arise because in the second division they fail both chromosome segregation and cytokinesis, hence they do not extrude the second polar body (Figure 1B, C). Spermatocytes in rec-8 germlines undergo spermatid budding or cytokinesis, but often segregate both sets of chromosomes to one of the spermatids to yield a diploid spermatid and a spermatid lacking chromatin (Figure 1D–F)27. Quantification of the proportion of rec-8 sperm lacking chromatin is shown in Figure 1F.
The formation of female and male diploid gametes in rec-8 mutants suggested that this mutant phenotype could be potentially used to generate full genome tetraploid strains.
Generation of Tetraploid C. elegans Strains:
The presence of a mutation in the rec-8 gene in all the generated tetraploid strains can be avoided by transiently knocking down rec-8 by RNAi29,37. This assumes that the reduction of rec-8 function by RNAi would generate diploid gametes that could potentially give rise to whole genome tetraploid animals, as rec-8 mutants do29,37. Essential to this protocol is that rec-8 mutants both give rise to diploid gametes and sire a reasonably large number of young, contrary to other meiotic mutants, which give rise to aneuploid gametes and are mostly sterile or embryonic/larval lethal.
Multiple tetraploid strains were generated by feeding C. elegans bacteria expressing rec-8 dsRNA using either one of two strategies (see Protocol, Figure 2, and Table 1)27. Tetraploids can be generated by self-fertilizing hermaphrodites for two to three generations in Petri dishes with bacteria expressing rec-8 dsRNA. By this strategy a hermaphrodite is placed in freshly made rec-8 RNAi plates and its progeny is transferred a few days later onto new plates with freshly induced rec-8 RNAi expressing bacteria (see Protocol). In addition, tetraploids can be generated through crossing the first generation of hermaphrodites fed bacteria expressing rec-8 dsRNA with untreated males of the same genotype in Petri dishes with bacteria expressing rec-8 dsRNA (Figure 2). In this case, the males in the cross are exposed to the rec-8 RNAi bacteria from the L4 stage onwards, during mating. Both schemes give rise to tetraploid animals27. Table 1 shows tetraploid strains obtained using the scheme presented here. Triploid and tetraploid C. elegans are larger in size than diploids, but triploid strains are unstable and tend to become diploid in one or two generations, whereas tetraploid strains are relatively stable22,23. Putative tetraploid strains were identified as larger than the original strain (Lon) hermaphrodites that sired only Lon progeny (Figure 3A). Lon animals are easily identified – wild type diploid animals are two thirds of the length of tetraploids and their bodies do not make an extra bend, which can be noticed during its forward sinusoidal movement (Figure 3A).
Tetraploid strains were confirmed by screening for the presence of 12 chromosome pairs in oocytes of the tetraploid hermaphrodites compared to six chromosome pairs in oocytes of diploid hermaphrodites (Figure 3B, C, and Movie 1).
Tetraploid Classes:
Two types of tetraploid hermaphrodites were identified: one sired males at similar frequencies as diploid hermaphrodites and the other sired males at much higher frequencies (Table 1). These two kinds of tetraploid hermaphrodites have been shown to differ in that the class producing frequencies similar to those of diploid males are tetraploid for all its chromosomes (4A, 4X), whereas the class producing high frequencies of males are hermaphrodites that are tetraploid for the autosomes but triploid for the sex chromosome (4, 3X). The later class of tetraploids are stable and produce 4A, 3X hermaphrodites and 4A, 2X males.
Tetraploid strains grow slower and produce reduced brood size compared to the diploids they were derived from, as seen for the strains generated with the previous method22,23. Madl and Herman22 suggested that the increased proportion of dead embryos in the tetraploid strains could be due to aneuploidy in the oocyte, however superficial inspection of tetraploid strains did not reveal sufficiently elevated numbers of aneuploid oocytes nor abnormal oocyte or spermatocyte divisions to account for the observed reduction in brood size (Movie 2, and Figure 3C, D).
