A PCR-based Genotyping Method to Distinguish Between Wild-type and Ornamental Varieties of Imperata cylindrica
Summary
We provide a cost-effective and rapid molecular genotyping protocol that employs variety-specific PCR primers that target DNA sequence differences within the chloroplast trnL-F spacer region to differentiate between varieties of Imperata cylindrica (cogongrass) that cannot be distinguished by morphology alone. These varieties include the federally listed noxious weed, cogongrass and closely-related, wide-spread ornamental variety, I. cylindrica var. koenigii (Japanese blood grass).
Abstract
Wild-type I. cylindrica (cogongrass) is one of the top ten worst invasive plants in the world, negatively impacting agricultural and natural resources in 73 different countries throughout Africa, Asia, Europe, New Zealand, Oceania and the Americas1-2. Cogongrass forms rapidly-spreading, monodominant stands that displace a large variety of native plant species and in turn threaten the native animals that depend on the displaced native plant species for forage and shelter. To add to the problem, an ornamental variety [I. cylindrica var. koenigii (Retzius)] is widely marketed under the names of Imperata cylindrica ‘Rubra’, Red Baron, and Japanese blood grass (JBG). This variety is putatively sterile and noninvasive and is considered a desirable ornamental for its red-colored leaves. However, under the correct conditions, JBG can produce viable seed (Carol Holko, 2009 personal communication) and can revert to a green invasive form that is often indistinguishable from cogongrass as it takes on the distinguishing characteristics of the wild-type invasive variety4 (Figure 1). This makes identification using morphology a difficult task even for well-trained plant taxonomists. Reversion of JBG to an aggressive green phenotype is also not a rare occurrence. Using sequence comparisons of coding and variable regions in both nuclear and chloroplast DNA, we have confirmed that JBG has reverted to the green invasive within the states of Maryland, South Carolina, and Missouri. JBG has been sold and planted in just about every state in the continental U.S. where there is not an active cogongrass infestation. The extent of the revert problem in not well understood because reverted plants are undocumented and often destroyed.
Application of this molecular protocol provides a method to identify JBG reverts and can help keep these varieties from co-occurring and possibly hybridizing. Cogongrass is an obligate outcrosser and, when crossed with a different genotype, can produce viable wind-dispersed seeds that spread cogongrass over wide distances5-7. JBG has a slightly different genotype than cogongrass and may be able to form viable hybrids with cogongrass. To add to the problem, JBG is more cold and shade tolerant than cogongrass8-10, and gene flow between these two varieties is likely to generate hybrids that are more aggressive, shade tolerant, and cold hardy than wild-type cogongrass. While wild-type cogongrass currently infests over 490 million hectares worldwide, in the Southeast U.S. it infests over 500,000 hectares and is capable of occupying most of the U.S. as it rapidly spreads northward due to its broad niche and geographic potential3,7,11. The potential of a genetic crossing is a serious concern for the USDA-APHIS Federal Noxious Week Program. Currently, the USDA-APHIS prohibits JBG in states where there are major cogongrass infestations (e.g., Florida, Alabama, Mississippi). However, preventing the two varieties from combining can prove more difficult as cogongrass and JBG expand their distributions. Furthermore, the distribution of the JBG revert is currently unknown and without the ability to identify these varieties through morphology, some cogongrass infestations may be the result of JBG reverts. Unfortunately, current molecular methods of identification typically rely on AFLP (Amplified Fragment Length Polymorphisms) and DNA sequencing, both of which are time consuming and costly. Here, we present the first cost-effective and reliable PCR-based molecular genotyping method to accurately distinguish between cogongrass and JBG revert.
Protocol
1. Specimen Collection and Preservation
This method was developed and tested using fresh, frozen, and recently dried leaf tissues.
