An Assay to Determine Polymicrobial Biofilm Initiation on Virus-Infected Cells
Abstract
Source: Plotkin, B. J. et al., Determination of Biofilm Initiation on Virus-infected Cells by Bacteria and Fungi. J. Vis. Exp. (2016)
This video illustrates the initiation of polymicrobial biofilm formation on virus-infected cells by bacteria and fungi. Herpes simplex virus infection impedes bacterial attachment to the cells, preventing bacterial-fungal co-localization. In contrast, uninfected cells display bacterial-fungal co-localization and initiation of biofilm formation.
Protocol
1. HSV Strains and Handling
NOTE: Recombinant non-spreading HSV-1(KOS) gL86 and HSV-2 (KOS) 333gJ– with beta-galactosidase reporter activity is used for the assay.
- Use virus from a single lot and store at -80 °C at a 1:1 ratio of Dulbecco's modified Eagle's medium (DMEM) with 20% fetal bovine serum (FBS) and skim milk until use. Before viral lot storage, determine virus concentration by o-nitrophenyl-β-D-galactopyranoside (ONPG) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) assay.
- Determine virus viability and multiplicity of infection (MOI) by X-Gal staining for each experimental assay run using reporter virus entry assay, as previously described (Figure 1).
- Dilute virus (Opt-MEM) to desired MOI. Fix monolayers (paraformaldehyde; 0.5 ml/well) before staining. Place virus viability controls in a separate microtiter plate in parallel with polymicrobial assay plates.
2. HeLa 299 Cell Handling
- Grow at 37 °C, 5% CO2 in DMEM with 4.5 g/L glucose, 10% heat-inactivated fetal bovine serum (FBS), gentamicin (50 µg/ml) and L-glutamine. Passage cells at 80% confluence with Trypsin solution (ethylenediaminetetraacetic acid, 0.53 M EDTA; 0.05% Trypsin; 5 ml/flask).
3. C. albicans Handling
NOTE: C. albicans obtained from a clinical laboratory source is stored at -80 °C in Remmel skim milk 2x medium.
- Culture frozen stock onto Sabouraud Dextrose Medium (37 °C). After 24 hr subculture the C. albicans onto Fungisel medium (37 °C; 48 hr) for use.
- Generate germ tube (GT) forms (pick representative colonies; 3 ml FBS; 3 hr; 37 °C; absorbance600 (Abs600), 0.3). After incubation, wash GT (Hanks' Balanced Salt Solution, HBSS; 2x; 4,000 x g). Add washed GT to warmed HBSS (37 °C; 0.32 Abs600). GT forms should be 99% of cells observed as determined by hemocytometer count.
- Make yeast form (YF) stock suspensions by picking representative Fungisel colonies (HBSS, 3 ml; 0.32 Abs600). Count the number of YF forms/ml microscopically using a hemocytometer. YF forms should be 99% of cells observed as determined by hemocytometer count.
- Make working fungal stock (250 µl of GT or YF stock in 25 ml HBSS; 37 °C; 105 colony forming unit, (CFU/ml)
4. S. aureus Handling
- Store S. aureus American Type Culture Collection, ATCC 25923 (-80 °C; Remmel skim milk 2x). Culture onto sheep blood agar (5%; 37 °C; 24 hr). Pick representative colonies and transfer to mannitol salts medium within 2 days for stock (37 °C; 18 hr).
- Make S. aureus stock suspension (3 ml HBSS; 1.32 Abs600 ;108 CFU/ml)
- Make working S. aureus stock (100 µl of the stock in 25 ml HBSS; 105 CFU/ml).
5. Candida and S. aureus Suspensions
- Make mixed C. albicans and S. aureus suspension (250 µl YF or GT stock and 100 µl S. aureus stock in 25 ml HBSS).
6. Polymicrobial Biofilm Assay
- Seed 96 well plates with 200 µl of 2 x 105 HeLa cells/ml (85% confluence level). Rock plates (30 – 45 min; 37 °C) before incubation (37 °C; 5% CO2 incubator; 18 hr). Wash monolayers (1x; Opt-MEM) then seed with HSV (HSV-1 (KOS) gL86 or HSV-2 (KOS) 33 gJ– in 100 µl Opt-MEM ; MOI 50 and 10). Incubate plates (3 hr; 37 °C; 5% CO2). Use only one viral strain per day.