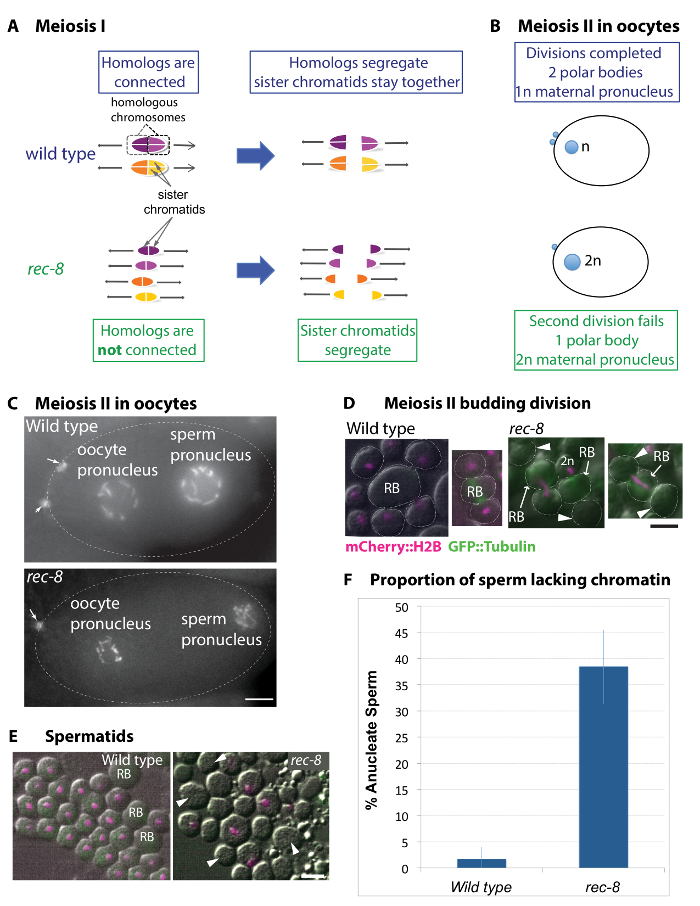
Figure 1: Gametogenesis in rec-8 mutant implies possible mechanisms for generating stable polyploid stains. (A) Diagram of the chromosome organization and segregation pattern in wild type and rec-8 mutant meiotic divisions. In wild type meiosis, homologous chromosomes separate in the first meiotic division. Sister chromatids in each homolog orient towards the same spindle pole and remain together until the second division. In rec-8 mutants, homologs do not form crossovers, and thus are not connected. In contrast to wild type, rec-8 sister chromatids orient away from one another and separate in the first meiotic division. (B) Diagram of wild type and mutant oocytes showing the female pronucleus and extruded polar bodies (male pronucleus is not depicted). In wild type oocytes, two asymmetric meiotic divisions result in the extrusion of two polar bodies. In rec-8 mutants however, the second polar body extrusion fails resulting in a diploid oocyte. (C) Images of wild type and rec-8 mutant oocytes expressing mCherry::histone H2B. Arrows indicate two polar bodies in the wild type oocyte and one in the rec-8 mutant. Scale bar is 5 µm. (D and E) Live Images of spermatids from wild type and rec-8 mutant animals expressing mCherry::histone H2B (magenta). Arrowheads indicate anucleate spermatids. Scale bar is 2 µm. (D) Spermatocytes undergoing the second (budding) division in the wild type and rec-8 mutant. The wild type spermatocytes undergo symmetric divisions resulting in four budding spermatids, each with a haploid chromosome complement. In rec-8 mutant spermatocytes, chromosome segregation is impaired in the second division. Most often a single mass of chromatin remains in the residual body (RB) or in one of the two sister spermatids in the second meiotic division. This gives rise to anucleate rec-8 mutant sperm (indicated by arrowheads) or diploid sperm. (E) Live images of post-budding spermatids in wild type and rec-8 mutants visualized using differential interference contrast (DIC) and fluorescence microscopy. Wild type spermatids all have similar chromatid masses. rec-8 mutant spermatids form anucleate sperm. (F) Quantification of wild type and rec-8 mutant anucleate sperm. rec-8 mutants produced 38.5% of anucleate sperm compared to less than 1.6% in wild type. Fisher's Exact test indicates that a rec-8 mutant has significantly higher incidence of anucleate sperm (p ≤0.0001) compared to the wild type. Error bars represent standard deviations. Please click here to view a larger version of this figure.
