Phospholipase Activity Assay: A Gel Diffusion Assay to Determine Phospholipase Activity in Crude Venom Extract from Anthopleura dowii
Abstract
Source: Ramírez-Carreto, S., et al., Identification of Hemolytic and Phospholipase Activity in Crude Extracts from Sea Anemones by Straightforward Bioassays. J. Vis. Exp. (2022)
In this video, we demonstrate an assay to establish the presence of phospholipase in the crude venom extract from Anthopleura dowii—a marine sea anemone. The assay utilizes an agarose gel diffusion technique, in which the enzyme phospholipase diffuses and breaks down phospholipids incorporated in the solid media in a radial manner.
Protocol
1. Organism collection
- Collect sea anemone A. dowii.
NOTE: Here, the organisms were collected during low tide periods in the intertidal zone off the coast of Ensenada Baja, California, Mexico (Figure 1A, B). - Transport the organisms to the laboratory in containers with seawater (Figure 1C).
- In the laboratory, clean the organisms with distilled water by hand to remove the big substrate particles adhering to the body.
- Immediately freeze the organisms at -80 °C in an ultra-freezer for 72 h.
- Place the organisms in special glasses for freeze-drying, with lyophilization conditions of -20 °C, 0.015 psi, for 48 h.
- Store the lyophilized organisms at -20 °C until use.
2. Tissue hydration
- Reconstitute 20 g dry weight of lyophilized organisms with 60 mL (1/3 w/v) of 50 mM sodium phosphate buffer, pH 7.5, and 1 inhibitor cocktail tablet (Table of Materials).
- In a magnetic stirrer, continuously stir the sample at ~800 rpm, 4 °C, for 12 h.
3. Toxin release
- To induce cell lysis and nematocyst discharge, freeze the sample at -20 °C for 12 h, and then place the container in water at room temperature (RT) until it thaws up to 90%. Repeat this procedure three times.
NOTE: The sample does not thaw 100%; it is crucial to maintain the sample at ~4 °C to prevent protein degradation. - Observe the nematocyst discharged under a confocal microscope. Take 10 µL of the sample on a slide and place a coverslip over it. Observe under a confocal microscope at 100x and 60x magnification. Here a dye was not necessary.
- Process the images captured in the confocal microscope in ImageJ software (version 1.53c, Wayne Rasband, National Institutes of Health, USA, https://imagej.nih.gov/ij).
- If the tubule inside the nematocyst uncoils and is outside, the nematocysts are discharged; then, continue with the next step. Otherwise, only conduct one more freeze-thaw cycle.
- To clarify the extract, centrifuge the sample at 25,400 x g for 40 min, 4 °C, recover the supernatant, and centrifuge it again. Repeat this step 3-4 times.
- Recover the supernatant and discard the pellet. The supernatant or crude extract contains the venom components (toxins) and other cell molecules, such as lipids, carbohydrates, and nucleic acids.
4. Quantitation of total protein
- Determine the protein concentration of the crude extract using a commercial Bradford colorimetric assay kit (Table of Materials).
- Dilute the dye reagent (one part of dye with four parts of distilled water).
- Prepare bovine serum albumin (BSA) Fraction V (Table of Materials) solution at 1 mg/mL concentration in phosphate buffer (50 mM sodium phosphate, pH 7.5).
- Prepare the solutions listed in Table 1 in triplicate.
- Vigorously mix each solution in a shaker for 4 s.
- Incubate the samples for 5 min at RT, without shaking.
- Measure absorbance (AU) at 595 nm.
- Plot the averages of the sample BSA absorbances (AU vs. protein concentration), and determine the equation of the graph (Equation 1).
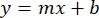
where y is the absorbance, m is the slope of the line, x is the protein concentration, and b is the y-intercept. To calculate crude extract concentration, solve for the x as follows:
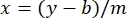
5. Determine polypeptide venom complexity
- Assemble the glass container for vertical gel electrophoresis according to the manual.
- Prepare acrylamide mix (30%): 29.2 g of acrylamide, 0.8 g of bis-acrylamide, the final volume of 100 mL. Filter the solution using a 0.45 µm membrane. Store the solution in a dark bottle at 4 °C. Then, prepare the mixture for the resolving gel (Table 2).
- The SDS-PAGE gel consists of a resolving gel and a stacking gel. Prepare the gels as per the solution volumes defined in Table 2. During preparation, ensure to maintain each mix at 4 °C to reduce polymerization time.
