Identifying Transcription Factor Olig2 Genomic Binding Sites in Acutely Purified PDGFRα+ Cells by Low-cell Chromatin Immunoprecipitation Sequencing Analysis
Summary
Here we present a protocol which is designed to analyze the genome-wide binding of the oligodendrocyte transcription factor 2 (Olig2) in acutely purified brain oligodendrocyte precursor cells (OPCs) by performing low-cell chromatin immunoprecipitation (ChIP), library preparation, high-throughput sequencing and bioinformatic data analysis.
Abstract
In mammalian cells, gene transcription is regulated in a cell type specific manner by the interactions of transcriptional factors with genomic DNA. Lineage-specific transcription factors are considered to play essential roles in cell specification and differentiation during development. ChIP coupled with high-throughput DNA sequencing (ChIP-seq) is widely used to analyze genome-wide binding sites of transcription factors (or its associated complex) to genomic DNA. However, a large number of cells are required for one standard ChIP reaction, which makes it difficult to study the limited number of isolated primary cells or rare cell populations. In order to understand the regulatory mechanism of oligodendrocyte lineage-specific transcription factor Olig2 in acutely purified mouse OPCs, a detailed method using ChIP-seq to identify the genome-wide binding sites of Olig2 (or Olig2 complex) is shown. First, the protocol explains how to purify the platelet-derived growth factor receptor alpha (PDGFRα) positive OPCs from mouse brains. Next, Olig2 antibody mediated ChIP and library construction are performed. The last part describes the bioinformatic software and procedures used for Olig2 ChIP-seq analysis. In summary, this paper reports a method to analyze the genome-wide bindings of transcriptional factor Olig2 in acutely purified brain OPCs.
Introduction
It is important to study the protein (or protein complex) DNA bindings and the epigenetic marks to build transcriptional regulatory networks involved in various biological processes. Particularly, the bindings of transcription factors to the genomic DNA can play an important role in the gene regulation, the cell differentiation, and the tissue development. A powerful tool for studying the transcriptional regulation and epigenetic mechanisms is chromatin immunoprecipitation (ChIP). Owing to the rapid advancements in the next generation sequencing technology, ChIP coupled with high-throughput DNA sequencing (ChIP-seq) is used for the analyses of protein-DNA bindings and epigenetic marks1. However, a standard ChIP-seq protocol requires about 20 million cells per reaction, which makes the application of this technique difficult when the cell number is limited, such as isolated primary cells and rare cell populations.
Oligodendrocyte lineage cells including oligodendrocyte precursor cells (OPCs) and oligodendrocytes are widely distributed throughout the brain and are essential for the development and function of the brain. As a type of precursor cells, OPCs are capable of both self-renewal and differentiation. OPCs not only serve as progenitors for oligodendrocytes but also play an important role in the propagation of neuronal signaling by communicating with other types of brain cells2. Previous studies have suggested that oligodendrocyte development is regulated by lineage-specific transcription factors such as Olig2 and Sox103,4. These transcription factors were found to bind to the promoter or enhancer regions of some crucial genes to influence their expression during oligodendrocyte specification and differentiation. However, it is challenging to identify DNA binding of protein (or protein complex) of interest in acutely purified primary OPCs with a very limited number of cells.
This protocol describes how to systematically investigate the genomic DNA immunoprecipitated by Olig2 in purified mouse OPCs at genome-wide scale using ChIP-seq technique. OPCs from mouse brains were acutely purified by immunopanning and used in a ChIP experiment without proliferation in vitro. A limited number of OPCs can be obtained by immunopanning and is insufficient for standard ChIP-seq experiments. Herein, a low-cell ChIP-seq protocol with as low as 20 thousand cells per ChIP reaction for transcription factors is described. In brief, cross-linked cells were lysed and sonicated by a sonication device to shear the chromatin. The sheared chromatin was incubated with Olig2 antibody as well as protein A-coated beads to precipitate Olig2 antibody bound genomic DNA. After elution from protein A-coated beads and reverse cross-linking, genomic DNA precipitated by Olig2 antibody was purified by phenol-chloroform extraction. The resulting product was quantified and subjected to T-tailing, primer annealing template switching and extension, the addition of adapters and amplification, library size selection and purification steps for ChIP-seq library construction.
After sequencing, the quality of the raw reads from both the sample prepared with Olig2 transcription factor antibody and the control sample was analyzed. Low-quality base pairs and adapter containing read fragments were trimmed. Next, trimmed reads were aligned to the mouse reference genome. The genomic regions that were significantly enriched for ChIP reads, compared to the control sample, were detected as peaks. Significant peaks, representing potential transcription factor binding sites, were filtered and visualized in a genome browser.
Notably, the method described in this protocol can be broadly used for ChIP-seq of other transcription factors with any cell type of limited numbers.
Protocol
All animal usage and experimental protocols were performed in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Institutional Biosafety Committee and Animal Welfare Committee at the University of Texas Health Science Center at Houston.
1. Purification of PDGFRα Positive Oligodendrocyte Lineage Cells from Mouse Brain (modified from previously described immunopanning protocols5,6,7)
- Preparation of an immunopanning plate for PDGFRα positive cell selection and 2 plates for depletion of endothelial cells and microglia.
NOTE: Please note that Petri dishes but not cell culture dishes work for immunopanning experiment; if the purified cells will be used for culturing, immunopanning steps must be carried out in the biosafety cabinet- Coat a 10 cm Petri plate with 30 µL goat anti-rat IgG in 10 mL of pH 9.5, 50 mM Tris-HCl overnight at 4 °C. Agitate the plate to make sure the surfaces of the plates are evenly and entirely covered by the coating solution.
- Prepare PDGFRα antibody solution by diluting 40 µL of rat anti-PDGFRα antibody with 12 mL of Dulbecco's phosphate-buffered saline (DPBS) containing 0.2% BSA.
- After 3 washes of IgG coated plate with 10 mL 1x DPBS each, incubate IgG-coated plate with PDGFRα antibody solution at room temperature for 4 h.
- Wash the rat anti-PDGFRα antibody coated plate 3 times with 10 mL 1x DPBS each. Gently add DPBS solution along the side wall of the plate and do not disturb the coated surfaces.
- Coat 2 new 15 cm Petri plates for depletion of endothelial cells and microglia with 20 mL of DPBS containing 2.3 µg/mL of Banderiaea simplicifolia lectin 1 (BSL-1) for 2 h.
- Wash the BSL-1 coated plates 3 times with 20 mL 1x DPBS. Gently add DPBS solution along the side wall of the plates and do not disturb the coated surfaces.
- Purification of PDGFRα positive oligodendrocyte lineage cells is modified from previously published methods6,7,8.
- Dissect the cortical tissues from 2 postnatal day 7 (P7) mouse brains according to previously published protocols5,6.
- Dissociate the tissues to generate a single-cell suspension with neural tissue dissociation Kit (P) according to detailed manufacturer's instructions.
- Briefly, cut the dissected cortical tissues into pieces with a scalpel and subject them to enzymatic digestion at 37 °C. After digestion, manually dissociate the pieces with Fire-polished glass Pasteur pipettes into a single-cell suspension.