- Identify cogongrass and/or JBG tissues with the help of a taxonomist that specializes in grass species identification. With its bright red leaves, ornamental JBG is easy to visually distinguish from wild-type cogongrass and JBG revert; however, cogongrass and JBG revert are almost indistinguishable from one another. Cogongrass and the JBG reverted phenotype have green, longer leaves, considerably larger and longer rhizomes, and more leaf area than JBG12 (Figure 1).
- Fresh leaf tissue provides the most abundant and highest quality DNA and can be collected from field or greenhouse grown I. cylindrica plants. If fresh tissue is to be used, extract DNA within 3 hours of collection to help prevent degradation. Otherwise, prepare tissue for storage, keeping the tissue cool and out of direct sunlight.
- To store tissues for DNA extraction at a later date, the most optimal method is to freeze and store the tissue at -80°C immediately. Do not allow the frozen tissue to thaw prior to DNA extraction. Transfer the frozen tissue to liquid nitrogen prior to the grinding steps of DNA extraction to prevent thawing.
- If a -80°C freezer is not available, dry the tissue immediately. For dry storage, place the tissue into a paper envelope and store the envelope in dehydrated silica gel or other active desiccants at room temperature. A small amount of indicator silica mixed in with non-indicating silica will ensure that silica is fully dehydrated and suitable for drying plant tissue.
- Use at least 10 times more silica gel than fresh leaf tissue by weight. Plant tissue should dry within 24 hours. The quality and quantity of DNA is reduced through dry storage over time (as in the case of herbarium specimens).
2. DNA Extraction
To extract DNA from plant tissue, follow the DNeasy Plant Mini Kit (Qiagen, Valencia, CA; Cat# 69104 or 69106) manufacturer’s instructions with one minor modification. Instead of using the suggested less than 100 mg fresh tissue or less than 20 mg dry tissue for each column, grind greater than 100 mg, and then transfer 100 mg from fresh or frozen tissue (or > 20 mg from dry tissue) to appropriate tubes for extraction. Nuclear and plastid DNA is extracted simultaneously.
- Before starting these procedures, verify that ethanol was added to buffers AP3/E and AW.
- Grind >100 mg of fresh or frozen leaf tissue (or >20 mg of dry leaf tissue) to a fine powder using three rounds of liquid nitrogen with grinding in a chilled mortar and pestle. Insufficient disruption of the starting material or insufficient lysis can also lead to lower yields of DNA. Carefully grind the tissue and do not overload the columns with too much tissue.
- Transfer 100 mg frozen powder from fresh or frozen tissue (or 20 mg of powder from dry tissue) to a 1.5 ml microcentrifuge tube containing 400 μl of Buffer AP1 and 4 μl of RNase A. Each tube can be placed in a small rack on a balance to monitor the correct weight of tissue per tube.
- Vortex or shake sample(s) to mix and incubate for 10 min at 65°C, inverting the tube(s) 2-3 times during incubation.
- Add 130 μl of Buffer AP2 to each sample. Mix by inverting the tube(s) several times, and incubate for 5 min on ice.
- Pipet each lysate into a separate QIAshredder Mini spin column, and place each column in a 2 ml collection tube (provided with the kit). Centrifuge column(s) for 2 min at 20,000 x g (~14,000 rpm), and transfer each flow-through fraction into a new tube (not supplied with the kit) without disrupting any formed pellet.
- Add 1.5 volumes of Buffer AP3/E, and mix by pipetting.
- Transfer 650 μl of mixture into a DNeasy Mini spin column in a 2 ml collection tube. Centrifuge column(s) for 1 min at 6,000 x g (~8,000 rpm), and discard the flow-through. Repeat this step with the remaining mixture for each sample.
- Place the spin column(s) into a new 2 ml collection tube(s), and add 500 μl of Buffer AW to the top of each column. Centrifuge column(s) for 1 min at 6,000 x g (~8,000 rpm), and discard flow-through.
- Add another 500 μl of Buffer AW to the top of each column. Centrifuge for 2 min at 20,000 x g (~14,000 rpm). This step will dry the column, thus removing any residual ethanol contained in the buffers that can inhibit PCR.
