Darkfield Microscopy to Visualize Diffusional Dynamics of Nanorods on Cell Membrane
Abstract
Source: Ge, F., et al. Visualizing Diffusional Dynamics of Gold Nanorods on Cell Membrane using Single Nanoparticle Darkfield Microscopy. J. Vis. Exp. (2021).
In this video, we describe the darkfield microscopy (DFM) technique combined with single-particle tracking (SPT) analysis to monitor the interaction of plasmonic gold nanorods with the cell membrane of a specific cell line of interest. Combined DFM and SPT analysis allows the visualization of the nanorod movement through the cell membrane with high spatiotemporal resolution.
Protocol
1. Microscope slide preparation
NOTE: U87 MG cells of the third to tenth generation with high activity are used in SPT experiments.
- Sterilize 22 mm × 22 mm coverslips already cleaned with Piranha solution by immersing in ethanol (99.9%).
- Use forceps to take out the cover slip from the ethanol solution (step 1.1) and sterilize by burning ethanol on the flame. Once all ethanol is burnt, place coverslips in a plastic cell culture dish (35 x 10 mm) filled with 2 mL of cell medium (no phenol red).
- Add 50 µL of the cell suspension on the coverslip and gently push the dish back and forth and left and right to evenly distribute the cells. Place it in a humidified atmosphere.
- When U87 MG cells on the coverslip reach 20%–40% confluency (~12 h), add 20 μL of CTAB-AuNRs (gold nanorods synthesized with cetyltrimethylammonium ammonium bromide molecule as a protective agent,138 pM) into the dish and disperse. Incubate in a humidified atmosphere for 5 min.
- Add 100 μL of the culture medium (no phenol red) from the dish in step 1.4 into the groove of the grooved glass slide (Figure 1) which is precleaned with Piranha solution.
- Take the coverslip out from the dish and invert it on the top of the groove of the microscope glass slide (Figure 1). Seal it with nail polish, let it dry, and place it on the stage to perform SPT experiments.
2. Performing single particle tracking experiments with darkfield microscopy (Figure 1).
- Place a drop of oil on the oil-immersed darkfield condenser (numerical aperture (NA) 1.43-1.20) and turn the knob to make the condenser contact the glass slide.
- Put a drop of oil on the top of the cover glass and turn the focusing knob to make the 60x oil immersion objective (NA 0.7–1.25) touch the oil.
- Turn on the light source and slightly turn the focusing knob to focus the imaging plane.
NOTE: In the field of view, the background is black, the cells are bright, and the CTAB-AuNRs (aspect ratio~2:1, Figure 2) are small colored (red, yellow, or green) scattering spots. - Capture sample scattering light by a color CMOS camera. Click the "Camera icon" in the software to record and export TIFF format to save images.
3. Data acquisition
- Extract a single long-term trajectory
- Operate as described in Figure 3 to convert the time-series dark-field images from "RGB color" mode to "8-bit" mode. In the Image J click Image | Type | 8 bit. To adjust the contrast, click Image | Adjust | Brightness | Contrast.
- Select a target particle and cut off the time-series backgrounds by boxing and deleting the background with "Ctrl+X".
- Open the particle detection and particle Linking window by clicking the "Plugins | Particle Tracker Classic | Particle Tracker".
- Set Radius to 6, Cutoff to 0, and Percentile to 0.01%.
NOTE: To detect the particle, adjust the above three parameters with the assistance of Preview. Ensure that the radius is slightly larger than the targeted particle and smaller than the smallest inter-particle separation. Percentile is the lower limit of intensity distribution that to be candidate particles. - Set the Link Range to 5 and Displacement to 10.
NOTE: To link the particle between consecutive adjacent frames, adjust the above two parameters. Displacement is the maximum pixels that a particle can move between two succeeding frames, and Link Range is the number of consecutive frames to consider when determining the best corresponding match. - Click "OK" to open the ParticleTracker Results window to see the results.
- Click "Visualize All Trajectories" to inspect the generated trajectories.
- Click the "Relink Particles" menu at the top to re-link the detected particles with different link ranges and percentile parameters, if the software-generated trajectory does not match the moving trajectory of the AuNR.
- Click "Save Full Report" to save results if the software-generated trajectory and the moving trajectory of the AuNR are matched.
NOTE: An example of extracting a single long-term trajectory with Image J is shown in Figure 4.
