Detection of Microregional Hypoxia in Mouse Cerebral Cortex by Two-photon Imaging of Endogenous NADH Fluorescence
Summary
Here we describe a method to directly visualize microregional tissue hypoxia in the mouse cortex in vivo. It is based on concurrent two-photon imaging of nicotinamide adenine dinucleotide (NADH) and the cortical microcirculation. This method is useful for high resolution analysis of tissue oxygen supply.
Abstract
The brain’s ability to function at high levels of metabolic demand depends on continuous oxygen supply through blood flow and tissue oxygen diffusion. Here we present a visualized experimental and methodological protocol to directly visualize microregional tissue hypoxia and to infer perivascular oxygen gradients in the mouse cortex. It is based on the non-linear relationship between nicotinamide adenine dinucleotide (NADH) endogenous fluorescence intensity and oxygen partial pressure in the tissue, where observed tissue NADH fluorescence abruptly increases at tissue oxygen levels below 10 mmHg1. We use two-photon excitation at 740 nm which allows for concurrent excitation of intrinsic NADH tissue fluorescence and blood plasma contrasted with Texas-Red dextran. The advantages of this method over existing approaches include the following: it takes advantage of an intrinsic tissue signal and can be performed using standard two-photon in vivo imaging equipment; it permits continuous monitoring in the whole field of view with a depth resolution of ~50 μm. We demonstrate that brain tissue areas furthest from cerebral blood vessels correspond to vulnerable watershed areas which are the first to become functionally hypoxic following a decline in vascular oxygen supply. This method allows one to image microregional cortical oxygenation and is therefore useful for examining the role of inadequate or restricted tissue oxygen supply in neurovascular diseases and stroke.
Protocol
1. Preparing the animal for imaging
Note: We use adult c57BL mice. Different transgenic strains can be used depending on the research question. Microvascular surgeries and pulse oximetry are facilitated in mice with white fur (e.g. FVB strain).
- Prepare the surgical site at the bench. All surgical instruments need to be autoclaved or disinfected with 70% ethanol.
- Insert a rectal probe, and continuously measure the animal’s body temperature throughout the procedure
- Anesthetize the mouse using initially 5% isoflurane. In 15-30 s, when the animal is not moving, place it on a heating pad (37 °C), and apply a face mask for the continuous delivery of air containing 1.3-1.5% isoflurane). Make sure that the animal is not responding to a pain stimulus (e.g. tail pinch).
- Use artificial tear gel to cover the mouse’s eyes, to prevent exposure to dry air.
- Using an electric razor, remove hair from the head and from both thighs. Apply hair remover (e.g. Nair) to the thigh for 2 min, and then wipe it carefully to remove the remaining hair. This thigh will be used for oximetry.
- Disinfect the scalp using a 10% povidone-iodine solution and 70% ethanol.
2. Preparing the open skull cranial window
- Starting 5 mm caudal to the skull, make an incision in the scalp with scissors, and advance one centimeter. Move skin to the sides to expose the skull.
- To remove the membranes on top of the skull apply 10% ferric chloride solution to the top of the skull. Wipe away the excess solution, and scrape the membranes away with 45° angle #5 tweezers. It is important to prepare the site carefully, in order to ensure that the head plate can be securely attached to the skull (steps #4 thru 6).
- Using a dissection microscope, identify the area of interest. Note that the cranial sutures should be avoided because the skull can break unpredictably along these lines.
- Apply a thin layer of rapid glue (e.g. Locktite 454) around the edges of the window of the head plate (Figure 1). Position the head plate in such a way that the area of interest is exposed in the window. Apply light pressure on the head plate. Apply a small amount of dental cement to polymerize the glue rapidly. Hold for 10 seconds while the glue is polymerizing.
- Apply a small amount of rapid glue to the edges of the window to seal the head plate and the skull. Screw the head plate to the animal holder under the dissection scope.
- Using the Microtorque II drill set at 6000 rpm and a IRF 005 drill bit, remove all extra glue from the skull and from the head plate. Any remaining glue on the head plate should be removed, since it will otherwise interfere with the attachment of the glass cover.
- Set the drill at 1000 rpm, and start drilling the skull by making a circle inside the head plate window, diameter ~5 mm. Use light sweeping movements as if you are drawing with a very soft pencil. Stop drilling every 20-30 seconds to remove bone dust using compressed air.
