Measurement of Natural Killer Cell Migration in Response to Chemotactic Stimuli
Published: November 30, 2023
Abstract
Source: Chava, S., et al. Measurement of Natural Killer Cell-Mediated Cytotoxicity and Migration in the Context of Hepatic Tumor Cells. J. Vis. Exp. (2020).
This video demonstrates a method to study natural killer (NK) cell migration through a porous membrane insert system in response to chemoattractants. This method enables quantitative analysis of NK cell migration using flow cytometry.
Protocol
1. NK Cell Migration Assay
- Grow NK92MI cells and centrifuge cells at 160 x g for 3 min in a 15 mL sterile centrifuge tube.
- Wash the cell pellet twice with 5 mL of 1x PBS and resuspend the cells in 3 mL of serum-free NK92MI cell medium. Count NK92MI cells using a hemocytometer or an automated cell counter.
- Plate NK92MI cells (2.5 x 105 cells/100 µL/well) in the upper compartment of the transwell permeable chamber (6.5 mm diameter insert and 5 µm pore size).
- In the bottom chamber add 0.6 mL of serum-free medium containing material to be tested for NK cell chemoattractant properties (e.g., conditioned medium, chemokines, cytokines).
NOTE: When preparing conditioned medium, use reduced-serum medium without added serum to eliminate interference from serum proteins in the migration assay. - Incubate the 24-well-permeable chambers at 37°C for 4 h. After 4 h, collect the non-adherent and migrated NK92MI cells from the bottom chamber and transfer them to fluorescence-activated cell sorting (FACS) tubes for further analysis.
NOTE: The time of culture may vary depending on the type of target cells, as well as the amount and kinetics of chemokines produced by the target cells. Therefore, this time should be empirically determined for each cell type, chemokine, and experiment. - Add a predetermined number of counting beads for flow cytometry in a volume of 50 µL to each tube containing migrated NK cells. Evaluate the volume of 300 µL/well cell suspension using any flow cytometer capable of automated FACS-based cell counting.
NOTE: Mix or vortex-mix the counting beads for flow cytometry thoroughly each time before use to ensure that a constant number of beads is used to minimize experimental variability. Reverse pipetting is recommended with count beads to maintain accuracy. Use only NK cells and counting beads for flow cytometry as FACS analysis controls. The authors recommend reading at least 10,000 beads + NK cells combined, an amount that has worked well. However, this number may vary depending on the experimental conditions. Therefore, the combined number of beads + NK cells should be empirically determined for each type of experiment. Additionally, it is important to perform experiments using biological triplicates to achieve statistically significant results and to account for variability between different cell counts. - Calculate the absolute number of migrated NK92MI cells using this formula:
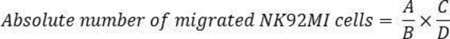
where A = number of cells, B = number of beads, C = assigned bead count of the lot (number of counting beads for flow cytometry/50 µL; in this example 49,500), and D = volume of sample (µL).
NOTE: If 300 µL of sample volume (migrated cells) is used for FACS analysis with 50 µL of counting beads for flow cytometry, the absolute number of migrated cells = 1,700 cells /3,300 bead events x 49,500 beads/300 µL = 84.975 cells/µL. The calculation should be corrected if the sample is diluted or if a different volume of FACS counting beads is used.
Divulgations
The authors have nothing to disclose.
Materials
| Absolute counting beads | Thermo Fisher Scientific | C36950 | |
| NK92MI cells | ATCC | CRL-2408 | |
| SKHEP-1 Cells | ATCC | HTB-52 | |
| Transwell permeable chambers | Costar | 3241 | |
| Opti-MEM | Gibco | 31985070 | |
| Phosphate Buffer Saline (PBS) | Sigma-Aldrich | P4417 | |
| Alpha-MEM | Sigma-Aldrich | M4256 |
Tags
Citer Cet Article
Measurement of Natural Killer Cell Migration in Response to Chemotactic Stimuli. J. Vis. Exp. (Pending Publication), e21784, doi: (2023).