- Transfer each spin column to a new 1.5 ml microcentrifuge sample tube. Add 100 μl Buffer AE to the top of each column for elution, and incubate column(s) for 5 min at room temperature. Centrifuge column(s) for 1 min at 6,000 x g (~8,000 rpm) to collect the DNA.
- Repeat these elution steps one time, eluting the DNA into the same 1.5 ml microcentrifuge tube to yield 200 μl of sample. Store DNA samples at -20°C until use. DNA concentrations depend on tissue type and storage conditions. Optimal yields are obtained when eluting DNA with a total of 200 μl of buffer AE; however, concentrations can be increased if elution volumes are reduced to as little as 50 μl.
3. Verification of DNA Quality and Quantity
- Test the quality and quantity of extracted DNA prior to PCR setup using a spectrophotometer or fluorometer and gel electrophoresis. This will help ensure the success of subsequent steps.
- Using a spectrophotometer, test DNA quality and quantity. Good DNA yields should be between 50 and 150 ng/μl with 260/280 and 230/280 ratios close to 2.0. As an example, Figure 2 shows good quality results using the NanoDrop spectrophotometer (ThermoScientific, Wilmington, DE).
- Conduct electrophoresis using a standard 1% agarose gel. Verify the presence of relatively large bands (>10 Kb) with no to little streaking from RNA contamination (Figure 3).
- If the DNA concentration is high, dilute DNA samples to 70 ng/μl for subsequent steps.
4. PCR Primers
The PCR primers used in this protocol are based on sequence differences between the plastid trnL-F spacer region of cogongrass and JBG genotypes. These differences come in the form of SNPs (Single Nucleotide Polymorphisms) and InDels (Insertions and Deletions) that allowed the development of variety-specific primers by locating the primers at the sites of unique sequences (Figure 4).
- The quality of the primers can have a significant impact on the PCR results. Order primers from a reputable company. We order our primers from Oblique Bio, Inc. (http://www.obliquebio.com/web/, Huntsville, AL), requesting only standard desalting procedures.
- trnL-F positive control primers: This primer set amplifies the plastid trnL-F spacer region of most grass taxa11-13 and serves as a good positive control (resulting in an 890 bp band).
Name Sequence trnF(GAA)-F 5′-ATTTGAACTGGTGACACGAG-3′ trnL(5′ exon)-C 5′-CGAAATCGGTAGACGCTACG-3′ - Wild-type cogongrass primers: This primer set is specific to cogongrass and does not amplify the trnL-F region of JBG genotypes. The set results in a band that is 595 bp.
Name Sequence trnLF-C-F1 5′-TCCACTTTTTTGAAAAAACAAGTGCAA-3′ trnLF-C-R1 5′-GCCGATACTCTAATAAATAAAAAAAAAAAAGAAAT-3 ‘ - JBG and JBG Revert primers: This primer set is specific to JBG genotype and does not amplify the trnL-F region of cogongrass. The set results in a band that is 594 bp.
Name Sequence trnLF-R-F2 5′-CCAAATCCACTTTTTTGAAAAAACAAGTGGTT-3′ trnLF-R-R2 5′-CGAGATTCCTTGCCGATACTCTAATAAAA-3′ - Resuspend each primer in enough volume of nuclease-free ddH2O to obtain a 100 mM stock solutions that can be stored at -20°C, long term.
- Dilute each primer stock to 12 mM prior to the PCR setup steps.
5. PCR Setup
DNA extractions are amplified using each of the above primer sets in PCR reactions. Include a positive control to ensure that all PCR reagents are working well and can generate a band. Include a negative control to ensure that none of the reagents are contaminated with unwanted DNA. The negative control contains no-template and should result in no band production.
- Prepare all reactions in thin-walled PCR tubes to allow better heat transfer between the thermocycler block and the sample. We recommend using commercially available aerosol-free pipette tips to help avoid contamination.