Representative Results
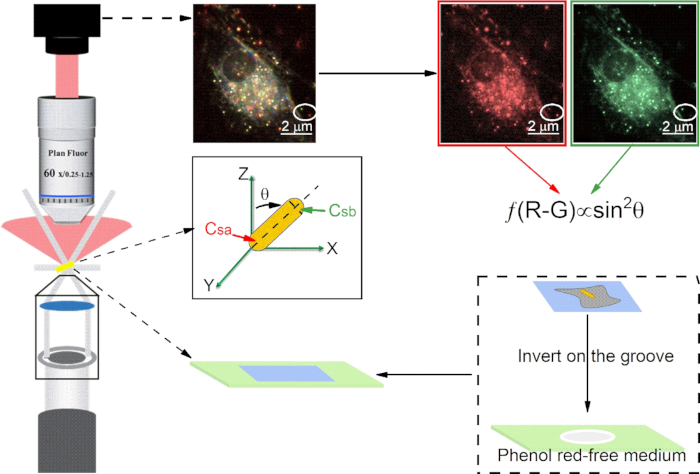
Figure 1: Schematic diagram of the optical path of darkfield microscopy, microscope slide preparation, and the dual-channel difference method for calculating polar angle (θ) of AuNRs. Scale bar in image is 2 μm.
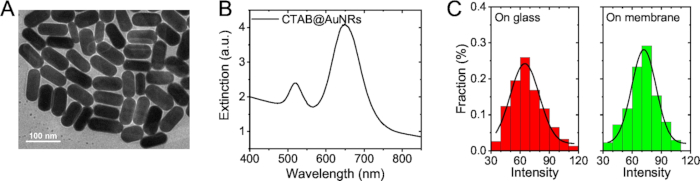
Figure 2: Characterization of CTAB-AuNRs. (A) TEM images of CTAB@AuNRs. (B) LSPR spectrums of CTAB@AuNRs. (C) Distribution of single particle intensity: CTAB@AuNRs on glass (left panel), CTAB@AuNRs on plasmon membrane (right panel).
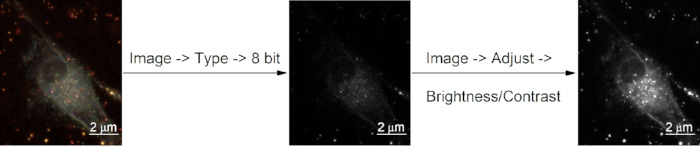
Figure 3: Pretreatment of darkfield images. Time-series color images were converted to “8 bit” mode and their contrast were adjusted.
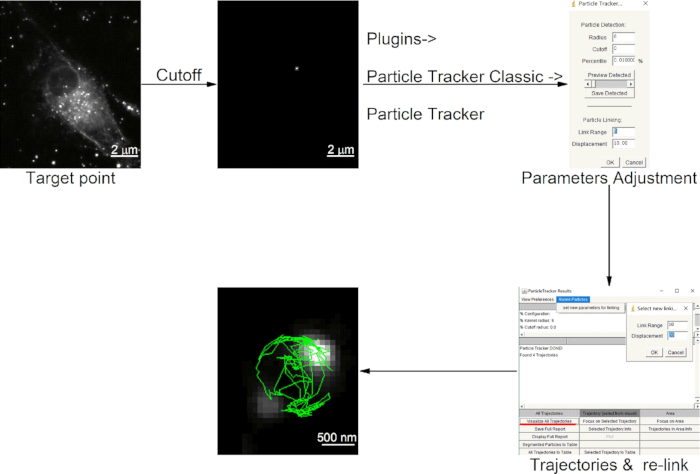
Figure 4: Extraction of single long-term trajectory with ImageJ.
Divulgations
The authors have nothing to disclose.
Materials
| CTAB-coated gold nanorods (CTAB-AuNRs) | Nanoseedz | NR-40-650 | 85 nm*40 nm |
| Color CMOS camera | Olympus | DP74 | Japan |
| Coverslips | Citoglas | z10212222C | 22*22 mm |
| Dark-field microscopy | Nikon | 80i | Upright microscope |
| Fetal bovine serum (FBS) | Gibco | 10099141 | |
| Fiji | National Institutes of Health | 2.0.0-rc-69/1.52 p | A distribution of ImageJ |
| Grooved glass slide | Sail brand | 7103 | Single concave |
| Image J | Image J | 1.52 j | |
| MATLAB | MathWorks | R2019b | |
| MATLAB Code | https://github.com/fenggeqd/ JOVE-2020 | ||
| Plastic cell culture dishes | Falcon | 353002 | |
| Plastic cell culture dishes | Falcon | 353001 | 35*10 mm |
| U87 MG cell | American Type Culture Collection | ATCC HTB-14 | A human primary glioblastoma cell line |
| Minimum essential medium (MEM) | Gibco | 51200038 | No phenol red |
| Penicillin-streptomycin | Gibco | 15140122 |