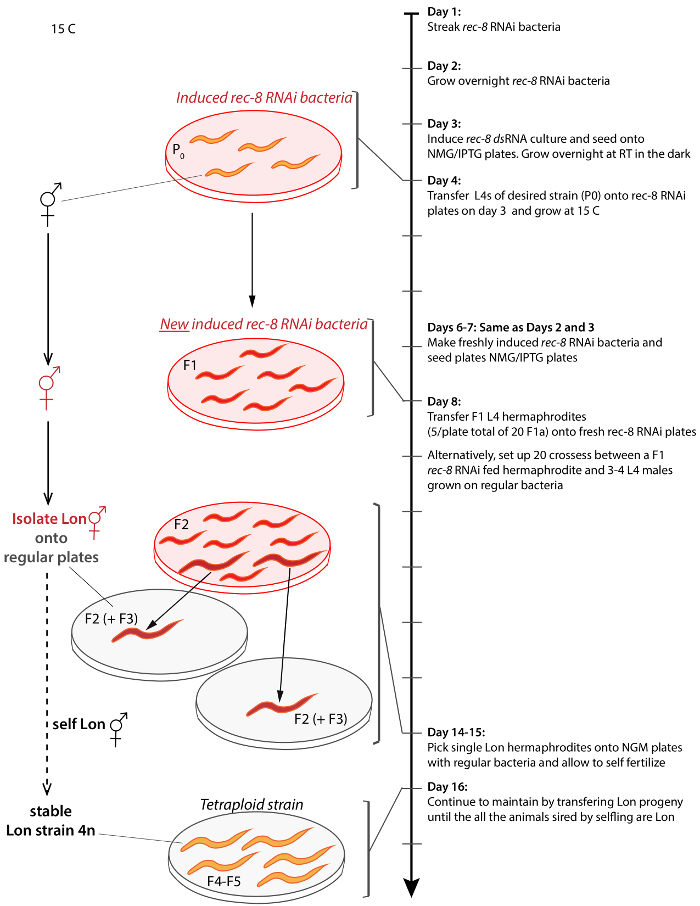
Figure 2: Scheme for generating and isolating tetraploid C. elegans from any strain. Arrow on the right represents the timeline of protocol starting from Day 1 and progressing to Day 17. Similar results can be achieved by crossing hermaphrodites to untreated males on Day 9. Brackets connect the depiction of a step to the timeline. Untreated animals are orange, treated animals are red, Lon animals are larger in size, rec-8 dsRNA expressing bacteria is depicted as transparent plates with transparent red background and regular OP50 bacteria is depicted on transparent grey background plates. Procedure is done at 15 °C unless otherwise stated. Please click here to view a larger version of this figure.
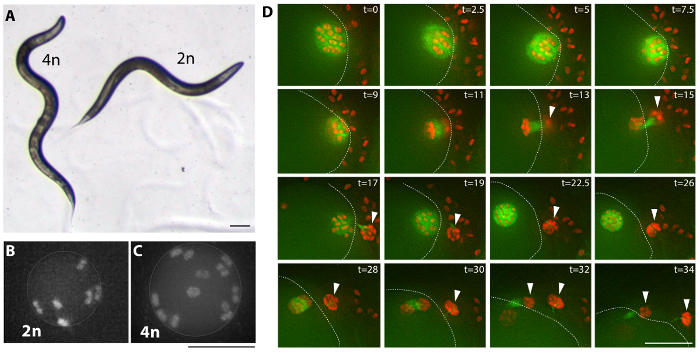
Figure 3: Tetraploid examples. (A) Bright field image of a tetraploid (MSC2) animal and the diploid strain it was derived from (AV740). Tetraploid C. elegans are overall larger and longer (Lon) than the diploid. This difference in body size results in an additional sinusoidal curve of the body of the moving tetraploid and can be used as a criterion to screen for tetraploid derivatives. Scale bar is 0.1 mm. (B, C) Fluorescence images of diploid (AV740) and tetraploid (MSC1) most mature unfertilized oocyte nulcei, prior to the meiotic divisions. Secondary screening is performed by observing the number of chromosome pairs in unfertilized oocytes of the established Lon strains, as shown in Movie 1 for an MSC1 strain expressing Mcherry::H2B and GFP::β-Tubulin in the germline. Scale bar is 5 µm. (D) Images from a time lapse (Movie 2) of tetraploid oocyte meiotic divisions depicting a generally normal meiosis. Arrow heads mark polar bodies and a dotted line marks the cortex of the oocyte inside the spermatheca and surrounded by sperm; t = time lapsed in minutes. Scale bar is 10 µm. Please click here to view a larger version of this figure.