Note: The shape of the opening in the head plate allows additional access for the drill bit (for left- and right-handed surgeon), which makes it easier to create a circular cranial window. In addition, the two odd shaped holes provide the required space to insert a Clark-electrode or glass-electrode into the brain tissue under the cranial window for additional polarographic or electrophysiological measurements. - As the drilling progresses, a burrow is formed around the intact skull in the center. Be careful not to poke through the thinned skull, but to make this burrow of even thickness. Be extra careful around large blood vessels -they should not be compromised and, if possible, not even touched.
- Use one “leg” of tweezers #3 to lift (“flick off”) the bone in the center of the window. Apply a drop of artificial cerebrospinal fluid (aCSF) on the window.
- Wipe aCSF gently using a corner of soft tissue paper (e.g. Kimwipe). Usually the dura is damaged in one or more places around the edge of the window. Starting at one of these sites, gently lift and then remove the dura from the brain surface, using 45° angle tweezers #5. When choosing the site at which to start removing the dura, avoid the areas adjacent to large blood vessels. If bleeding occurs, apply a drop of aCSF and wait for 2-3 min for minor bleeding to stop.
- Prepare 0.07% low-melting agarose dissolved in aCSF) Bring the melted agarose to body temperature. Wipe away the excess of aCSF from the surface of the brain. Pour the agarose in the window, and cover with a glass slide. Press the glass gently to make a contact between the glass and the head plate, and wait 10-20 sec until the agarose is solid.
- Remove excess agarose from the glass cover using tweezers and soft tissue paper.
- Put a small amount of rapid glue around the glass to glue the glass to the head plate. Apply a small amount of dental cement to solidify the glue (Figure 1).
3. Intravenous injection and blood oxygenation monitoring
Note: We routinely used the femoral vein for intravenous application of Texas red rather than the tail vein. This is because femoral vein injections provide safe and reproducible bolus injections of the dye.
- Turn the animal partially over to expose the inside of the left thigh. Use tape to secure the animal in this position.
- Apply antiseptic scrub, and then 100 % ethanol to the skin.
- Make an incision along the medial thigh from the knee to the pubic symphysis. Separate soft tissues by blunt dissection using tweezers #5.
- Fill 1ml syringe with 130 μl of Texas Red (2.5 mg/ml). Attach a new needle (gauge 30), and bend it carefully at 30°, keeping the bevel up. Fill the needle with Texas Red solution.
- Under a dissection microscope, insert the needle into the femoral vein and inject 100 μl of Texas Red. Remove needle and lightly press the vein with gauze to stop bleeding.
- Close the skin using 4-0 suture.
- For continuous monitoring of blood SpO2 (to ensure adequate blood oxygenation) place the oximeter probe on the right thigh (from which hair was previously removed).
4. Two-photon imaging
Note: We use an Olympus Fluoview1000 multiphoton imaging system (upright) with a Spectra-Physics MaiTai HP DeepSee femtosecond Ti:Sa laser as an excitation source. For imaging, we use × 10 NA 0.45 (Zeiss C-Apochromat), or × 25 NA 1.05 (Olympus XLPlan N) water-immersion objectives. We take images at 12-bit depth at a resolution of 512 × 512 pixels with a pixel dwell time of 2 μs. The NADH and Texas-Red-dextran are concurrently excited at 740 nm, and fluorescence is separated from the excitation light using a dichroic mirror/near-IR-blocking filter combination (FF665-Di01; FF01-680/SP, Semrock, Rochester, NY, USA) divided into two channels using a dichroic mirror (505DCXRU, Chroma, Rockingham, VT, USA), and bandpass filtered (NADH-Semrock FF460/80Texas-Red-dextran-Semrock FF607/36). Laser power measured after the objective was 10-20 mW for imaging in layer I and 20-50 mW for imaging in layer II.
- Transfer the surgically prepared mouse to the microscope stage. Take an initial picture of the cranial window site using bright field illumination at 4x magnification to use as a reference map for registration with higher magnification two-photon angiographies.
- Zoom in on the area of interest using × 10 magnification. Select an area of interest in cortical layers I or II (up to 150 μm below the pial surface). If higher magnification is desired, use × 25 objective
- Start record the time series (Figure 2). We recommend using an average of 3-5 frames to increase the quality of the image.