- Centrifuge the single-cell suspension at 300 x g for 10 min at room temperature and suspend cell pellet using 15 mL of immunopanning buffer (immunopanning buffer is DPBS with 0.02% BSA and 5 µg/mL insulin).
- Incubate the single-cell suspension from 2 mouse brains sequentially on 2 BSL-1 coated plates for 15 min at room temperature with gentle agitation of the plate every 5 min to ensure a better depletion of microglia and endothelial cells.
- Gently swirl the plate to collect the non-adherent cells in the cell suspension and incubate them on the rat-PDGFRα antibody-coated plate for 45 min at room temperature.
- After the incubation of the cell suspension on the rat-PDGFRα antibody-coated plate, gently swirl the plate to collect the cell suspension, and rinse the plate 8 times with DPBS to get rid of non-adherent cells. Gently add wash solution along the side wall of the plate and agitate the plate several times to get rid of non-adherent cells.
- Detach the cells from the rat-PDGFRα antibody coated plate using a 4 mL of cell detachment solution treatment for 10 min at 37 °C. Shake the plate to dislodge adherent cells.
- Collect the purified OPCs by centrifugation at 300 x g at room temperature, suspend the cell pellet with 2 mL OPC cell culture medium and count the cells by using trypan blue and a hemocytometer (500 mL cell culture medium is DMEM/F12 medium containing 5 mL penicillin-streptomycin solution (P/S), 5 mL N2, 10 mL B27, 5 µg/mL insulin, 0.1% BSA, 20 ng/mL bFGF and 10 ng/mL PDGFRα).
- Validation of the purity of OPCs after immunopanning.
- In order to evaluate the enrichment of OPCs after immunopanning, use some of the purified OPCs for RNA extraction with a guanidium thiocyanate based extraction according to manufacturer's instructions.
- Perform qRT-PCR by using fluorescent green dye master mix to check for the enrichment of PDGFRα expression in purified OPCs as compared with dissociated brain cells according to the previously published materials4.
- Additionally, seed some purified OPCs into the Poly-D-Lysine coated 24 well plates for immunostaining with anti-NG2 chondroitin sulfate proteoglycan (NG2) antibody as previously published materials9.
2. Low-cell ChIP Preparation and ChIP Library Construction for High-Throughput Sequencing
- Olig2 Low-cell ChIP preparation with a commercially available sonication system and a commercially available low cell number ChIP kit (see Table of Materials) by following the detailed standard procedures from manufacturer's instructions.
- After detaching from the rat anti-PDGFRα antibody-coated plate and the cell counting with trypan blue and a hemocytometer put 20,000 purified OPCs in 1 mL OPC cell culture medium for each ChIP reaction.
- Add 27 µL of 36.5% formaldehyde to fix cell suspension for 10 min at room temperature.
Caution: Please note that formaldehyde must be used in the chemical fume hood for safety reasons. - Stop DNA-protein cross-linking with 50 µL 2.5 M glycine for 5 min at room temperature.
NOTE: All steps must be carried out on ice or in 4 °C cold room from this point. - Wash the cross-linked cell pellets with 1 mL of ice-cold Hanks' Balanced Salt Solution (HBSS) with protease inhibitor cocktail and get cells pelleted by a pre-cooled centrifuge at 300 x g at 4 °C.
- Lyse the cell pellet in 25 µL complete Lysis Buffer for 5 min on ice.
NOTE: Agitate the tube to suspend the cells in Lysis Buffer. - Supplement the cell lysate with 75 µL ice-cold HBSS containing protease inhibitor cocktail and shear the chromatin of cell lysate by pre-cooled sonication system with 5 cycles of 30 s ON and 30 s OFF program. Always sonicate 6 tubes together and make sure the balance tubes contain 100 µL water.
- After shearing, centrifuge at 14,000 x g for 10 min 4 °C and collect supernatant in a new tube after centrifugation.
- Dilute 100 µL of sheared chromatin with the equal volume of ice-cold complete ChIP Buffer with the protease inhibitor.
- Save 20 µL of the diluted sheared chromatin as Input control sample.
NOTE: Input control sample is also required as the comparison to Olig2 immunoprecipitated sample to identify Olig2 antibody immunoprecipitated DNA. - Add 1 µL of rabbit anti-olig2 antibody to 180 µL of the diluted sheared chromatin and incubate the reaction tube on a rotating wheel at 40 rpm for 16 h at 4 °C.
NOTE: Perform this step in a cold room. - For each ChIP reaction, wash 11 µL magnetic protein A-coated beads with 55 µL Beads Wash Buffer and put the bead pellet in 11 µL of bead wash buffer after 2 washes.
NOTE: Perform this step in a cold room. - For each ChIP reaction, add 10 µL of pre-washed Protein A-coated beads to ChIP reaction tube. Perform this step in a cold room.
- Incubate the ChIP reaction tube at 4 °C for another 2 h on a rotating wheel.
- Put the ChIP reaction tube on the magnetic rack for 1 min.
NOTE: Perform this step in a cold room. - Remove the supernatant and don't dislodge the bead pellet.
- Wash the bead pellet with 100 µL for each of 4 wash buffers respectively for 4 min on a rotating wheel at 4 °C.
- After washes, add 200 µL of elution buffer to the bead pellet and incubate the ChIP reaction tube at 65 °C for 4 h to reverse cross-link protein-DNA.
- Additionally, add 180 µL of elution buffer to 20 µL Input control sample and incubate at 65 °C for 4 h to reverse cross-link protein-DNA as well.
- Add 200 µL 25:24:1 (v/v) mixture of phenol, chloroform, and isoamyl alcohol to each ChIP reaction tube.
- Vortex vigorously for 1 min and centrifuge at 13,000 x g for 15 min at room temperature. Transfer the upper phase to a new tube.
- Add 40 µL 3 M sodium acetate solution, 1,000 µL of 100% ethanol and 2 µL of glycogen precipitant covalently linked to a blue dye to each ChIP reaction tube overnight at -20 °C.
- Centrifuge at 13,000 x g for 20 min at 4 °C.
- Discard the supernatant, wash with 500 µL cold 70% ethanol and centrifuge at 13,000 x g for 20 min at 4 °C.
- Discard the supernatant, keep the pellet air dry for 10 min and dissolve the pellet with 50 µL water.
- ChIP library construction for high-throughput sequencing
- Clean and concentrate the ChIP DNA with a clean-up kit according to the manufacturers' instructions.
- Quantify the ChIP sample by a pico-green according to the manufacturer's instructions.
- Use a ChIP-seq kit for T-tailing, replication and tailing, template switching and extension, the addition of adapters and amplification, library size selection and purification according to the manufacturer's instructions.
- After denaturation of dsDNA, dephosphorylate the 3' end of ssDNA by shrimp alkaline phosphatase and add a poly (T) tail to the ssDNA by terminal deoxynucleotidyl transferase.
- Anneal the DNA Poly (dA) Primer to the ssDNA template for the DNA replication and template switching.