| Genotype | Tetraploid derivative (sets of Autosomes : # of X-chromosomes)* |
Parental Strain (diploid) |
| meIs16 [pie-1p::mCherry::his-58 + unc-119(+)]; | MSC1 (4A:4X) | MSC0 |
| ruIs57 [pie-1p::GFP::b-tubulin + unc-119(+)] | MSC2 (4A:3X) | |
| MSC3 (4A:4X) | ||
| MSC5 (4A:3X) | ||
| MSC6 (4A:4X) | ||
| MSC8 (4A:4X) | ||
| unc-119(ed3) III; ddIs6; ddIs6[tbg-1::GFP + unc-119(+)]; ltIs37; ltIs37 [pAA64; pie-1p::mCherry::HIS-58 + unc-119(+)] IV | MSC14 (4A:3X) | TMR17 |
| MSC15 (4A:3X) | ||
| MSC16 (4A:4X) | ||
| spo-11(me44)/nT1 IV; +/nT1[ qIs51[myo-2::gfp Ppes-10::gfp, PF22B7.9::gfp]] V | AV800& | AV776 |
| meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)] II; | AV809& | AV727 |
| ltIs37 [pie-1p::mCherry::his-58 + unc-119(+)] IV; | ||
| ltIs38 [pie-1p::gfp::ph(PLC1delta1) + unc-119(+)] | ||
| meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)] II; mnT12 (X; IV) | AV826& | AV695 |
| mIn1[ dpy-10(e128) mIs14[myo-2::gfp pes-10::gfp]] / | AV810& | DR2078 |
| bli-2(e768) unc-4(e120) II | ||
| ruIs32 [pie-1::GFP::H2B + unc-119(+)] III | AV822& | AZ212 |
| AV823& | ||
| C. briggsae – mfIs42[Cel-sid-2 + Cel-myo-2::DsRed] | AV824& | JU1018 |
Table 1: Tetraploid strains generated. *Deduced by scoring the number of bivalents (connected homologous pairs) and univalents (individual homologs) in oocytes prior to the meiotic divisions in addition to the proportion of males sired.
Movie 1: Screening to confirm whether stable Lon strains are full or partial tetraploids. Movie starts with a diagram of a wild type hermaphrodite highlighting the region imaged (unfertilized oocytes) to identify tetraploids by counting chromosome pairs. Following the diagram, a series of Z-stack movies and projections show gonads of diploid and tetraploid animals fixed and DAPI stained or live images of strains expressing Mcherry::H2B histone. Screening is done by counting the number of connected homologous chromosome pairs in unfertilized oocytes. Counting of either MCherry or DAPI stained bodies is done in the unfertilized oocyte closest to the sperm storing spermatheca (the "-1 oocyte"). Chromosomes in this oocyte are most condensed and separated from one another, allowing for more accurate homolog pairs counts. More than 10 animals per strain were screened to ensure the -1 oocyte chromosome counts were accurate. Stack thickness is 0.2 µm. Please click here to view this video. (Right-click to download.)
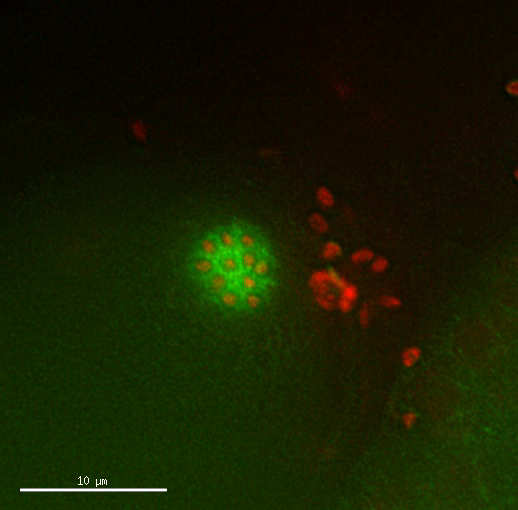
Movie 2: Tetraploid oocyte divisions appear normal. Time lapse of dividing oocyte of a tetraploid strain expressing Mcherry::H2B histone and GFP::β-Tubulin. Timing and pattern of the divisions appear normal. Images taken every 2.5 min. Please click here to view this video. (Right-click to download.)
Discussion
Production of haploid (n) gametes is key to generating a diploid (2n) zygote at fertilization. This reduction of the genome is accomplished during meiosis with two consecutive cell divisions after a single genome duplication. To generate haploid gametes, C. elegans, as with most other metazoans, segregate maternally- and paternally-derived homologs in the first division, whereas sister chromatids from each homolog segregate in the second division. One way that whole genome polyploidy arises in nature is through the generation of gametes that fail to half their genome size during meiosis.