NOTE: The volume of each mixture varies depending on the brand of equipment used; for this protocol, the volumes correspond to the preparation of 15% acrylamide gel, 0.75 mm thick, for a vertical electrophoresis chamber (Table of Materials). - After the acrylamide has polymerized, ~10 min, add the mixture of stacking gel.
- Prepare the mixture for the stacking gel, immediately add it to the glass plates, and place a comb to form the wells for loading the samples.
- Once the stacking gel has polymerized (~15 min), remove the comb and place the glass container in a chamber for vertical electrophoresis.
- Place 200 mL of electrode buffer (0.25 M Tris, 0.192 M glycine, 0.1% SDS) in the electrophoresis chamber.
- Analyze the crude extract: Mix 30 µg of crude extract with 5 µL of protein loading buffer (25% glycerol, 15% sodium dodecyl sulfate, 25% 2-mercaptoethanol, 0.125 mM Tris pH 7, bromophenol blue 0.1 mg/mL) to denature proteins. The final volume must be <50 µL.
- Heat at 90 °C for 5 min, cool, and centrifuge for 3 s at 1,400 x g. 10. Load the samples in the wells of the stacking gel. Load a standard molecular weight ladder.
- Load the samples in the wells of the stacking gel. Load a standard molecular weight ladder.
- Run electrophoresis at 25 mA constant for 1 h 30 min.
- Remove the glass containers from the electrophoresis chamber to proceed with the staining of the gel proteins.
6. Protein staining
- Rinse the gel in 50 mL of distilled water to remove debris from the buffer electrode.
- Discard the water.
- To avoid the diffusion of the gel proteins, add the fixation solution (60 mL of ethanol, 20 mL of acetic acid, and 20 mL of distilled water, final volume 100 mL). Incubate at RT for 40 min with shaking at 80 rpm. Decant the solution.
- Add the protein crosslinking solution (15 mL of ethanol, 0.25 mL of glutaraldehyde, 50 mL of distilled water, final volume 65.25 mL), and incubate for 30 min at RT with shaking at 80 rpm. Remove the solution.
- Wash the gel with 100 mL of distilled water for 5 min, and remove the water. Repeat this procedure four times.
- Carry out protein staining with 50 mL of Coomassie brilliant blue R-250 filtered solution (40% methanol, 10% acetic acid, 50% distilled water, 0.1% dye, final volume 100 mL), stirring at 60 rpm in a laboratory rotary oscillator at RT for 15 min.
- Recover the solution, which can be used to stain other gels.
- To eliminate the excess dye, add a destaining solution (40 mL of methanol, 10 mL of acetic acid, 50 mL of distilled water). Keep stirring (80 rpm) at RT until the bands (stained proteins) visualize.
- Keep the gel in 50 mL of distilled water and scan the image in a Gel documentation system.
7. Phospholipase assay
- Wash one chicken egg with 1% SDS in distilled water.
- Separate the egg yolk from the egg white under sterile conditions.
- Prepare 50 mL of 0.86% NaCl solution, and filter through a 0.22 µm filter. Then, prepare Solution A: Add 12 mL of egg yolk and 36 mL of 0.86% NaCl solution.
- Prepare Solution B: Mix 300 mg of agarose in 50 mL of buffer (50 mM Tris, pH 7.5), filter through a 0.22 µm filter. Heat the solution in a microwave until boiling. Cool down in warm water to reach 43-45°C.
- Prepare Solution C: Prepare 10 mM CaCl2 and filter it with a 0.22 µm filter.
- Prepare Solution D: 10 mg of rhodamine 6G in 1 mL of distilled water.
- Under sterile conditions (laminar flow hood), add 500 µL of solutions A and C to solution B and 100 µL of solution D, mix, and pour 25 mL into each Petri dish (90 x 15 mm).
- Wait for the solution to solidify under sterile conditions (30 min).
- Make wells (~2-3 mm diameter) with a thin tube.
- Add a total of 20 µL of phosphate buffer (negative control) in one well and 20 µL of a determined phospholipase (positive control) in another well.
- Place different amounts of the crude extract protein, 5, 15, 25, 35, 45 µg, in the remaining wells, each in a final volume of 20 µL.
- Wait for the agar to adsorb all the samples (30 min).
- Incubate at 37 °C for 20 h.
- If a halo forms around the well, this indicates phospholipase activity. Measure the halo formed with a vernier caliper.
NOTE: Perform the experiment in triplicate (steps from 7.1-7.14).
Representative Results

Figure 1: Sea anemone collection. (A) Intertidal zone in El Sauzal, Baja California Norte, Mexico. (B) Sea anemone collected. (C) Anthopleura dowii Verrill, 1869.