- After template switching, amplify the ChIP-seq library by PCR with the forward and reverse primers for indexing.
- Select the PCR amplified ChIP-seq library with fragments ranging from 250 to 500 bp by paramagnetic beads by option 2 for double size selection.
- Examine the quality of the selected ChIP-seq library using a microfluidic chip-capillary electrophoresis device.
3. Data Analysis
- Prepare directory structure.
- Create a directory for performing the ChIP-seq data analysis. Inside the newly created directory, create six subdirectories with the following names: raw.files, fastqc.output, bowtie.output, homer.output, macs2.output, motif.analysis, and reports.
- Prepare data and perform quality control analysis of raw sequence data.
- Move to the "raw.files" directory and download the ChIP-seq data files.
- Verify the filenames. If the sequencing was paired-end, there will be 4 files (two for treatment and two for input) with similar base names: PDGFRα_Olig2.read1.fastq, PDGFRα_Olig2.read2.fastq, PDGFRα_input.read1.fastq, and PDGFRα_input.read2.fastq. If the filenames are different, rename them using the "mv" command.
- Move to the "fastqc.output" directory.
- Obtain quality control metrics from the raw reads using a fastq file quality control software10. Use the command in Figure S1A to run the quality control process separately for each fastq file. The software will display several quality control metrics in an html file.
- Open the html quality control file and verify the number of reads, read length, per base sequence quality, per sequence quality scores, sequence GC content, sequence duplication levels, adapter content and kmer content.
- Trim trailing low-quality reads and adapter content.
- Observe the "Per base sequence quality" and "Adapter Content" plots. Determine the trimming length for both the head and tail of each read. An adequate length for trimming is where the base quality falls below 30 and there is evidence of adapter content.
- Move to the "raw.files" directory.
- Download and install a software for trimming reads11. Use the command in Figure S1B for trimming both the treatment and the input separately. Parameter 'CROP' indicates the remaining read length after trimming bases from the end and 'HEADCROP' specifies the number of bases to be eliminated from the start of the read.
- The trimming command also includes the minimum read length to be accepted after filtering in the parameter 'MINLEN'. If reads are at least 50 bp, use 35 bp as the read length threshold.
- Move to the "fastqc.output" directory.
- Perform the fastq file quality control analysis after trimming the reads. Verify that the quality control issues have been resolved. Use the command in Figure S1C to obtain the quality control metrics.
- Align the single-end or paired-end ChIP-seq reads to the mouse reference genome.
- Move to the "bowtie.output" directory for mapping.
- Download the GENCODE mm10 mouse reference genome. Rename the mm10 mouse reference genome as "mm10.fa".
- Download a read mapper12 and install it in the system.
- Create an index file of the downloaded reference genome using the command in Figure S2A. Six files are automatically created: mm10.1.bt2, mm10.2.bt2, mm10.3.bt2, mm10.4.bt2, mm10.rev.1.bt2, and mm10.rev.2.bt2.
- Use the command in Figure S2B to execute the alignment and adjust parameter "-p" to the number of processing cores according to your system settings. If running paired-end mode, parameters "-1" and "-2" indicate the names of the trimmed fastq files.
NOTE: Mapping is performed separately for the treatment and input samples and output metric log files are created for each sample. - Download a software for manipulating files in SAM format13 and install it in the system. Convert the aligned SAM file into a BAM file using the command in Figure S2C.
- Obtain quality control metrics of mapped reads before peak calling for both paired-end treatment and control samples.
- One of the most important quality control metrics to address is the sequencing depth. Open the files "log.bowtie.PDGFRα_Olig2.txt" and "log.bowtie.PDGFRα_input.txt " in the bowtie.output directory and verify that the number of uniquely mapped read pairs in each sample is greater than 10 million.
- Move to the "homer.output" directory.
- Download a software for motif discovery and next-generation sequencing analysis14 and install it in the system.
- Create a directory named "tagDir".
- To verify tag clonality, use the command depicted in Figure S3A. The "makeTagDirectory" command will generate four quality control output files:tagAutocorrelation.txt, tagCountDistribution.txt, tagInfo.txt, and tagLengthDistribution.txt.
- Move to the "tagDir" directory.
- Copy the "tagCountDistribution.txt" file into the "reports" directory.
- Move to the "reports" directory.
- Open the tagCountDistribution.txt file using a spreadsheet program and create a bar plot of the number of tags per genomic position. Store the histogram file with the name "tag.clonality.xlsx".
- Install R programming language15 in the system.
- In the terminal, type 'R' and press ENTER to access the R programming environment.
- Download an R package for processing ChIP-seq data16 and install it using the command depicted in Figure S3B.
- Use the R script in Figure S3C to plot the strand cross-correlation.
- Detection of peaks using a peak mapper.
- Move to the "macs2.output" directory. Download and install a peak mapper17 in your system.
- Use the function "callpeak" with the treatment and control BAM files generated in Step 3.4.6.
NOTE: Other parameters include "-f" for input file format, "-g" for genome size, "-name" for indicating the base name of all output files, and "-B" for storing the fragment pileup in bedgraph format. The complete peak calling command is included in Figure S4A. Use BEDPE for paired-end samples. - Verify that the peak mapper generated six files: PDGFRα_Olig2_vs_PDGFRα_input_peaks.xls, PDGFRα_Olig2_vs_PDGFRα_input_peaks.narrowPeak, PDGFRα_Olig2_vs_PDGFRα_input_summits.bed, PDGFRα_Olig2_vs_PDGFRα_input_model.r, PDGFRα_Olig2_vs_PDGFRα_input_control_lambda.bdg, and PDGFRα_Olig2_vs_PDGFRα_input_treat_pileup.bdg.
- Open file PDGFRα_Olig2_vs_PDGFRα_input_peaks.xls to view the called peaks, location, length, summit position, pileup, and the peak enrichment metrics: -log10(pvalue), fold enrichment, and -log10(q-value).
- Filter and annotate called peaks.
- Using the file opened in the previous step, filter resulting peaks according to the fold enrichment, the p-value and/or the q-value. To decide adequate filtering thresholds, make a histogram plot to determine the density of each metric. Filter out values below a selected threshold to obtain significant peaks.
- Download the mm10 blacklist (regions are known to have artificially high signal) and filter out the ChIP-seq peaks which are inside any of those artifact regions.
- Create a bed file with the filtered peaks containing the following columns: chromosome, start, end, peakID, mock, and strand. Fill the mock column with a dot "." since it is not used. In the strand column, set all values to "+". Save the bed file as "filtered.peakData2.bed".
- Copy the "filtered.peakData2.bed" file in the "reports" directory.
- Move to the "reports" directory.
- To annotate filtered peaks to a specific gene region, use the function "annotatePeaks" with the bed file created in the previous step. The command used is shown in Figure S5A.
- Open the "filtered.annotatedPeaks.txt" file using a statistical software.
- Filter resulting peaks lying in an intergenic region at a distance greater than 5 kb from an annotated TSS. Use the "Distance to TSS" column in the file as the distance for filtering. Store the filtered peaks in an excel file named "5kbup.geneBody.peakData.txt".