It has been known for over 50 years that tetraploid C. elegans nematodes are viable and fertile. Nigon23, and later Madl and Herman22, generated and identified a handful of C. elegans tetraploids by disrupting meiotic chromosome segregation using heat-shock treatments and using genetic markers, respectively. A single additional C. briggsae tetraploid strain was derived using this protocol over 30 years later24. These tetraploids were utilized to investigate how C. elegans determine whether to become male or hermaphrodite, how ploidy regulates growth and size, and to analyze pairing and synapsis in meiosis25,26,38,39,40. Yet these and other studies requiring the use of specific tetraploids or triploid strains were limited by the difficulty in generating tetraploid strains by this method.
The protocol described here enables the generation of stable full 4A, 4X and partial 4A, 3X tetraploid Caenorhabditis nematode strains from any initial diploid genetic background or karyotype without the use of genetic markers.
Tetraploidy May Arise by More than One Mechanism in C. elegans:
Madl and Herman suggested that the tetraploid strains they generated likely derived from a triploid intermediate state. Their strains were obtained through selection over multiple generations or by crossing the putative triploid intermediate with diploid males22. The defects in chromosome partitioning in rec-8 mutants that gives rise to diploid oocytes and sperm suggest another possible mechanism by which tetraploid animals may arise with the rec-8 RNAi scheme27.
The rec-8 RNAi treated hermaphrodites could produce diploid oocytes and spermatocytes, which would give rise to tetraploid animals upon fertilization. Consistent with this possibility is the fact that some of the cloned F2 hermaphrodites gave rise to stable Lon strains in the next generation, which suggests that the cloned Lon F2 hermaphrodites where already tetraploid. In the crossing scheme, the first generation of rec-8 RNAi treated hermaphrodites is crossed with untreated males. Diploid spermatocytes could still arise in males because the cross is done in the presence of bacteria expressing rec-8 dsRNA and thus, males are exposed to rec-8 RNAi during mating for at least 3 days. Therefore, the Lon polyploids in the cross-fertilizing scheme could also have formed from fertilization of diploid oocytes by diploid sperm. Stable tetraploid strains may arise from either fertilization between diploid gametes or from crossing triploid animals containing oocytes of variable ploidy with diploid animals producing haploid sperm.
Important Considerations:
Self- versus cross-fertilization schemes:
The scheme of self-fertilizing rec-8 RNAi treated F1 hermaphrodites and the scheme involving crossing of treated hermaphrodites with untreated males both gave rise to 4A, 4X and 4A, 3X tetraploid strains. Although more polyploids were initially isolated from the self-fertilizing scheme, more of these polyploid animals were sterile and thus both schemes are similarly efficient at producing tetraploid stable strains. The reason for the increased success and sterility in the self-fertilizing scheme remains unknown. Although both schemes are similarly efficient, the self-fertilizing scheme is easier as it does not require the isolation of males for the mating. In addition, when the strain of interest is inefficient or defective at mating, the self-fertilizing scheme would be preferable. The cross-fertilizing scheme may be used for generating complex tetraploids containing more than two versions of a single chromosome.
Modifications and limitations:
Currently, this protocol involves rec-8 RNAi treatment by feeding bacteria expressing dsRNA for the rec-8 gene. Thus, this protocol does not work in Caenorhabditis species unresponsive to RNAi by feeding, or in mutants defective in environmental or systemic spread of RNAi between tissues41,42,43. This problem could potentially be solved by introducing RNAi treatment by direct injection of the dsRNA of interest directly into the germline.
Triploid animals must be generated by crossing a tetraploid to a diploid animal from which it was derived, because this scheme does not generate stable triploid strains44,45. Only 15% of the eggs sired by triploids hatch, and their progeny are mostly sterile progeny due to aneuploidy. In addition, the few surviving fertile progeny tend to be complete or near diploids within a couple of generations. This is likely, at least in part, because oocytes are partially correcting trisomy by segregating the third chromosome into the polar body in the first meiotic division.
An insuperable limitation of this protocol is that it does not work for making tetraploids of mutants that affect components of the RNAi machinery because they are resistant to RNAi treatment46.