- Induce hypoxemia by adding 50 % N2 to the air that the animal is breathing. This will bring the O2 level to 10 %. Verify hypoxemia by monitoring oximeter data.
- Continue to record the time series through hypoxemia (for example, for 30 seconds), and after normal O2 level is restored.
- Collect data for calculation of the oxygen distribution by detecting, measuring and quantifying the area of hypoxia and oxygenated tissue cylinders around the blood vessels.
5. Data processing
- Determine the Krogh tissue cylinder radius R. This can be done manually (Figure 3) by measuring the distance between center of the blood vessel and the boundary at which high NADH fluorescence is detected.
- Alternatively, use the computational investigator-independent semi-automated determination of the tissue cylinder radius R and the central blood vessel r that has been described previously1 in supplementary form and summarized in the discussion section of this protocol.
- Determine the area of hypoxia using open-access image analysis software, e.g. ImageJ. To do this, use NADH signal, define a threshold that delineates the high NADH intensity area, and then measure the area with elevated NADH tissue fluorescence.
6. Representative Results
Figure 2 Shows a video of NADH/angiography images during transient hypoxemia (for the PDF version of this Figure, selected still images have been used). The inspired oxygen concentration was lowered from 21 % to 10 % for a period of 3 min. This level of experimentally induced hypoxemia is sufficient to induce severe hypoxia in the cerebral cortex. Hypoxia led to an increase in NADH fluorescence, initially in the areas which were furthest from the arterial blood supply. Note the sharp NADH tissue boundaries, which represent observable tissue boundaries of oxygen diffusion from the cortical microcirculation. The image in Figure 3 shows the geometry of oxygen diffusion boundaries surrounding cerebral vessels. It is possible to deduce Krogh cylinder diameters from these boundaries, as described previously 1.
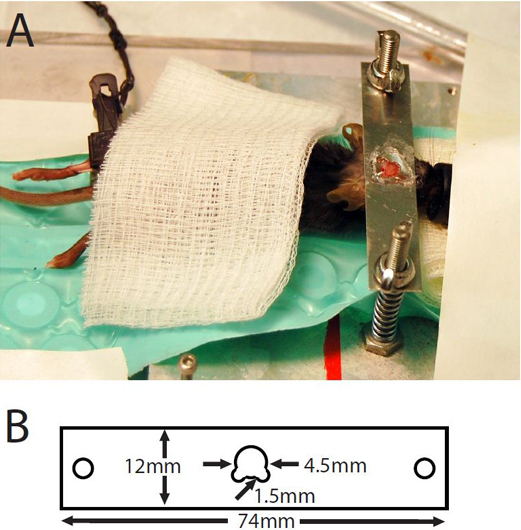
Figure 1. (A) Mouse with cranial window prepared for imaging. (B) Dimensions of head plate.
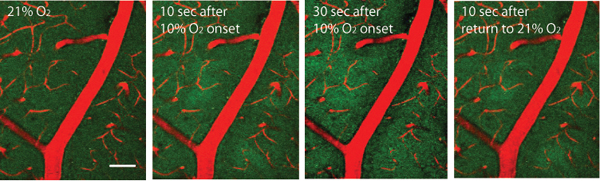
Figure 2. Endogenous NADH green fluorescence visualized with two-photon imaging through cranial window, approximately 50 μm below the pial surface. Blood vessels are visualized with Texas Red dextran. Oxygen in inhaled air was reduced to 10% for 3 min, and then restored for 21%. Scale bar: 50μm.
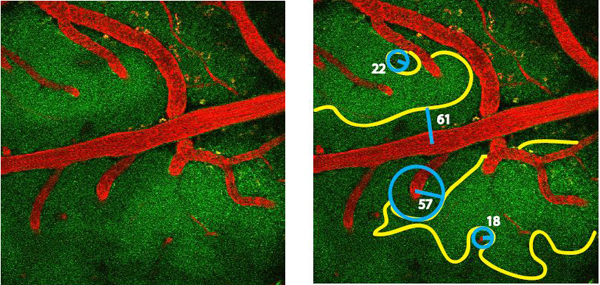
Figure 3. Geometry of NADH fluorescence boundaries. Yellow outline shows boundary of functional hypoxia; blue circles show projected oxygenated (Krogh) cylinders. Blue lines show cylinder radii; numbers show radii in micrometers.