- Store the resulting filtered peaks in a bedGraph file with columns: chromosome, start, end, and -log10(q-value). Name the file "5kbup.geneBody.peakData.bedGraph".
- Generate bigwig files for browser visualization.
- Download and install a software for genome arithmetic18.
- Download a tool for converting bedgraph to bigwig and install in the system. Download the 'mm10.chrom.sizes' file.
- Use the command in Figure S6A to generate a bigwig file.
- Download and install a genome browser19 in the system.
- Open the genome browser and load the file "5kbup.geneBody.peakData.bigwig" to visualize the filtered significant peaks and their location with respect to known genes.
- Motif Search.
- Move to the "motif.analysis" directory.
- Download a de novo motif scanning and motif enrichment program and install it into your system20.
- Download a comprehensive motif database that includes motifs of Olig2.
- Obtain the genomic sequences of 500 bp regions centered at significant peak summits. Store the sequences as peak.sequences.txt. Use the command in Figure S7 to search for Olig2 motifs within each of the 500 bp peak regions.
- Filter the resulting motifs using an adequate E-value (default = 0.05).
Representative Results
Low-cell ChIP-seq was performed and bioinformatic analyses were done to investigate the potential interactions of transcriptional factor Olig2 with genomic DNA in acutely purified brain OPCs. Figure 1 shows a general workflow of both the experimental and the data analysis procedures. In this protocol, postnatal mice brains were dissociated into a single-cell suspension. After tissue dissociation, immunopanning was performed to purify OPCs by using PDGFRα antibody. Figure 2 shows the significant enrichment of PDGFRα, little expression of neuron marker Tuj1 (neuron-specific class III beta-tubulin), astrocyte marker GFAP (glial fibrillary acidic protein), microglia marker ionized calcium binding adaptor molecule 1 (Iba1), myelinating mature oligodendrocytes marker myelin basic protein (Mbp) and myelin-oligodendrocyte glycoprotein (Mog) from purified OPCs as evaluated by RT-qPCR. Additionally, immunostaining result indicates the majority of cells are NG2 positive as shown in Figure 2. Subsequently, 20 thousand purified OPCs per reaction were fixed by formaldehyde and chromatin was sheared by using a sonication system. Olig2 low-cell ChIP was performed and the library was constructed. After ChIP library preparation, the quality of the generated product was validated by a microfluidic chip-capillary electrophoresis device. Figure 3 shows an example electropherogram of an Olig2 low-cell ChIP library. A clear peak of Oligo2 library product ranging from 250 to 500 bp should be visible after size selection of library PCR product. The ChIP-seq libraries of both the sample prepared with Olig2 transcription factor antibody and the control sample were subjected to high-throughput sequencing to produce paired-end 50 cycle sequencing reads. Longer read lengths will improve the mapping rates and reduce the probability of multi-mapping, at the expense of sequencing costs. With 50 bp ChIP-seq paired-end sequencing libraries, good results are usually achieved.
After the initial quality control assessment of raw reads and trimming of adapters and trailing low-quality base pairs, the quality control plots are depicted in Figure 4. Trimmed reads should have a base quality greater than 30, no adapter sequences, and have a low duplication rate (Figure 4B-D). Good quality trimmed reads will increase the overall alignment rate. Other parameters such as GC content are also important and should be considered for trimming.
Once the sequencing samples are aligned, it is necessary to verify the sequencing depth. It is known that the number of called peaks will increase with a higher sequencing depth since weak sites will become statistically more significant21. A saturation analysis is required to determine a sufficient sequencing depth for each specific transcription factor used, at the expense of time and budget. A consensus sequencing depth for transcription factor binding has been suggested by the ENCODE consortium regarding mammalian samples: a minimum of 10 million uniquely mapped reads for each of at least two biological replicates22. The number of uniquely mapped reads and the overall alignment rate were calculated by the read mapper. An example of good mapping metrics is shown in Figure 5A. Aligned reads should be mapped to distinct genomic locations, indicating an adequate library complexity, also referred to as tag clonality. The ENCODE consortium suggests that at least 80% of the 10 million of aligned reads should be mapped to different genomic regions22. Figure 5B shows an adequate tag clonality in which more than 90% of the reads are mapped to only one genomic location. Low-complexity libraries generally occur when not enough DNA is obtained, and the PCR amplified fragments are sequenced repeatedly. A low-complexity library will yield a high false peak detection rate.
When using paired-end ChIP-seq data, fragments bind around the transcription factor with a certain distance of separation. Paired-end sequencing will allow for a more accurate estimation of the mean fragment length yielding a better estimation of the genomic regions where more binding occurred. The strand correlation plot depicts a peak of enrichment corresponding to the predominant fragment length. As observed in Figure 5C, the calculated fragment length is 130 bp whereas the read length is 50 bp. A ChIP-seq dataset where the fragment length is longer than the read length is indicative of high quality16.
The output of peak calling is a list of genomic regions likely to be bound by the transcription factor (or its complex). The list of genomic regions or peaks is included in a "bed" file which may be viewed in a genome browser. For each peak, this file contains a peak ID, the genomic location of the peak summit, and the FDR as -log10 (q-value). The significantly enriched peaks are those with higher fold-enrichment and significance metrics. An example of the output peak file is shown in Figure 6A. After selecting significant peaks using custom thresholds, filtering peaks within ENCODE's blacklist23, and identifying peaks within a gene's promoter region (5 kb upstream and whole gene body), a "bigwig" file is useful for viewing peaks in a genome browser. Most enriched peak regions have a higher probability of transcription factor binding. It is important to confirm peak presence in known transcription factor regulatory regions as well as peak absence in regions where transcription factor binding is unlikely. Figure 6B-E shows examples of gene regions with and without peak enrichment as well as their location with respect to ENCODE promoter regions.
In silico complementary methods to ChIP-seq experiment are motif enrichment analysis and de novo motif search. Since Olig2 binding in OPCs has not been studied before, Olig2 ChIP-seq derived peaks in motor neuron progenitor cells and embryonic stem cells were used for de novo motif identification4,24. Olig2 de novo identified motifs were added to the comprehensive ENCODE motif database25 and motif enrichment analysis was performed. High enrichment of the de novo identified OLIG2 motifs and known transcription factor (HDAC2, SP1, FOXP1, NR3C1, NFKB2/4, SMAD2/3, PAX5, and ASCL1) motifs were found in ChIP-seq peaks of PDGFRα purified cells. An enrichment of 30 de novo identified motifs with an E-value < 0.05 was discovered. Figure 7 shows the top two de novo identified motifs of Olig2. These two de novo identified Olig2 motifs were found in >60% of the ChIP-seq peaks obtained from PDGFRα purified cells, in addition to known motifs.