Troubleshooting:
rec-8 RNAi:
A few considerations are crucial for this protocol to be successful. The first is to use fresh IPTG and freshly made IPTG NMG plates for induction of rec-8 dsRNA production in the bacteria HT115 bacteria carrying the rec-8 (W02A2.6) clone. IPTG is light sensitive and it is important to reduce light exposure to plates. IPTG plates can be stored up to one month at 4 °C in the dark. Second, the rec-8 (W02A2.6) RNAi bacterial HT115 strains from the Ahringer library (Kamath and Ahringer 2003) yielded a stronger rec-8 phenotype than other available rec-8 HT115 clones.
Tetraploid strain maintenance:
Tetraploid strains grow very slowly and produce at most 50 progenies per generation47. All identified tetraploid strains are relatively stable, but they can break down and become diploid when stressed (Jonathan Hodgkin personal communication and our unpublished observations). Therefore, it is important to note that when tetraploid strains are grown at 25 °C, heat-shocked, starved, or frozen and defrosted, they can rapidly revert to diploidy, so it is important to continue to pick the Lon animals when defrosting these strains or when exposing these strains to stressful conditions that may cause them to revert.
Possible Applications:
Investigation of the effect or the role of whole genome polyploidization in evolution, cell cycle, gene expression, and development in multicellular organisms has relied on comparisons between: cells in an organism that contain different ploidy, the same cell types in closely related species with different ploidy, or species that have undergone evolutionarily recent polyploidization events and physical isolation3,5,6,7,8,11,18,19,20,48,49,50. Although tetraploids can be derived from zebrafish and mouse model systems, their progeny are sterile or embryonically lethal16,17. In addition, these model systems have long life cycles compared to C. elegans, and the available methods to generate polyploid animals are complex and inefficient. Therefore, C. elegans strains derived by this method will be instrumental for furthering any investigation on the effects and roles of whole genome polyploidy in multicellular organisms.
Since a triploid can be derived from a tetraploid by crossing the tetraploid to the original diploid, a comparison between diploids, triploids, and tetraploids that only differ in the number of genome copies, provides a unique and unprecedented opportunity to evaluate equivalent animals/organs/cells with different genome size (or gene dose). The flexibility and ease of the scheme described here has allowed us to generate dozens of tetraploid strains from different genetic diploid backgrounds or karyotypes. Some of these strains were already used to query mechanisms of homologous chromosome pairing and synapsis during meiosis27.
Wild type and mutant tetraploid strains carrying fluorescent markers will provide new avenues of inquiry to understand relationships between genome size and nuclear/cytosol ratio on intracellular/cellular/organ and whole animal scaling, whole genome polyploidization on adaptation, speciation, gene dose and expression, and tissue and organ development. In addition to the study of biological scaling, the generation of tetraploid strains will significantly further queries of fundamental biological questions relevant to extracellular signaling, genome instability, endoreduplication, whole genome duplication, gene dosage, adaptation to stress, development of resistance to drugs, and mechanism speciation.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
We thank the Caenorhabditis Genetics Center (funded by National Institute of Health (NIH) Office of Research Infrastructure Programs P40 OD010440) for strains. The authors would like to thank Baptiste Roelens and Eli Lessman, for providing us with constructive feedback and the Mano Lab at CCNY, CUNY for the use of their laboratory for part of the filming and for their assistance. This work was supported by a PSC-CUNY award TRADB-46-113 and NIH grant 1SC2GM118275-01. M.S. was partially supported by a Canadian Institute of Health (CIHR) postdoctoral fellowship, and E.K. was supported by the NIH/NIGMS RISE grant GM062981.
