Annotation of Plant Gene Function via Combined Genomics, Metabolomics and Informatics
Summary
Combination of genomics, co-expression gene analysis and the identification of target compounds via metabolism give gene functional annotation.
Abstract
Given the ever expanding number of model plant species for which complete genome sequences are available and the abundance of bio-resources such as knockout mutants, wild accessions and advanced breeding populations, there is a rising burden for gene functional annotation. In this protocol, annotation of plant gene function using combined co-expression gene analysis, metabolomics and informatics is provided (Figure 1). This approach is based on the theory of using target genes of known function to allow the identification of non-annotated genes likely to be involved in a certain metabolic process, with the identification of target compounds via metabolomics. Strategies are put forward for applying this information on populations generated by both forward and reverse genetics approaches in spite of none of these are effortless. By corollary this approach can also be used as an approach to characterise unknown peaks representing new or specific secondary metabolites in the limited tissues, plant species or stress treatment, which is currently the important trial to understanding plant metabolism.
Protocol
1. Sample Preparation
- Plant materials are harvested and frozen immediately.
- Frozen plant materials are powdered by mixer mill (or mortar) and stored in Falcon tube or Eppendorf tube at -80 °C.
2. Extraction for Metabolite Profiling
- Aliquot frozen plant material in a 2 ml Eppendorf tube.
- Add 5 μl of extraction buffer per milligram of fresh weight of frozen plant material.
- Add one metal (or gilconia) ball and homogenize of frozen powder with the Mixer Mill for 2 min at 25 l s-1.
- Centrifuge 10 min at 12,000 rpm.
- Transfer the supernatant to NANOSEP centrifugal filter.
- Centrifuge 2 min at 4,000 rpm.
- Transfer the supernatant to new Eppendorf tube, and store at -20 °C until use.
3. Metabolite Profiling by LC-MS
- Set up HPLC and check temperature of column oven and sample tray.
- Set up MS condition and check the state of vacuum and heating capillary.
- Perform m/z calibration of MS detector.
- Transfer of 50 μl of extracts to glass vial for LC-MS.
- Inject 5 μl of extracts to LC-MS.
4. Data Analysis
- Configure Xcalibur or Metalign 4 and select the data analysis to be processed.
- Prepare a table of detected peaks of your interest in accordance with compound class in Table I.
- Identify peaks by co-elution of standard compounds.
- Annotate detected peaks using MS2 analysis, literature survey, metabolite database search (Figure 2, 12,13).
5. Prediction of Metabolic Pathway
- Construct a possible pathway with detected compounds. Prediction of pathway using peak annotations should be based on the chemical structure of detected compounds by predicting linking enzymatic functions on the metabolic pathway 13. Structuring biosynthetic steps should be conducted with precise peak annotation. But determination of detailed chemical structure, for example sugar moiety, is not indispensable in this step, because prediction of sugar moiety and adducted position is very difficult to identify by MS analysis. Determination of sugar type such as hexoside and pentoside will be identified by enzymatic assay of sugar donor at the last step of project. Mostly constructing of prediction of pathway should be performed as small molecule is intermediate of larger molecule except in the some cases such as dehydration reaction. List of atomic molecular weight, for example 16 m/z for difference between -H and -OH moiety (oxidation), 14 m/z (Carbon atom) for difference between -OH and -OMe (methylation) and 162 m/z (MW-H2O) for hexose (glycosylation), is useful for prediction. Determination of modification type with correlation analysis of tissues specificities helps prediction of metabolic pathway. Database of general metabolic pathway such as KEGG database (http://www.genome.jp/kegg/) and PlantCyc (http://plantcyc.org/), is very effective for prediction of metabolic pathway of your interest.
6. Preparation of Gene List with Arabidopsis Orthologous Gene ID
- Download gene ID list from genomic database.
- Add Arabidopsis gene ID of orthologous gene, in case of your target plant is not Arabidopsis.
- Prepare a list of genes in your pathway-of-interest. Annotation of Arabidopsis pathway data and gene family data are available in TAIR website (http://www.arabidopsis.org/). If you prepared a list of Arabidopsis orthologous genes, you can subsequently combine them.
7. Co-expressed Gene Analysis
- Test using the prepared gene ID list to search best database for your pathway by checking the correlation using well known gene pairs in your pathway-of-interest (Table II). If co-expression database or gene expression database are not available in the plant of your interest, Arabidopsis co-expression database should be used with a list of Arabidopsis orthologous genes. In case of barley, rice, poplar, wheat, medicago and soybean, co-expression analysis of plant species can be used (Table II).
- Construct the framework for your target co-expression network based on the connections of the well-known genes in your pathway-of-interest.
- Add correlated candidate genes (r<0.4~0.90, within approximate value of coefficient value between the connections of the well-known genes in your pathway-of-interest) and check their gene annotation in your predicted families to the connections of this network for finding best candidate genes (Figure 3). Threshold of coefficient value should be coordinated according to network structure and density of correlated genes.
- Make list of genes which you are able to narrow down as being specialized to your target pathway.
- Check gene expression of the organ specificities and stress responses of your candidate genes.
8. Integration of All Information to Predict New Pathways
- Add well-characterised genes which have been used for query of co-expression analysis to predicted metabolic pathway.
- Check uncharacterized parts in this pathway, for example uncharacterized enzymatic steps, transport proteins and transcription factors.
- Predict most suitable gene annotation for these missing uncharacterized steps.
- Combine the results of metabolite profiling and candidate genes of in silico gene expression based on the predicted pathway.
- Arrange your candidate genes on the predicted pathway according to gene function, for example, acetyltransferase for acetylated metabolite, glycosyltransferase for glycoside, P450 for oxidised compound. Phylogenetic tree analysis of amino acid sequences is useful for some wide gene family such as P450 and glycosyltransferase.