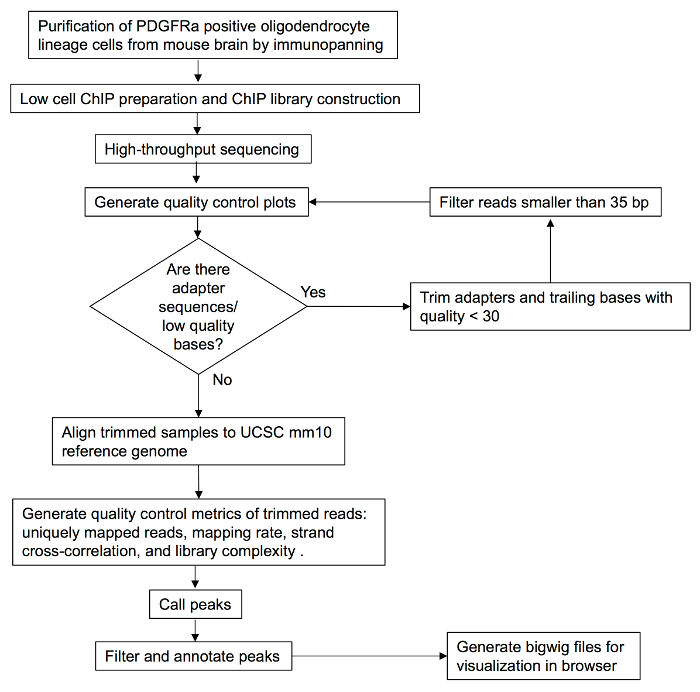
Figure 1: Overview of the protocol workflow. PDGFRα positive OPCs were isolated from postnatal mouse brains and were subjected to Olig2 ChIP experiment. The precipitated DNA fragments were used for preparing ChIP library. After assessment of library quality, samples were used for sequencing. Data sets were analyzed, and peaks were identified, indicating potential Olig2 binding sites in OPC purified cells. Please click here to view a larger version of this figure.
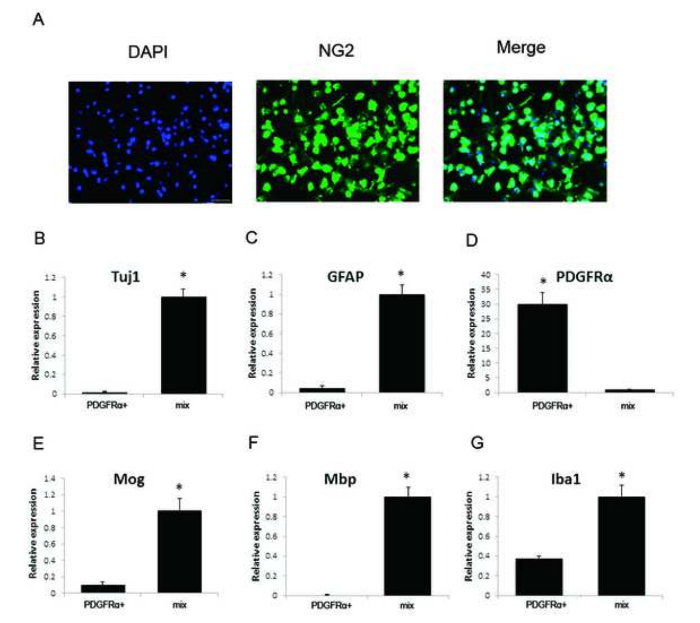
Figure 2: Evaluation of the purity of immunopanned OPCs by qPCR and immunostaining. (A) PDGFRα antibody immunopanned cells were seeded and immunostaining was performed by using OPC lineage marker NG2 antibody. Blue: DAPI, Green: NG2. Scale bar = 10 µm. (B) The relative expression level of neuron marker Tuj1 in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) were evaluated. Tuj1 expression in dissociated brain cells was set to 1. (C) The relative expression level of astrocyte marker GFAP in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) was evaluated. GFAP expression in dissociated brain cells was set to 1. (D) The relative expression level of OPC marker PDGFRα in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) was evaluated. PDGFRα expression in dissociated brain cells was set to 1. (E) The relative expression level of myelinating mature oligodendrocytes marker Mog in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) was evaluated. Mog expression in dissociated brain cells was set to 1. (F) The relative expression level of myelinating mature oligodendrocytes marker Mbp in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) was evaluated. Mbp expression in dissociated brain cells was set to 1. (G) The relative expression level of microglia marker Iba1 in purified OPCs (PDGFRα+) and in dissociated brain cells (mix) was evaluated. Iba1 expression in dissociated brain cells was set to 1. Data in B-G represent triplicate experiments and error bars indicate standard error. T-test analysis * P <0.05. Please click here to view a larger version of this figure.
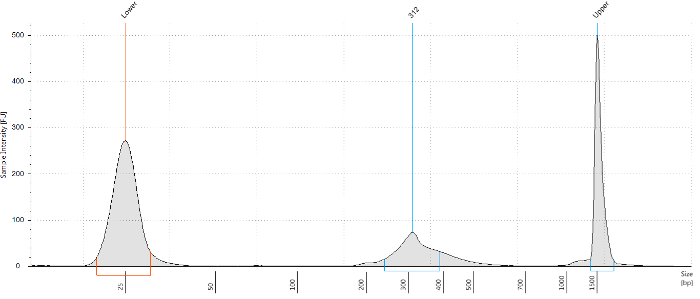
Figure 3: An example electropherogram of a library from Olig2 low-cell ChIP. After double size selection, the quality of Olig2 ChIP library was analyzed by a microfluidic chip-capillary electrophoresis device. Please click here to view a larger version of this figure.
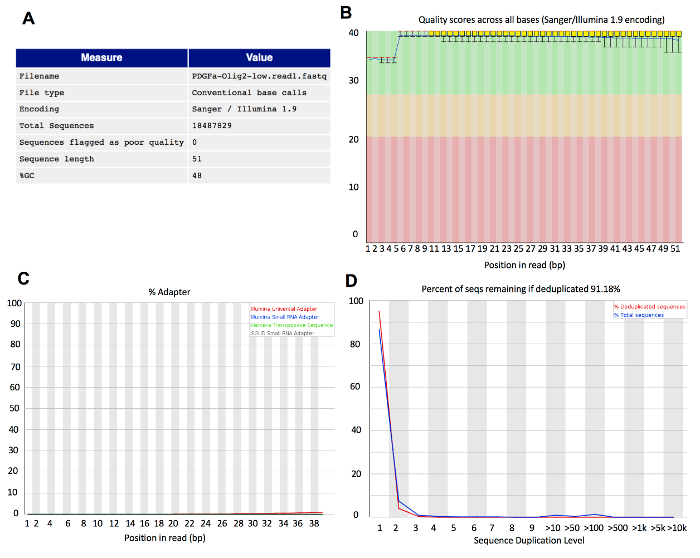
Figure 4: Representative quality control results for a 50 bp read length low-cell ChIP-seq sample. (A) Summary table depicting the total number of reads, number of poor quality reads, sequence length, and overall GC content. (B) Plot indicating the distribution of base quality scores at different positions in the reads. (C) Plot showing the potential adapter content at different positions in the reads. (D) Line plot indicating the percent of duplicated sequences. The majority of reads originate from sequences which only occur once within the library, therefore indicating a low duplication rate or high library complexity. Please click here to view a larger version of this figure.