Materials
| Dissecting Microscope | Motic Microscopy | SMZ171 | |
| Name | Company | Catalog Number | Comments |
| NGM agar plates | |||
| 60 mm x 15 mm Petri Dishes, Sterile | Tritech Research | T3305 | |
| 35 mm x 15 mm Petri Dishes, Sterile | Tritech Research | T3500 | |
| Bacteriological Agar, 5 kg | VWR | 89140-850 | |
| Sodium Chloride, Biotechnology Grade | VWR | 97061-278 | |
| Peptone, BD Bacto | VWR | 90000-368 | |
| Ampicillin Trihydrate | VWR | 97062-300 | |
| IPTG, dioxane-free | Thermo Scientific | R0391 | |
| Name | Company | Catalog Number | Comments |
| Growth Culture | |||
| LB Lennox Broth | IBI Scientific | 89126-176 | |
| Name | Company | Catalog Number | Comments |
| LB Agar plates | |||
| LB Agar Lennox | IBI Scientific | 89126-182 | |
| Name | Company | Catalog Number | Comments |
| Antibiotics | |||
| Ampicillin Trihydrate | VWR | 97062-300 | |
| Tetracycline Hydrochloride | Fisher Scientific | BP912-100 | |
| Name | Company | Catalog Number | Comments |
| Staining | |||
| Hoechst 33258, Pentahydrate | Biotium | 40045 | |
| DAPI | Biotium | 40011 | |
| Name | Company | Catalog Number | Comments |
| Ethanol Fixation | |||
| Ethanol, Pure, 190 Proof (95%), USP | Koptec, VWR | 89125-166 | |
| Name | Company | Catalog Number | Comments |
| M9 Buffer | |||
| Sodium chloride, Biotechnology Grade | VWR | 97061-278 | |
| Potassium phosphate, Monobasic Anhydrous Grade | VWR | 97062-346 | |
| Sodium phosphate, monobasic dihydrate | Fisher Scientific | AC271750025 | |
| Name | Company | Catalog Number | Comments |
| RNAi Clones | |||
| HT115 bacteria expressing rec-8 dsRNA | Source Bioscience | W02A2.6 | |
| HT115 bacteria expressing rec-8 dsRNA | Dharmacon | RCE1182-202299820 |
Referencias
- Ricke, R. M., van Ree, J. H., van Deursen, J. M. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 24, 457-466 (2008).
- Frawley, L. E., Orr-Weaver, T. L. Polyploidy. Curr Biol. 25, R353-R358 (2015).
- Orr-Weaver, T. L. When bigger is better: the role of polyploidy in organogenesis. Trends Genet. 31, 307-315 (2015).
- Otto, S. P. The evolutionary consequences of polyploidy. Cell. 131, 452-462 (2007).
- Davoli, T., de Lange, T. The Causes and Consequences of Polyploidy in Normal Development and Cancer. Annu Rev Cell Dev Biol. 27, 585-610 (2011).
- Adams, K., Wendel, J. Novel patterns of gene expression in polyploid plants. Trends Genet. 21, 539-543 (2005).
- Adams, K. L., Wendel, J. F. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 8, 135-141 (2005).
- Berman, J. Ploidy plasticity: a rapid and reversible strategy for adaptation to stress. FEMS Yeast Res. 16, (2016).
- Levy, D. L., Heald, R. Mechanisms of Intracellular Scaling. Annu Rev Cell Dev Biol. 28, 113-135 (2012).
- Breneman, A., Cande, J., Dunn, J., Burbank, K., O’toole, E. Genome-wide genetic analysis of polyploidy in yeast. Nature. 443, 541-547 (2006).
- Kuznetsova, A. Y., et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 14, 2810-2820 (2015).
- Taylor, J. S., Van de Peer, Y., Meyer, A. Genome duplication, divergent resolution and speciation. Trends Genet. 17, 299-301 (2001).
- Younis, A., Hwang, Y. -. J., Lim, K. -. B. Exploitation of induced 2n-gametes for plant breeding. Plant Cell Rep. 33, 215-223 (2014).
- Ihssen, P. E., McKay, L. R., McMillan, I., Phillips, R. B. Ploidy Manipulation and Gynogenesis in Fishes: Cytogenetic and Fisheries Applications. Trans Am Fish Soc. 119, 698-717 (2011).
- Stanley, J. G., Allen, S. K., Hidu, H. Polyploidy induced in the American oyster, Crassostrea virginica, with cytochalasin B. Aquaculture. 23, 1-10 (1981).
- Eakin, G. S., Behringer, R. R. Tetraploid development in the mouse. Dev Dyn. 228, 751-766 (2003).
- Heier, J., Takle, K. A., Hasley, A. O., Pelegri, F. Ploidy manipulation and induction of alternate cleavage patterns through inhibition of centrosome duplication in the early zebrafish embryo. Dev Dyn. 244, 1300-1312 (2015).
- Arnold, B., Kim, S. -. T., Bomblies, K. Single Geographic Origin of a Widespread Autotetraploid Arabidopsis arenosa Lineage Followed by Interploidy Admixture. Mol Biol Evol. 32, 1382-1395 (2015).
- Session, A. M., et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 538, 336-343 (2016).