- For each isolated DNA sample, set up 50 μl PCR reactions using each of the above primer sets in 0.2-ml thin-walled PCR tubes by adding the following reagents on ICE in the order listed below. If multiple samples are being prepared, make a cocktail containing all reagents with the exception of the DNA template to establish uniform conditions between all reactions.
PCR Reagent Volume Used Final Concetration Nuclease-free ddH2O 40.5 μl 10X Advantage 2 PCR Buffer (Clonetech, CA) 5.0 μl 10% (v/v) Advantage UltraPure PCR dNTP Mix (10 mM each, Clonetech, CA) 1.0 μl 0.2 mM Primer 1 (12μM in ddH2O) 1.0 μl 0.24 μM Primer 2 (12μM in ddH2O) 1.0 μl 0.24 μM Advantage 2 Polymerase Mix (Clonetech, CA) 0.5 μl 1% (v/v) DNA extraction (70 ng/reaction; adjust ddH2O volume as needed) 1.0 μl 1.4 ng/μl Total: 50.0 μl - To ensure that all PCR reagents are working well, setup the positive control using the positive control primers. This primer set works equally well for cogongrass, JBG, JBG revert and other grasses and will result in a band that is 890 bp.
- Setup the negative control using the same control primer set as the positive control, using ddH2O instead of the DNA extraction. If all reagents are free of DNA contaminates, this reaction will result in no band.
- If the DNA concentration is low, more DNA can be added to each reaction, adjusting the amount of Nuclease-free ddH2O used to bring the total reaction volume to 50 μl. Do not use more than 5 μl of DNA (10% of the total volume) per reaction, as possible impurities contained in the DNA samples can inhibit PCR reactions.
6. PCR Cycling
- Carry out PCR amplifications in a thermocycler equipped with a heated lid using the following PCR cycling parameters. We use the Mastercycle pro S thermocycler (Eppendorf, Hauppauge, NY) set to operate with standard temperature ramping conditions. Any quality thermocycler should perform well.
Cycle Denaturation Annealing Polymerization 1 2 min at 95°C 2 30 sec at 95°C 30 sec at 61°C 90 sec at 68°C 35 Cycles 3 5 min at 68°C Hold at 4°C until the sample is removed - Optimize the conditions for PCR (including primer annealing temperature, extension times, and number of cycles) as needed depending on the quality of the DNA, primers, Taq polymerase or type of thermocycler used. We recommend using a gradient capable thermocycler when determining the optimal annealing temperatures.
- If the thermocycler being used does not have a heated lid, add 1 drop of mineral oil to the top of each sample to prevent evaporation during PCR cycling.
7. Gel Electrophoresis of PCR Products
To visualize the results of the analysis, separate PCR products on a 1% agarose gel using standard electrophoresis.
- Combine 2 μl of a standard DNA loading buffer (typically a 5x or 6x solution) with 5 μl of each amplified PCR product.
- Load samples onto a 1% agarose gel containing EtBr (ethidium bromide for DNA staining) made with either TAE or SB (sodium borate) buffer systems16. We use 1 μl of a 10 mg/ml EtBr stock solution per 100 ml 1% agarose (0.1 μg/ml).
- Run samples at ~120V until the dye front reaches ¾ of the total length of the gel.
- Under UV light (e.g. a short wave UV light box), inspect the resulting bands to see if an appropriate fragment was amplified.
- Document the gel and resulting bands using an available photo-documentation system or camera.
8. Representative Results
Upon visualization of the PCR products, cogongrass has a unique banding pattern compared to that of JBG or reverted JBG (Figure 5). For each DNA sample, the trnL-F positive control primer set should result in a single high-intensity band at ~890 bp. This verifies that all PCR reagents are working well. Similarly, the negative control (no template) reaction should contain no bands for any primer set used. This verifies that none of the reagents were contaminated.