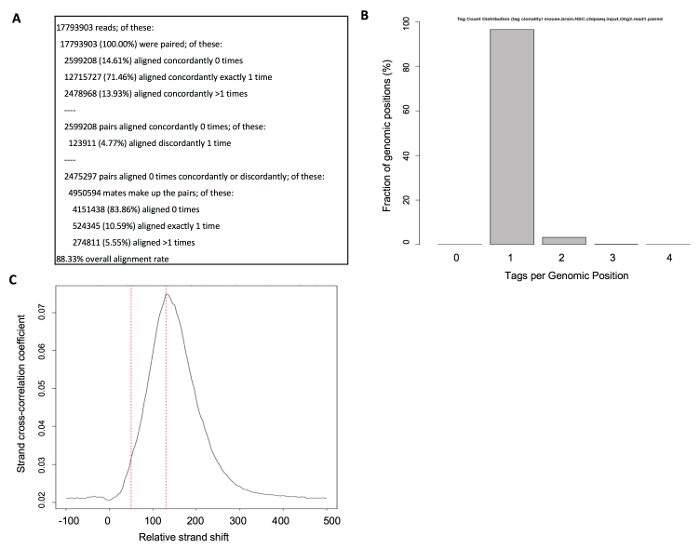
Figure 5: Quality control metrics obtained before peak calling: bowtie2 alignment metrics, strand cross-correlation, and tag clonality. (A) Alignment metrics obtained from the read mapper. The most important metrics are the number of uniquely mapped read pairs, indicated as "aligned concordantly exactly 1 time", and the overall alignment rate. (B) Histogram showing the tag clonality. Bars indicate the percent of genomic positions where tags were found. (C) An example of a cross-correlation plot indicating good quality data. Red lines depict the read length at 50 bps and the predominant fragment length at 130 bps. Please click here to view a larger version of this figure.
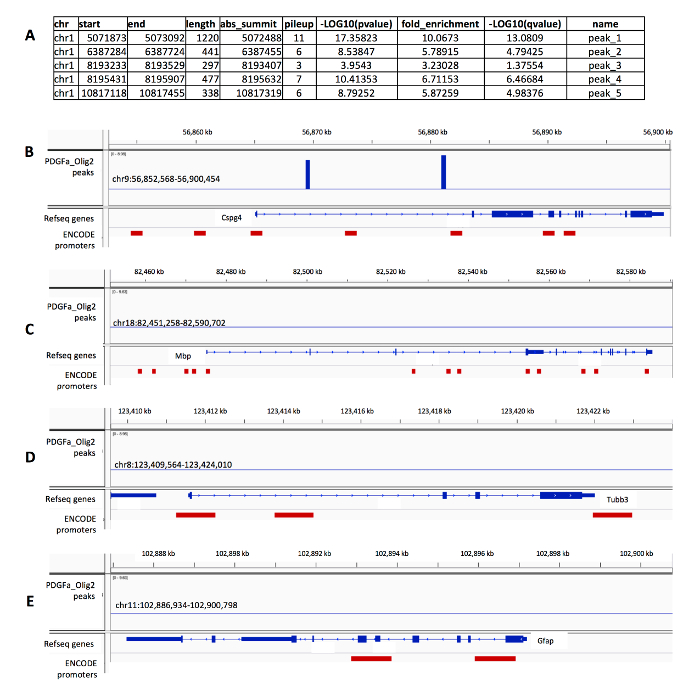
Figure 6: ChIP-seq peak regions and genome browser views of significant ChIP-seq peaks at different genomic locations. (A) Example of the peak regions identified with the peak caller. (B) Significant ChIP-seq peaks found in the gene body of Cspg4, a known OPC marker gene. (B) No ChIP-seq peaks were found either in the promoter regions or within the gene bodies of Mbp gene, a marker for mature oligodendrocytes, (C) Tubb3, a neuronal cell marker, or (D) GFAP, a cell marker of astrocytes. Please click here to view a larger version of this figure.
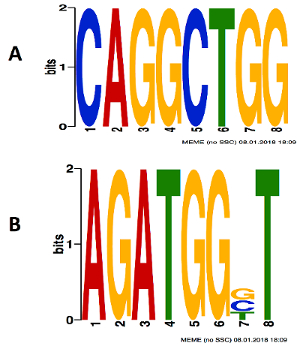
Figure 7: Highly enriched de novo identified motifs of Olig2 found in PDGFRα-Olig2 ChIP-seq peaks. Please click here to view a larger version of this figure.
Figure S1: Preparation of ChIP-seq data sets. (A) Directory architecture definition. (B) Calculating raw read quality control metrics. (C) Trimming of reads. Please click here to download this file.
Figure S2: Alignment of reads to the reference genome. Please click here to download this file.
Figure S3: Quality control of aligned reads, to address strand cross-correlation and determine tag clonality. Please click here to download this file.
Figure S4: Peak calling. Please click here to download this file.
Figure S5: Annotation of significant peaks. Please click here to download this file.
Figure S6: Conversion of bedGraph file format to bigwig for peak visualization in the genome browser. Please click here to download this file.
Figure S7: De novo motif search and motif enrichment. Please click here to download this file.
Figure S8: Flow-chart describing the bioinformatics analysis of ChIP-seq data. Please click here to download this file.
Discussion
Mammalian gene regulation networks are very complex. ChIP-seq is a powerful method to investigate genome-wide protein-DNA interactions. This protocol includes how to perform Olig2 ChIP-seq by using a low number of purified OPCs from mouse brains (as low as 20 thousand cells per reaction). The first key step for this protocol is the purification of OPCs from mouse brains by immunopanning with PDGFRα antibody. For positive selection of OPCs with PDGFRα coated plates, OPCs often weakly bind to the PDGFRα coated plates. Even after several washes, there are still some non-adherent cells left. It is important to check the non-adherent cells after each D-PBS wash under a microscope. If more D-PBS wash does not help to reduce the number of non-adherent cells, it is time to collect the selected cells, since excess washes might dislodge attached OPCs and reduce the yield of purified cells. However, insufficient washes may result in the contamination of other brain cells types. Therefore, every time after isolation, it is necessary to evaluate the purity of the isolated OPCs by immunostaining with OPCs marker NG2 qPCR for the expression of known cell-type specific genes such as Tuj1 for neurons, GFAP for astrocytes, PDGFRα for OPCs, Iba1 for microglia, and MOG and MBP for myelinating mature oligodendrocytes. As shown in Figure 2, immunostaining result indicates that the majority of the purified cells are positive for NG2 staining. In addition, some classic cell-type specific markers for neurons, astrocytes, microglia and myelinating mature oligodendrocytes exhibit undetectable or extremely low expression levels in purified OPCs when compared to the unpurified brain cell mixture. PDGFRα exhibits high expression level in purified OPCs when compared to the unpurified brain cell mixture. Using immunopanning method, microglia are greatly reduced in the purified OPCs compared to the unpurified brain cell mixture, but a small amount of contamination exists. This observation is consistent with previous publications by others7,8,26. Additionally, when compared with previously published reports, a commercially available kit is used in this protocol for the standardized, efficient and convenient dissociation of brain tissues into single-cell suspensions.