- Wash infected monolayers (1x; phosphate-buffered saline (PBS) with Mg+2 and Ca+2; 100 µl). Replace PBS with warm HBSS leaving 25 µl in each well.
- Add YF, GT and/or S. aureus working suspensions (100 µl; target to cell ratio =5:1; n=16) as indicated in Table 1. Incubate plates (static; 30 min; 37 °C; 5% CO2).
- After incubation, aspirate one column at a time immediately refilling with 300 µl PBS with Mg+2 and Ca+2. Repeat this step twice then add radio-immunoprecipitation assay lysis buffer (RIPA; filter sterilized; 200 µl of a 1:50 dilution).
- Rapidly triturate the HeLa cell lysate then place 50 µl onto mannitol salts (MS) and/or Fungisel (F) media (Figure 2). Spread the lysate using a glass rod bent at a 90° angle. Incubate the plates (18 hr at 37 °C). Manually count the number of colonies per plate. Controls consist of S. aureus and/or C. albicans adherence to HSV-uninfected HeLa cells.
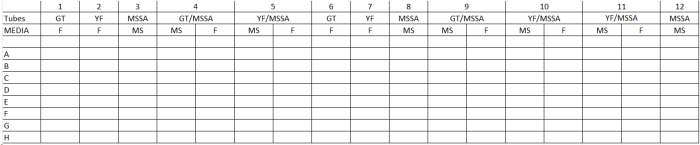
Table 1. General Template Determination of Microbial Adherence in Polymicrobial Interactions. GT= Candida albicans germ tube phenotype; YF= C. albicans yeast form phenotype; MSSA= methicillin-sensitive Staphylococcus aureus; MS= mannitol salts medium; F= Fungisel medium.
Representative Results
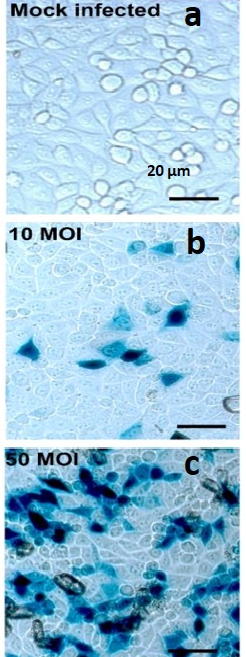
Figure 1. X-gal Staining Pictures of Dosage Dependent HSV-1 Infection in HeLa Cells. HeLa cells infected with HSV-1 at various MOI and X-Gal stained. (A) HeLa cells in well of 96 well plate with X-Gal stained mock-infected HeLa cell control; 20x initial magnification; (B) HeLa cells in well of 96 well plate with X-Gal stained HeLa cell infected with HSV-1 at an multiplicity of infection (MOI) of 10; 20x initial magnification; (C) HeLa cells in well of 96 well plate with X-Gal stained HeLa cell infected with HSV-1 at an multiplicity of infection (MOI) of 50; 20x initial magnification; scale bar applies to all.