Table 1: Quantitation of protein by Bradford assay.
| Tube | Distilled Water | BSA | Dye (µL) | Final volume | |
| (µL) | (µg) | (µL) | (µL) | ||
| 1 | 800 | – | – | 200 | 1000 |
| 2 | 799 | 1 | 1 | 200 | 1000 |
| 3 | 797 | 3 | 3 | 200 | 1000 |
| 4 | 795 | 5 | 5 | 200 | 1000 |
| 5 | 793 | 7 | 7 | 200 | 1000 |
| 6 | 790 | 10 | 10 | 200 | 1000 |
| Crude extract (µL) | |||||
| 7 | 798-790 | from 2 to 10 | 200 | 1000 | |
Table 2: Solutions for preparing a 15% acrylamide SDS-PAGE electrophoresis gel.
| Solution | Resolving gel | Stacking gel |
| Distilled water | 1.1 mL | 1.4 mL |
| Acrylamide mix (30%) | 2.5 mL | 0.33 mL |
| Tris (1.5 M, pH 8.8), adjust pH with HCl. | 1.3 mL | |
| Tris (0.5 M, pH 6.8), adjust pH with HCl. | 0.25 mL | |
| Sodium dodecyl sulfate (SDS) 10% (w/v) | 0.5 mL | 0.1 mL |
| Ammonium persulfate (APS) (10%) | 0.5 mL | 0.1 mL |
| N,N,Nˈ,Nˈ-tetramethylethylenediamine (TEMED) | 0.005 mL | 0.005 mL |
Divulgations
The authors have nothing to disclose.
Materials
| 15 mL conical centrifuge tube | Corning | 430766 | |
| 2-Bromophenol blue | Sigma | B75808 | |
| 2-mercaptoetanol | Sigma-Aldrich | M6250-100ML | |
| 50 mL conical centrifuge tubes | Corning | 430828 | |
| Acetic Acid Glacial | J.T. Baker | 9515-03 | |
| Acrylamide | Promega | V3115 | |
| Agarose | Promega | V3125 | |
| Bisacrylamide | Promega | V3143 | |
| Bovine Serum Albumin Fraction V | Sigma | A3059-100G | |
| Bradford Protein Assays | Bio-Rad | 5000006 | |
| Calcium chloride | Sigma-Aldrich | C3306 | |
| Cell culture plates 96 well, V-bottom | Corning | 3894 | |
| Centrifuge | Eppendorf | 5804R | |
| Centrifuge tubes | Corning | CLS430829 | |
| ChemiDoc MP system | Bio-Rad | 1708280 | |
| Citric acid | Sigma-Aldrich | 251275 | |
| Clear flat.bottom 96-Well Plates | Thermo Scientific | 3855 | |
| Coomassie Brilliant Blue G-250 | Bio-Rad | #1610406 | |
| Coomassie brilliant blue R-250 | Bio-Rad | 1610400 | |
| Dextrose | J.T. Baker | 1916-01 | |
| Ductless Enclosure | Labconco | Vertical | https://imagej.nih.gov/ij ImageJ 1.53c |
| Gel Doc EZ | Bio Rad. | Gel Documentation System | |
| Glycerol | Sigma-Aldrich | G5516-4L | |
| Hemocytometer | Marienfeld | 650030 | |
| ImageJ (Software) | NIH, USA | Version 1.53c | |
| Incubator 211 | Labnet | I5211 DS | |
| Methanol | J.T. Baker | 9049-03 | |
| Mini-PROTEAN tetra cell | Bio-Rad | 1658000EDU | |
| Na2HPO4 | J.T. Baker | 3824-01 | |
| NaCl | J.T. Baker | 3624-01 | |
| NaH2PO4.H2O | J.T. Baker | 3818-05 | |
| Origin software | version 9 | To design the plot with sigmoidal adjustments | |
| Petridish | Falcon | 351007 | |
| Pipetman kit | Gilson | F167380 | |
| Precast mini gel | BioRad | 1658004 | |
| Prestained Protein Ladder | Thermo Scientific | 26620 | |
| Protease Inhibitor Cocktail | Roche | 11836153001 | |
| Protein Assay Dye Reagent Concentrate | Bio-Rad | 5000006 | |
| Rhodamine 6G | Sigma-Aldrich | 252433 | |
| SDS | Sigma-Aldrich | L4509 | |
| Sodium citrate dihydrate | JT Baker | 3646-01 | |
| Spectrophotometer | THERMO SCIENTIFIC | G10S UV-VIS | |
| Tris Base | Sigma-Aldrich | 77-86-1 | |
| Volt Power Supply | Hoefer | PS300B |