If the DNA sample is derived from wild-type cogongrass, a PCR reaction using the cogongrass-specific primer set will result in a single band at ~595 bp while the JBG-specific primers will result in no band. Likewise, if the DNA sample is derived from JBG or reverted JBG, a PCR reaction using the JBG-specific primer set will result in a single band at ~594 bp while the cogongrass-specific primers will result in no band. Because JBG and reverted JBG have identical nucleic acid sequence, they will hence have identical banding patterns. If many samples are to be compared on a gel at the same time, we recommend running all samples derived from each primer set next to one another, thus making it easier to scan the samples for positive results.
Morphological differences between JBG and JBG reverts are fairly obvious (e.g. red color of the leaves and smaller stature of JBG vs. the green coloration, larger stature and aggressive growth of the JBG revert), so while PCR results will be the same, the JBG varieties are easy to distinguish using plant morphology.

Figure 1. Comparison of greenhouse grown Imperata cylindrica var. koenigii (Japanese blood grass), Reverted I. cylindrica var. koenigii (JBG Revert) and I. cylindrica (Wild-type cogongrass).
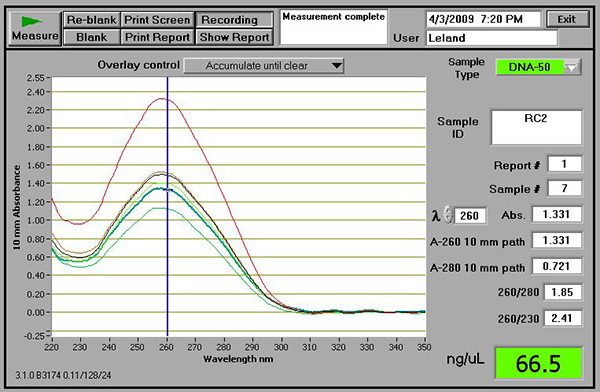
Figure 2. An example of DNA samples verified using a NanoDrop spectrophotometer. Note that, irrespective of the spectrophotometer used, the 260/280 ratio should be close to 1.8 and the 260/230 ratio should close to 2.0.
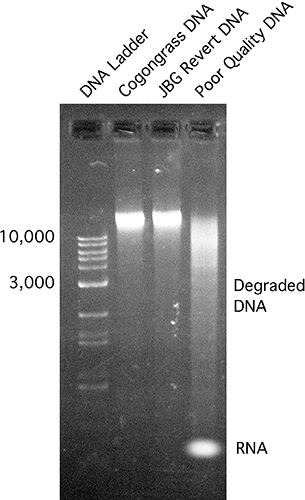
Figure 3. DNA samples verified using standard gel electrophoresis on a 1% agarose gel. A commercial DNA marker was used for size analysis. Lane #4 is an example of poor quality DNA sample, showing smearing and some RNA contamination.
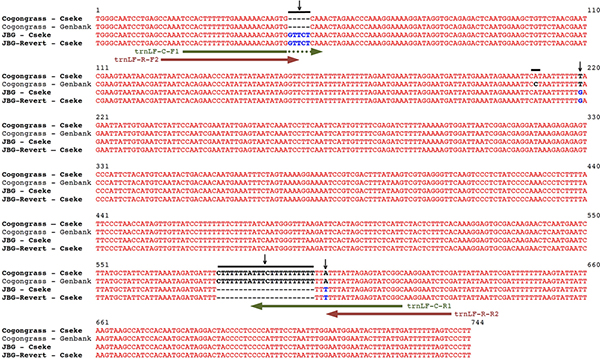
Figure 4. Sequence alignments of the trnL-F regions of Imperata cylindrica var. koenigii (Japanese blood grass), Reverted I. cylindrica var. koenigii (JBG Revert) and I. cylindrica (Wild-type cogongrass). Vertical black arrows indicate differences in sequences resulting from SNPs and InDels. Horizontal green arrows indicate the positions of the Wild-type cogongrass primers used for cogongrass-specific PCR. Horizontal red arrows indicate the positions of the JBG and JBG Revert primers used for JBG-specific PCR. Please click here to see a larger version of this figure.