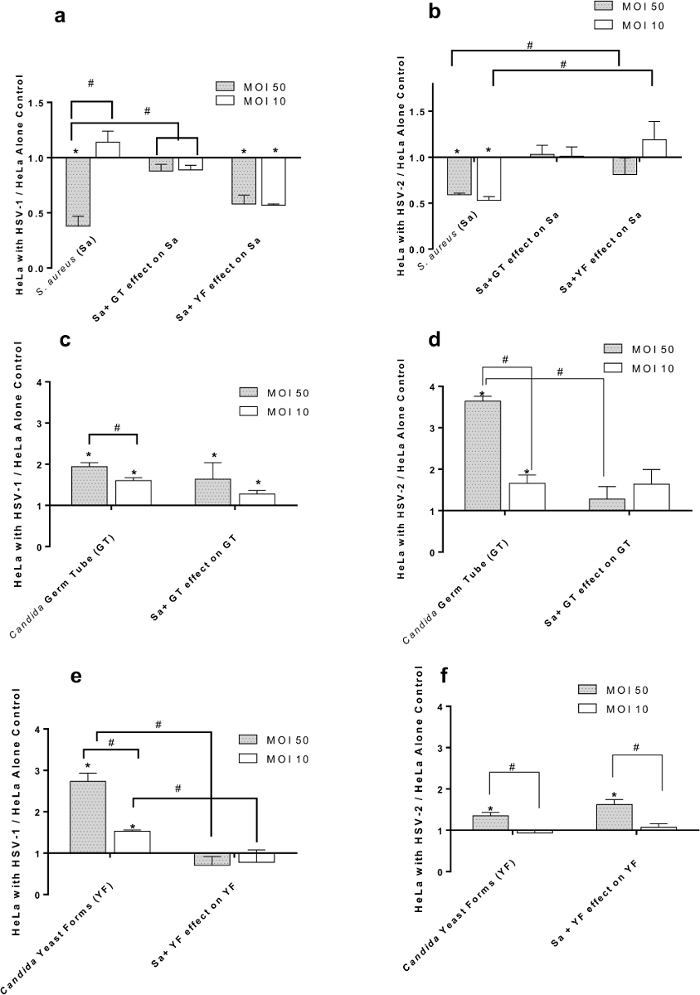
Figure 2. Effect of HSV-1 (panels A, C, E) and HSV-2 (panels B, D, F) at Multiplicities of Infection (MOI) of 50 and 10 on Adherence of S. aureus and/or C. albicans to HeLa Cells. (A) S. aureus (Sa) binding to HSV-1 infected cells in the presence of C. albicans germ tubes (GT) or yeast forms (YF); (B) S. aureus (Sa) binding to HSV-2 infected cells in the presence of C. albicans germ tubes (GT) or yeast forms (YF); (C) C. albicans germ tubes (GT) binding to HSV-1 infected cells in the presence of S. aureus (Sa); (D) C. albicans germ tubes (GT) binding to HSV-2 infected cells in the presence of S. aureus (Sa); (E) C. albicans yeast forms (YF) binding to HSV-1 infected cells in the presence of S. aureus (Sa); (F) C. albicans yeast forms (YF) binding to HSV-2 infected cells in the presence of S. aureus (Sa). All data points are Mean +/- SEM, n= 16 normalized to virus-free control. * = significantly different (p< 0.05) from uninfected HeLa cell control. # = significantly different (p< 0.05) from paired point indicated by bracket; scale bar applies to all.
Divulgations
The authors have nothing to disclose.
Materials
| C.albicans | |||
| BBL Sabouraud Dextrose | BD | 211584 | |
| Fungisel Agar | Dot Scientific | 7205A | |
| S.aureus | |||
| Mannitol Salt Agar | Troy Biologicals | 7143B | |
| Sheep blood agar | Troy Biologicals | 221239 | |
| Hela cells | |||
| 1xDMEM (Dubelcco's Modified Eagle Medium, with 4.5 g/L glucose and L-glutamine, without sodium pyruvate | Corning | 10-017-CM | |
| Gentamicin 50mg/ml | Sigma | 1397 | 50µg/ml final concentration in the complete DMEM |
| Trypsin EDTA (0.05% Trypsin, 0.53M EDTA)Solution 1X | Corning | 25-052-CI | |
| Fetal Bovine Serum | Atlanta Biologicals | S11150 | 10% final concentration in the complete DMEM |
| Other medium and reagents | |||
| ONPG | Thermo Scientific | 34055 | |
| Ultra-Pure X gal | Invitrogen | 15520-018 | |
| 1x HBSS (Hanks' Balanced Salt Solution) | Corning | 20-021-CV | |
| 1XPBS | Dot Scientific | 30042-500 | |
| RIPA Lysis | Life Technologies | 89901 | |
| Supplies | |||
| 96-Well plates | Evergreen Scientific | 222-8030-01F | |
| Culture tubes 100×13 | Thomas Scientific | 9187L61 |