After cross-linking of purified OPCs, the cross-linked cells should be washed with ice-cold HBSS solution and the remaining ChIP steps should be carried out at 4 °C, otherwise the antibody cannot precipitate the genomic DNA properly.
The next key step in this protocol is library preparation. The PCR amplification of ChIP material was used for library construction. The number of PCR cycles for library preparation depends on the amount of starting DNA. A good library preparation requires accurate quantification of input DNA amount. Too many or too few PCR cycles can influence library concentration as well as the complexity, leading to PCR artifacts.
It is challenging to construct ChIP-seq library when using a limited number of cells for immunoprecipitation27. The previously used standard ChIP protocol requires 10 ng of immunoprecipitated DNA for ChIP-seq library preparation1,28. However, the protocol described here normally generates 2 ng of immunoprecipitated DNA by Olig2 antibody from 20,000 OPCs. DNA is used to prepare the ChIP-seq library by a single-step adapter addition method, which enables enhanced sensitivity allowing picograms of immunoprecipitated DNA to be amplified. By combining commercial kits, this protocol provides a practical solution to perform ChIP-seq for transcriptional factors based on a small number of cells.
Importantly, the low cell ChIP-seq protocol described is applicable for ChIP-Seq of other transcription factors in primary cells or rare cell populations as well. Antibodies used in this protocol need to be experimentally validated to be specific by IP experiment first. Non-specific binding of antibodies will lead to poor results. Additionally, it is preferable to carry out this low cell ChIP-seq experiment in cells with a high expression level of the transcription factor of interest.
For data analysis, it may be challenging to decide which values to use as filtering cut-offs after peak calling. Depending on the objectives of the analysis, stringent or more relaxed parameters may be used. Generally, a combination of fold-enrichment and either p-value or FDR is used. If some binding sites of the transcription factor under study were previously known, this may help in determining the filtering thresholds. Transcription factor binding site databases derived from publicly available ChIP-seq experiments in different cell lines and conditions have been compiled29,30. One could search for a gene and check if transcription factor binding sites have been previously found. However, it is important to be aware that transcription factor binding sites differ depending on cell types and conditions.
In silico complementary methods to ChIP-seq experiment are motif enrichment analysis and de novo motif search using MEME-ChIP20 or similar programs. The motif search requires a comprehensive known and de novo derived motif database such as the ENCODE motifs25 or the Hocomoco database31. Hocomoco is a database of motifs discovered from publicly available human and mouse ChIP-seq experiments. If motifs for the assayed protein are unknown, the de novo motif search could reveal repetitive sequence patterns in a significant fraction of the peaks as new motifs. The potential combinatorial action of transcription factors may be unveiled when motifs of other transcription factors are also found to be enriched in the resulting peaks.
The enriched peak regions may be experimentally validated by using the chromatin of mutant or knockout cells as controls. Performing ChIP-seq using cells not expressing the transcription factor are used for identifying false positive peaks, also denoted as "phantom peaks"32. False positives should be filtered out.
It is also interesting to compare the resulting ChIP-seq peaks with transcriptomic data. The PDGFRα-Olig2 ChIP-seq peaks were compared with the expression of genes in OPCs from a previous publication18. This strategy may be limited since expressed genes in OPCs may be controlled by mechanisms other than Olig2 transcription factor binding. Additionally, Olig2 transcription factor may bind to a gene's regulatory region but the gene might have low gene expression level due to possible inhibitory mechanisms or the lack of co-factors necessary for gene transcription.
ChIP-seq is a method used for identifying genome-wide DNA binding sites of transcription factors and other proteins. However, through analysis of co-occurrence of transcription factors, researchers found that transcription factors tend to associate with other proteins forming co-regulatory modules33. This means some bindings between transcriptional factors and genomic DNA revealed by ChIP-seq are indirect and bridged by other proteins. Therefore, to achieve more meaningful results researchers should consider studying more than one transcription factor simultaneously.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
JQW, XD, RCDD, and YY were supported by grants from the National Institutes of Health R01 NS088353; NIH grant 1R21AR071583-01; the Staman Ogilvie Fund-Memorial Hermann Foundation; the UTHealth BRAIN Initiative and CTSA UL1 TR000371; and a grant from the University of Texas System Neuroscience and Neurotechnology Research Institute (Grant #362469).
Materials
| Reagent/ Equipment | |||
| Banderiaea simplicifolia lectin 1 | Vector Laboratories | # L-1100 | |
| Rat anti-PDGFRa antibody | BD Bioscience | # 558774 | |
| Neural tissue dissociation Kit (P) | MACS Miltenyi Biotec | # 130-092-628 | |
| Accutase | STEMCELL technologies | # 07920 | |
| TRIzol | Thermo Fisher | # 15596026 | |
| Anti-NG2 Chondroitin Sulfate Proteoglycan Antibody | Millipore | # AB5320 | |
| Bioruptor Pico sonication device | Diagenode | # B01060001 | |
| True MicroChIP kit | Diagenode | # C01010130 | |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | Thermo Fisher | # 15593031 | |
| NucleoSpin Gel and PCR Clean-Up kit | MACHEREY-NAGEL | # 740609 | |
| Quant-iT PicoGreen dsDNA Assay Kit | Thermo Fisher | # P11496 | |
| DNA SMART ChIP-Seq kit | Clontech Laboratories | # 634865 | |
| Agencourt AMPure XP | Beckman Voulter | # A63880 | |
| GlycoBlue | Thermo Fisher | # AM9516 | |
| pico-green | Thermo Fisher | # P11496 | |
| D-PBS | Thermo Fisher | # 14190-144 | |
| SYBR Green master mix | Bio-Rad Laboratories | # 1725124 | |
| DMEM/F12 | Fisher Scientific | # 11-320-033 | |
| Penicillin-Streptomycin (P/S) | Fisher Scientific | # 15140122 | |
| N-2 Supplement | Fisher Scientific | # 17502048 | |
| B-27 Supplement | Fisher Scientific | # 17504044 | |
| insulin | Sigma | # I6634 | Prepare 0.5 mg/ml insulin solution by dissolving 5 mg insulin in 10 ml water and 50 μl of 1 N HCl. |
| Bovine Serum Albumin, suitable for cell culture (BSA) | Sigma | # A4161 | Prepare 4% BSA solution by dissolving 4 g BSA in 100 ml D-PBS and adjust the pH to 7.4 |