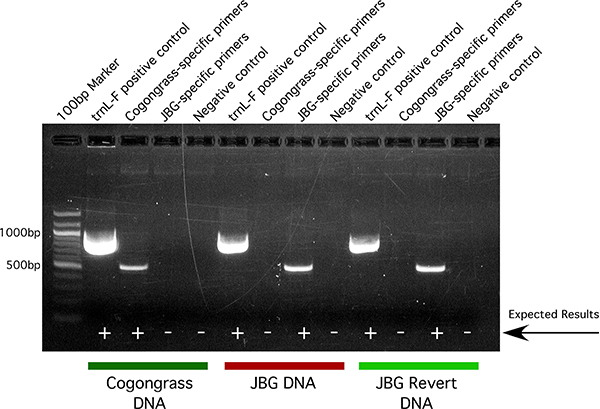
Figure 5. Representative result of gel electrophoresis of PCR products derived from cogongrass, JBG and JBG revert DNA samples combined with cogongrass- and JBG-specific primers as well as the trnL-F positive control and a no template negative control.
Discussion
The U.S. nursery and landscape industries thrive on cultivating and selling exotic and novel plant species. This, coupled with the growing globalization of trade, increases the chances that an invasive plant species will enter, establish, and spread in the U.S. The ability to federally regulate such plants relies on information that is often not available, including the potential to become invasive, correct taxonomy, and genetic distinctness from native and naturalized taxa. Because our knowledge of invasive plants is often limited, imported plants with hidden invasive characteristics have been willingly introduced only to learn later that they invade our agricultural and natural resources. This protocol aims to address such problems associated with I. cylindrica varieties by providing the first simplified molecular method that can accurately distinguish between wild-type cogongrass and the reverted form of its ornamental JBG counterpart.
For the development of this protocol, wild-type cogongrass was collected from naturalized populations at the Pond Creek Forestry Unit in Santa Rosa County near Jay, FL in June of 2008. JBG was procured from a commercial nursery (Bluebird Nursery, Inc.) in June 2008 as well as from a homeowner’s collection in Columbia, MO. JBG reverts were obtained from the courtyard of the Campbell Geological Museum at Clemson University, SC in June 2008; from University Park in Riverdale, MA in June 2009; and from the front yard of a homeowner in Columbia, MO in 2009 (identified by Leland Cseke). All plants were maintained in a greenhouse located at the University of Alabama in Huntsville (Huntsville, AL).
Genetic sequencing of DNA collected from these plants included the in depth comparisons of 9 independent DNA regions commonly used to barcode plants2. In all cases, the sequences of JBG were a 100% match to those of the JBG revert, thus helping to verify that JBG does indeed revert to a green, invasive form. Only the nuclear ITS and the chloroplast trnL-F regions have differences that can be used to genetically distinguish between cogongrass and JBG. The ITS region has a total of 3 SNPs (single nucleotide polymorphisms) between cogongrass and JBG, while the trnL-F region has 2 SNPs and 2 InDels (insertions and deletions). These genetic differences allowed variety-specific PCR primers to be developed that can distinguish between wild-type cogongrass and JBG reverts. The most reliable results have come from primers derived from the plastid trnL-F region. Thus, this protocol is based on the sequence differences between the trnL-F regions of the chloroplast genome of cogongrass, JBG and reverted JBG (Figure 4).