Discussion
High spatial resolution information about oxygen diffusion is important to understanding how blood flow in the brain is regulated to provide oxygen to brain cells, and to meet metabolic demand. Traditional polarographic oxygen measurements using Clark-style glass electrodes are highly invasive and have low spatial resolution2-3 and significant (second range) response time. So far the only non-invasive method for measuring pO2 in brain tissue is phosphorescence quenching, where the rate of decay of the excited probe is proportional to oxygen concentration4. This method provides accurate oxygen concentrations, but requires a proprietary dye and a technically sophisticated phosphorescence lifetime imaging system. Here, we demonstrate a straightforward, simpler approach that can be conducted on a standard two-photon imaging system with two flurescence channels. Our approach takes advantage of an intrinsic tissue signal5 and only requires contrasting visualization of the cortical microcirculation. Because of the non-linear, essentially binary increase of NADH fluorescence at functionally limiting oxygen concentrations1, increased intrinsic NADH fluorescence is observed only in areas with significant, metabolically limiting hypoxia. An important implication is that tissue boundaries of oxygen diffusion from cortical microvessels are directly observable by cylindrically shaped intensity changes of endogenous NADH fluorescence. We refer to these structures as Krogh cylinders, because the concept of cylindrically shaped structures that define the oxygenated volume of tissue surrounding a blood vessel was introduced by August Krogh and has been recently experimentally observed using two-photon NADH imaging1. Krogh cylinders images can be collected in 3D by taking a z-stack of image frames. They are especially prominent in the vicinity of penetrating arterioles and they are congruous with capillary depleted periarteriolar tissue cylinders1,4.
To provide an objective determination of the Krogh tissue cylinder radius R (see Section 5.2) we measured their radial pixel intensity values within a well-defined segment between the center of the cylinder and the outer boundary using the Matlab function “improfile”. The outer boundary of the segment should be chosen to extend with a safety margin beyond the visible boundary. To improve the signal-to-noise level we averageed over all radial lines needed to cover the visible cylinder segment at 1° steps. The resulting mean radial intensity profile within the segment exhibited a steep increase which corresponded to the visible tissue boundary R. The we fit a sigmoidal function (e.g. Boltzmann function) to the averaged radial intensity profile and used its inflection point (also known as x0) as a definition of R. The corresponding two-photon microangiography (Texas-red) revealed the cross-section of a solitary central blood vessel in the center of cylinder. The diameter of the central blood vessel can be directly applied to determine r.
Two-photon NADH imaging provides the same spatial resolution as the concurrent high-resolution imaging of the cortical microangiography. An important feature for the quantitative application of this method is that p50 of the NADH fluorescence increase has been measured to be of 3.4 ± 0.6 mm Hg 1 and that the NADH fluorescence intensity as a function of microregional tissue pO2 can be mathematically described with a sigmoidal function. . We show that this technique allows one to identify brain areas which are most vulnerable to hypoxia (by decreasing oxygen content in the air to 10 %). We also show that oxygen diffusion follows a simple geometric perivascular pattern.
One critical step for this method is the quality of the cranial window preparation. The surgery should produce minimal damage in order not to disturb blood flow to the exposed area. A concern is that in a surgically compromised preparation, the cortex under the window may be hypoxic to begin with, precluding any meaningful experiments. A well-prepared cranial window should have intact major and minor blood vessels with vivid blood flow in all vessel types and no significant bleeding along the edges. Under normoxic conditions (PaO2 80-100 mmHg, Sp O2 97-99%) the brain parenchyma should exhibit uniform, homogeneous NADH fluorescence without conspicuous, bright tissue patches with elevated NADH fluorescence.
A fundamental physical constraint of our approach is limited depth penetration. The blue-green NADH fluorescence in brain is rapidly attenuated by hemoglobin absorption and tissue scattering at these wavelengths. Even with high numerical aperture (e.g. 1.05) water immersion objectives two-photon NADH imaging is currently limited to cortical layers I and II. This limitation is scientifically relevant because energy metabolism in or in proximity to white matter will likely differ from gray matter. However, the investigation of deep cortical structures such as layers IV-VI or subcortical structures such as white matter tracts or the striatum would require the use of specialized microlenses as described in the mouse cortex in vivo6.