| Fibroblast Growth Factor basic Protein, Human recombinant (bFGF) | EMD Millipore | # GF003 | |
| HUMAN PDGF-AA | VWR | # 102061-188 | |
| poly-D-lysine | VWR | # IC15017510 | Prepare 1 mg/ml poly-D-lysine solution by dissolving 10 mg poly-D-lysine in 10 ml water and dilute 100 times when using. |
| Falcon Disposable Petri Dishes, Sterile, Corning, 100x15mm | VWR | # 25373-100 | |
| Falcon Disposable Petri Dishes, Sterile, Corning, 150x15mm | VWR | # 25373-187 | |
| HBSS, 10X, no Calcium, no Magnesium, no Phenol Red | Fisher Scientific | # 14185-052 | |
| Trypan Blue | Stemcell Technologies | # 07050 | |
| protease inhibitor cocktail | Sigma | # 11697498001 | |
| Software | |||
| FastQC | [10] http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | Obtaining quality control metrics of raw and trimmed reads | |
| Trimmomatic 0.33 | [11] http://www.usadellab.org/cms/?page=trimmomatic | Trimming and filtering raw reads | |
| GENCODE mm10 | ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_mouse/release_M15/GRCm38.primary_assembly.genome.fa.gz | Mouse reference genome | |
| Bowtie2 2.2.4 | [12] http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | Aligning reads to a reference genome | |
| SAMTools 1.5 | [13] http://samtools.sourceforge.net/ | Converting SAM file into BAM format | |
| HOMER 4.9.1 | [14] http://homer.ucsd.edu/homer/ | Creating a tag directory and annotating enriched genomic regions with gene symbols | |
| R 3.2.2 | [15] https://www.R-project.org/ | Programming scripts and running functions | |
| SPP 1.13 | [16] https://github.com/hms-dbmi/spp | Creating a strand cross-correlation plot | |
| MACS2 2.2.4 | [17] https://github.com/taoliu/MACS | Finding regions of ChIP enrichment over control | |
| BEDTools 2.25 | [18] http://bedtools.readthedocs.io/en/latest/ | Genome arithmetics | |
| bedGraphToBigWig | http://hgdownload.cse.ucsc.edu/admin/exe/ | Converting bedGraph file into bigwig | |
| IGV browser 2.3.58 | [19] http://software.broadinstitute.org/software/igv/ | Visualization and browsing of significant ChIP-seq peaks | |
| Microsoft Excel | Spreasheet program | ||
| ENCODE's blacklist | https://sites.google.com/site/anshulkundaje/projects/blacklists | Filtering peaks | |
| mm10.chrom.sizes | http://hgdownload.cse.ucsc.edu/goldenPath/mm10/bigZips/mm10.chrom.sizes. | Converting bedGraph file into bigwig | |
| ENCODE's motif database | [25] http://compbio.mit.edu/encode-motifs/ | Comprehensive motif database required for motif enrichment | |
| MEME-ChIP | [20] http://meme-suite.org/index.html | Motif enrichment analysis and motif discovery | |
| Primer names used for qPCR | Primer sequences used for qPCR | ||
| Mbp-F: | CTATAAATCGGCTCACAAGG | ||
| Mbp-R: | AGGCGGTTATATTAAGAAGC | ||
| Iba1-F: | ACTGCCAGCCTAAGACAACC | ||
| Iba1-R: | GCTTTTCCTCCCTGCAAATCC | ||
| Mog-F: | GGCTTCTTGGAGGAAGGGAC | ||
| Mog-R: | TGAATTGTCCTGCATAGCTGC | ||
| GAPDH-F | ATGACATCAAGAAGGTGGTG | ||
| GAPDH-R | CATACCAGGAAATGAGCTTG | ||
| Tuj1-F | TTTTCGTCTCTAGCCGCGTG | ||
| Tuj1-R | GATGACCTCCCAGAACTTGGC | ||
| PDGFRα-F | AGAGTTACACGTTTGAGCTGTC | ||
| PDGFRα-R | GTCCCTCCACGGTACTCCT |
Referencias
- Wu, J. Q., et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS Genet. 8 (3), e1002565 (2012).
- Zuchero, J. B., Barres, B. A. Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 23 (6), 914-920 (2013).
- Liu, Z., et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 302 (2), 683-693 (2007).
- Dong, X., et al. Comprehensive Identification of Long Non-coding RNAs in Purified Cell Types from the Brain Reveals Functional LncRNA in OPC Fate Determination. PLoS Genet. 11 (12), e1005669 (2015).
- Hilgenberg, L. G., Smith, M. A. Preparation of dissociated mouse cortical neuron cultures. J Vis Exp. (10), e562 (2007).
- Emery, B., Dugas, J. C. Purification of oligodendrocyte lineage cells from mouse cortices by immunopanning. Cold Spring Harb Protoc. 2013 (9), 854-868 (2013).
- Cahoy, J. D., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 28 (1), 264-278 (2008).
- Zhang, Y., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34 (36), 11929-11947 (2014).
- Cheng, X., et al. Bone morphogenetic protein signaling and olig1/2 interact to regulate the differentiation and maturation of adult oligodendrocyte precursor cells. Stem Cells. 25 (12), 3204-3214 (2007).
- Bolger, A. M., Lohse, M., Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30 (15), 2114-2120 (2014).
- Langmead, B., Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9 (4), 357-359 (2012).
- Li, H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25 (16), 2078-2079 (2009).
- Heinz, S., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 38 (4), 576-589 (2010).
- Team, R. C. A language and environment for statistical computing. R Foundation for Statistical Computing. , (2015).
- Kharchenko, P. V., Tolstorukov, M. Y., Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 26 (12), 1351-1359 (2008).
- Zhang, Y., et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9 (9), R137 (2008).
- Quinlan, A. R., Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26 (6), 841-842 (2010).
- Robinson, J. T., et al. Integrative genomics viewer. Nat Biotechnol. 29 (1), 24-26 (2011).
- Bailey, T. L., et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server issue), W202-W208 (2009).
- Sims, D., Sudbery, I., Ilott, N. E., Heger, A., Ponting, C. P. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 15 (2), 121-132 (2014).
- Landt, S. G., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22 (9), 1813-1831 (2012).
- Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature. 489 (7414), 57-74 (2012).
- Mazzoni, E. O., et al. Embryonic stem cell-based mapping of developmental transcriptional programs. Nat Methods. 8 (12), 1056-1058 (2011).
- Kheradpour, P., Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42 (5), 2976-2987 (2014).
- Dugas, J. C., Tai, Y. C., Speed, T. P., Ngai, J., Barres, B. A. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 26 (43), 10967-10983 (2006).
- Gilfillan, G. D., et al. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 13, 645 (2012).
- Raskatov, J. A., et al. Modulation of NF-kappaB-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci U S A. 109 (4), 1023-1028 (2012).
- Cheneby, J., Gheorghe, M., Artufel, M., Mathelier, A., Ballester, B. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. , (2017).
- Yevshin, I., Sharipov, R., Valeev, T., Kel, A., Kolpakov, F. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 45 (D1), D61-D67 (2017).
- Kulakovskiy, I. V., et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 41 (Database issue), D195-D202 (2013).
- Jain, D., Baldi, S., Zabel, A., Straub, T., Becker, P. B. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 43 (14), 6959-6968 (2015).
- Gerstein, M. B., et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 489 (7414), 91-100 (2012).