To help generate primers that are more specific to the varieties in question and to help avoid false positives from closely-related species, all known trnL-F sequences of I. cylindrica varieties were compared with trnL-F sequences from related grass species (43 independent sequences from 29 species, e.g., Cymbopogon citratus, Sorghastrum incompletum, Coix lacryma-jobi, Miscanthus sinensis, Saccharum officinarum, Sorghum halepense). Although, we examined variety-specific primer sequences across 29 species of grass, the specificity of the variety-specific primers has not been comprehensively examined for its ability to amplify DNA from most other grass species. Consequently, tissue used for DNA extraction should be carefully identified as I. cylindrica prior to starting this protocol. If the grass cannot be identified as either cogongrass or JBG, then we suggest sequencing the PCR product to make sure that sequences are an exact match to cogongrass or JBG. Currently, the most accurate method to verify the identity of a given grass specimen is to perform PCR on both the trnL-F and ITS regions, followed by sequence verification of PCR products and comparison of the sequences to known sequences from accurately identified taxa. DNA can be amplified using control primers detailed in this protocol (for the trnL-F region) or other primers that available in other publications13-15. Sequencing is much more labor and cost intensive than using our simplified procedure.
The quality of the primers used for PCR is critical to the success of the procedure. We have made the primers for this procedure available from Oblique Bio, Inc. (http://www.obliquebio.com/web/, Huntsville, AL). The advantage to ordering the primers from Oblique Bio is that they store large numbers of aliquots of each primer from the identical sample production series as the primers we used for optimization in our laboratory. Consequently, primers not only have the same sequence, but they come from the exact same production lot that was used in this protocol. Using primers from the same lot, can help avoid extraneous variables in the procedure that may result from differences in the quality of PCR primers. Likewise, while other Taq polymerases should work fine for PCR, the quality of the Taq polymerase used will have an impact on the quality of PCR results. To allow for better consistency in PCR reagents, we have optimized the protocol using reagents from Clontech. The Advantage 2 Polymerase (Clontech, Mountain View, CA, Cat# 639201 or 639202) is a mixture of a robust, hot-start Taq polymerase and a proof-reading enzyme that helps to provide high specificity and more accurate amplifications.
Because this protocol relies on chloroplast DNA, which is maternally inherited in grasses, hybridization events between cogongrass and JBG genotypes may not be captured with our molecular identification procedure. In cases where hypridization is suspected, we recommend using nuclear regions that are inherited from both parents. The most commonly used non-plastid variable region to consider in plant genotyping is the nuclear ribosomal ITS region13-15,17. Currently, we are making progress towards multiplexing the amplification of the chloroplast trnL-F region with that of the nuclear ITS region in the same PCR tube. Multiplexing plastid with nuclear DNA regions would potentially circumvent the limitations of using either alone; however, such methods require optimization and additional evaluation to determine feasibility on a case by case basis. The use of quantitative real-time PCR (qPCR) and newer technologies, such as molecular beacons (fluorescent primer probes), are also being evaluated as fail-safe and accurate plant genotyping methods.
The protocol presented here provides a fast and reliable approach to distinguish the JBG revert from that of wild-type cogongrass. We encourage users of this protocol to contact us to report results derived from the use of this protocol. Such shared information will help provide information on the distribution of JBG reverts. This will also help regulators at the USDA make informed decisions on actions that may be need to circumvent the spread and potential hybridization of the highly invasive cogongrass varieties.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Alan Tasker (USDA-APHIS, Riverdale, MD), Stephen Compton (Clemson), Sherry Aultman (Clemson), Craig Ramsey (USDA-APHIS, Fort Collins, CO), and Betty Marose (UMD) for assistance in obtaining specimens. We thank students Andrew Adrian (UA-Huntsville) and Derek Thacker (UA-Huntsville) for their assistance in testing this protocol, and Joseph Herdy for his work in the filming of the video. This work was funded by National Fish and Wildlife Foundation.