- Gallardo, M. H., Bickham, J. W., Honeycutt, R. L., Ojeda, R. A., Köhler, N. Discovery of tetraploidy in a mammal. Nature. 401, 341 (1999).
- Hodgkin, J. Primary sex determination in the nematode C. elegans. Development. 101, 5-16 (1987).
- Madl, J. E., Herman, R. K. Polyploids and sex determination in Caenorhabditis elegans. Genética. 93, 393-402 (1979).
- Nigon, V. Polyploidie experimentale chez un nematode libre, Rhabditis elegans maupas. Bull Biol Fr Belg. 95, 187-225 (1951).
- Woodruff, G. C., Eke, O., Baird, S. E., Félix, M. -. A., Haag, E. S. Insights Into Species Divergence and the Evolution of Hermaphroditism From Fertile Interspecies Hybrids of Caenorhabditis Nematodes. Genética. 186, 997-1012 (2010).
- Mlynarczyk-Evans, S., Roelens, B., Villeneuve, A. M. Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis. PLoS Genet. 9, e1003963 (2013).
- Lozano, E., Sáez, A. G., Flemming, A. J., Cunha, A., Leroi, A. M. Regulation of Growth by Ploidy in Caenorhabditis elegans. Curr Biol. 16, 493-498 (2006).
- Roelens, B., Schvarzstein, M., Villeneuve, A. M. Manipulation of Karyotype in Caenorhabditis elegans Reveals Multiple Inputs Driving Pairwise Chromosome Synapsis During Meiosis. Genética. 201, 1363-1379 (2015).
- Martinez-Perez, E., et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22, 2886-2901 (2008).
- Severson, A. F., Ling, L., van Zuylen, V., Meyer, B. J. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23, 1763-1778 (2009).
- Kamath, R. S., Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30, 313-321 (2003).
- Wood, W. B. . The Nematode Caenorhabditis Elegans. , (1988).
- MacQueen, A. J., et al. Chromosome Sites Play Dual Roles to Establish Homologous Synapsis during Meiosis in C. elegans. Cell. 123, 1037-1050 (2005).
- Martinez-Perez, E., et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22, 2886-2901 (2008).
- Schvarzstein, M., Wignall, S. M., Villeneuve, A. M. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24, 219-228 (2010).
- Kim, S., Spike, C., Greenstein, D. . Germ Cell Development in C. elegans. , 277-320 (2013).
- Peters, N., et al. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J Cell Sci. 123, 795-805 (2010).
- Pasierbek, P. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15, 1349-1360 (2001).
- Hodgkin, J. Primary sex determination in the nematode C. elegans. Development. 101, 5-16 (1987).
- Flemming, A. J., Shen, Z. -. Z., Cunha, A., Emmons, S. W., Leroi, A. M. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc Natl Acad Sci U S A. 97, 5285-5290 (2000).
- Meneely, P. M. Sex determination in polyploids of Caenorhabditis elegans. Genética. 137, 467-481 (1994).
- Whangbo, J. S., Hunter, C. P. Environmental RNA interference. Trends Genet. 24, 297-305 (2008).
- Imae, R., Dejima, K., Kage-Nakadai, E., Arai, H., Mitani, S. Endomembrane-associated RSD-3 is important for RNAi induced by extracellular silencing RNA in both somatic and germ cells of Caenorhabditis elegans. Sci Rep. 6, 28198 (2016).
- Tijsterman, M., May, R. C., Simmer, F., Okihara, K. L., Plasterk, R. H. A. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol. 14, 111-116 (2004).
- Cortes, D. B., McNally, K. L., Mains, P. E., McNally, F. J. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. Elife. 4, e06056 (2015).
- Vargas, E., et al. Autosomal Trisomy and Triploidy Are Corrected During Female Meiosis in Caenorhabditis elegans. Genética. 207, 911-922 (2017).
- Tabara, H., et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 99, 123-132 (1999).
- Hodgkin, J. . Karyotype, ploidy, and gene dosage. , (2005).
- Schoenfelder, K. P., Fox, D. T. The expanding implications of polyploidy. J Cell Biol. 209, 485-491 (2015).
- Levy, D. L., Heald, R. Biological Scaling Problems and Solutions in Amphibians. Cold Spring Harb Perspect Biol. 8, a019166 (2015).
- Taylor, J. S., Van de Peer, Y., Braasch, I., Meyer, A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans Royal Soc B. 356, 1661-1679 (2001).