NADH-based measurement of oxygen diffusion boundaries can be especially useful when combined with other measurements such as analyses of functional hyperemia, and detection of capillary flux rates7. For example, this technique can be adapted to visualize hypoxia in stroke and Alzheimer’s disease (AD) models. The simple geometry of oxygen diffusion allows one to predict the oxygen gradient in microvascular beds under circumstances where capillary density is decreased8 (e.g., AD9) and to examine whether brain tissue regions with reduced capillary density are at increased risk for hypoxia damage due to microstrokes. The ability to image microregionally also allows one to examine the geometry and size of tissue microstrokes and determine the volume of tissue in which hypoxia occurs, as well as the relationship between tissue hypoxia and subsequent neuronal death or capillary remodeling10.
Finally, since increases in endogenous NADH fluorescence are the direct consequence of acute mitochondrial dysfunction, this method creates the opportunity to use NADH imaging as a specific reporter for neural energy metabolism11 and a proxy for mitochondrial dysfunction.
In conclusion, two-photon imaging of endogenous NADH fluorescence is a simple, undemanding tool that can be used to understand oxygen delivery and consumption in the brain under both normal and in pathologic states.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Maiken Nedergaard (University of Rochester Medical Center) for the head plate design. The work has been supported by NIH awards to S.D (R01DA026325 and P30AI078498 and foundation grants to KK (DANA foundation Brain and Immunoimaging program, American Heart Association 0635595T and the ALS Association [#1112)]).
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| Heating pads | Beyond Bodi Heat | ||
| Ophthalmic ointment (Artificial tears) | Pfizer | ||
| Povidone-iodine 10% solution | Betadine | ||
| Ferric chloride 10% solution | |||
| Cement | Stoelting Company | 51456 | |
| Cyanoacrylate 454 | Loctite | ||
| aCSF | Harvard Apparatus | 597316 | |
| Microtorque II handpiece kit | Pearson | R14-0002 | |
| IRF 007 drill bits | Fine Science Tools | 19008-07 | |
| Forceps #5 | Fine Science Tools | 11295 | |
| Forceps #5/45 | Fine Science Tools | 11251-35 | |
| #0 glass coverslip | Electron Microscopy Sciences | 63750-01 | |
| Photomultiplier tube | Hamamatsu | HC125-02 | |
| Ti:Sapphire laser Mai-Tai | Spectra-Physics |
References
- Kasischke, K. A. Two-photon NADH imaging exposes boundaries of oxygen diffusion in cortical vascular supply regions. J. Cereb. Blood. Flow. Metab. 31, 68-81 (2011).
- Ndubuizu, O., LaManna, J. C. Brain tissue oxygen concentration measurements. Antioxid. Redox. Signal. 9, 1207-1219 (2007).
- Sharan, M., Vovenko, E. P., Vadapalli, A., Popel, A. S., Pittman, R. N. Experimental and theoretical studies of oxygen gradients in rat pial microvessels. J. Cereb. Blood. Flow. Metab. 28, 1597-1604 (2008).
- Sakadzic, S. Two-photon high-resolution measurement of partial pressure of oxygen in cerebral vasculature and tissue. Nat. Methods. 7, 755-759 (2010).
- Vishwasrao, H. D., Heikal, A. A., Kasischke, K. A., Webb, W. W. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J. Biol. Chem. 280, 25119-25126 (2005).
- Kleinfeld, D., Mitra, P. P., Helmchen, F., Denk, W. Fluctuations and stimulus-induced changes in blood flow observed in individual capillaries in layers 2 through 4 of rat neocortex. Proc. Natl. Acad. Sci. U. S. A. 95, 15741-15746 (1998).
- Brown, W. R., Thore, C. R. Review: cerebral microvascular pathology in ageing and neurodegeneration. Neuropathol. Appl. Neurobiol. 37, 56-74 (2011).
- de la Torre, J. C. Impaired brain microcirculation may trigger Alzheimer’s disease. Neurosci. Biobehav. Rev. 18, 397-401 (1994).
- Brown, C. E., Boyd, J. D., Murphy, T. H. Longitudinal in vivo imaging reveals balanced and branch-specific remodeling of mature cortical pyramidal dendritic arbors after stroke. J. Cereb. Blood. Flow. Metab. 30, 783-791 (2010).
- Wilson, D. F., Erecinska, M., Drown, C., Silver, I. A. The oxygen dependence of cellular energy metabolism. Arch. Biochem. Biophys. 195, 485-493 (1979).