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| DNeasy Plant Mini Kit | Qiagen, Valencia, CA | 69104 or 69106 | Any reputable genomic plant DNA kit that yields good quality DNA should work fine for these procedures. |
| trnF(GAA)-F | Oblique Bio, Inc. | 3-0578 | Forward positive control primer |
| trnL(5′ exon)-C | Oblique Bio, Inc. | 3-0579 | Reverse positive control primer |
| trnLF-C-F1 | Oblique Bio, Inc. | 3-0864 | Forward wild-type cogongrass primer |
| trnLF-C-R1 | Oblique Bio, Inc. | 3-0865 | Reverse wild-type cogongrass primer |
| trnLF-R-F2 | Oblique Bio, Inc. | 3-0866 | Forward JBG and JBG revert primer |
| trnLF-R-R2 | Oblique Bio, Inc. | 3-0867 | Reverse JBG and JBG revert primer |
| Advantage UltraPure PCR dNTP Mix | Clontech, Mountain View, CA | 639125 | |
| Advantage 2 Polymerase | Clontech, Mountain View, CA | 639201 or 639202 | A good proof-reading, hot-start Taq polymerase |
References
- Holm, L. G., Pancho, J. V., Herberger, J. P., Plucknett, D. L. . A Geographical Atlas of World Weeds. , (1979).
- CABI, Crop Protection Compendium. Commonwealth Agricultural Bureau International (CABI). , (2007).
- MacDonald, G. E. Cogongrass (Imperata cylindrica) – Biology, ecology, and management. Critical Reviews in Plant Sciences. 23, 367-380 (2004).
- Talley, S. M., Cseke, L. J., Zink, R. Molecular diagnostic technologies for invasive plants. CPHST Fort Collins Laboratory 2009 Annual Report. , 27-28 (2009).
- Dozier, H., Gaffney, J. F., McDonald, S. -. K., Johnson, E. R. R. L., Shilling, D. G. Cogongrass in the United States: History, ecology, impacts, and management. Weed Technology. 12, 737-743 (1998).
- Chikoye, D., Ekeleme, F. Weed flora and soil seedbanks in fields dominated by Imperata cylindrica in the moist savannah of West Africa. Weed Research. 41, 475-490 (2001).
- Weber, E. . Invasive Plant Species of the World: A Reference Guide to Environmental Weeds. , (2003).
- Patterson, D. T. Shading effects on growth and partitioning of plant biomass in Cogongrass (Imperata cylindrica) from shaded and exposed habitats. Weed Science. 28, 735-740 (1980).
- Cole, J. T., Cole, J. C. Ornamental grass growth response to three shade intensities. Journal of Environmental Horticulture. 18, 18-22 (2000).
- Bryson, C. T., Koger, C. H., Byrd, J. D. Effects of temperature and exposure period to heat on cogongrass (Imperata cylindrica) viability. Weed Technology. 21, 141-144 (2007).
- Capo-chichi, L. J. A., Faircloth, W. H., Williamson, A. G., Patterson, M. G., Miller, J. H., van Santen, E. Invasion Dynamics and Genotypic Diversity of Cogongrass (Imperata cylindrica) at the Point of Introduction in the Southeastern United States. Invasive Plant Science and Management. 1 (2), 133-141 (2008).
- Talley, S. M., Ramsey, C. L., Zink, R. Experimentally assessing the invasive potential of plants. CPHST Laboratory 2009 Annual Report. , 29-30 (2009).
- Hodkinson, T. R., Chase, M. W., Lledó, M. D., Salamin, N., Renvoize, S. A. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnLintron and trnL-F intergenic spacers. J. Plant Res. 115, 381-392 (2002).
- Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., Janzen, D. H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 102 (23), 8369-8374 (2005).
- Roodt-Wilding, R., Spies, J. J. Phylogenetic relationships in southern African chloridoid grasses (Poaceae) based on nuclear and chloroplast sequence data. Systematics and Biodiversity. 4, 401-415 (2006).
- Brody, J. R., Kern, S. E. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques. 36, 214-216 (2004).
- Chou, C. -. H., Tsai, C. C. Genetic variation in the intergenic spacer of ribosomal DNA of Imperata cylindrica (L.) Beauv. var. major (Cogongrass) populations in Taiwan. Botanical Bulletin of Academia Sinica (Taipei). 40, 319-332 (1999).