- Check the consistency of tissue specificities or stress responses between metabolite accumulation and gene expression level of candidate genes.
- Check the connections to other metabolism for providing substrate and stress responsive genes.
9. Experiments for Gene Identification Using Bio-resources
- Check the availability of bioresources for facilitation of experiment for candidate gene identification.
- Perform an experiment for identification of gene function using bio-resources, such as KO mutant library and full-length cDNA library. The experiments for functional identification of genes with preparation of overexpression plants and knockout mutants, enzymatic assay and promoter binding assay, have to be performed for the best candidate genes in your prediction. Recombinant protein assay for characterisation of protein properties and preparation of overexpression plants better to be carried out after confirmation of metabolite profile using KO mutant since it takes considerably longer to prepare recombinant protein and gene cloning for transformation.
10. Representative Results
The procedure of integrated analysis described in this protocol has many possibilities depending on specified experimental purpose and choice of biological and analytical combinations. Choice of procedures and experimental design should be carried out properly on the basis of your target pathway, compounds and plant species. The integration strategy described in this protocol is focused on annotation of plant gene function and the discovery of novel gene functions with an efficient usage of several bio- and data-resource. Expected outcome is promised to provide with the only case of conclusive prediction. This fact indicates that if enough evidences can not be given by combination profiles, experiment should not be started. For this reason, in any cases, additional preliminary experiments such as targeted gene expression profiling by RT-PCR, can support your prediction of gene function. Accuracy and correctness of prediction correlates higher depending on qualitative difference and number of variation of combination. In addition, good candidates and valid outcomes can only come from accurate prediction of pathways. Peak annotation should be conducted by combination of several approaches, for example literature survey, reference plant extract, MSn analysis, organ specificity and mutant analysis 13.
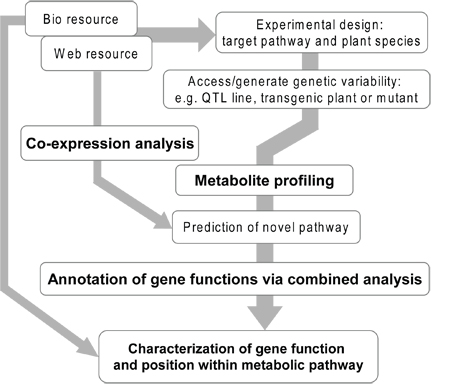
Figure 1. Overview of the experimental flow of gene annotation via combined approach. In some cases, projects start with the discovery of a novel peak which is detected in special conditions or tissues, and the desire to understand its role within its metabolism. In other instances the purpose of the project is gene identification or discovery of key regulatory factors such as transcription factors. Design of experiment should be planed with an data set which shows clear differences of metabolite levels in your target pathway, using a wide range of tissue samples from different organs, and for differentially grown plants or plants exposed to stress conditions, and subjecting the material to metabolite profiling. Mutant and transgenic plants as well as QTL harbouring breeding material also represent suitable genetic material for these studies. Prediction of novel pathway should be performed carefully with accurate peak annotation and combination approach with different type of metabolotype such as organ securities and stress responses according to gene expression data of your pathway-of-interest. In the last step, metabolite and transcript profiling should be performed which will eventually, when combined with in silico analysis of web-resources and in vitro characterisation of gene expression via heterologous expression, lead to the confirmation of the gene candidate and elucidation of its function and position within a metabolic pathway. Abbreviations: QTL, Quantitative Trait Loci.
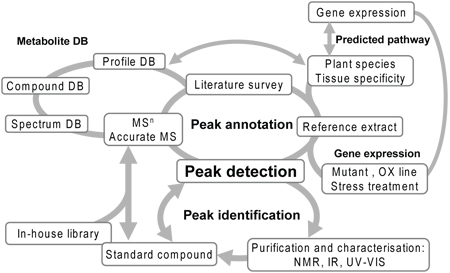
Figure 2. Work flow of combinational approach for peak annotation. An procedure for peak identification and annotation by the standard compound, comparison of wild type and knock out mutants, multi-dimensional mass spectrometry of the target peak referring to mass spectra of pure compounds from the databases 12. Abbreviations: DB, database; KO, knock-out; 1-D, one- dimensional; 2-D, two-dimensional; NMR, nuclear magnetic resonance; IR, infra-red; MSn, mass-mass spectrometries.
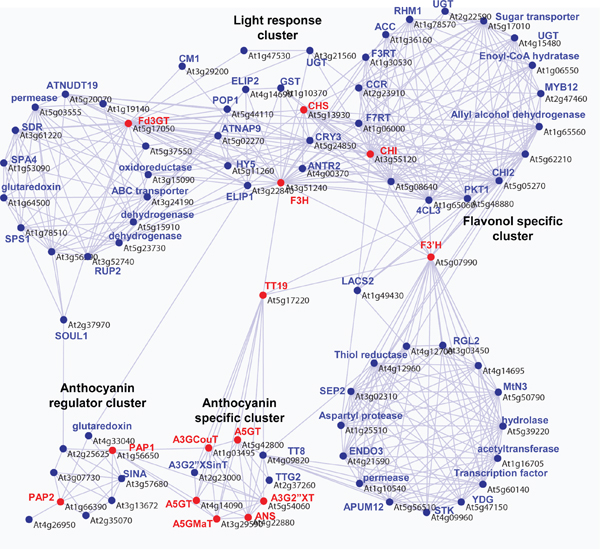
Figure 3. Example co-regulation network analysis of the anthocyanin pathway. Coexpression analyses were performed using the PRIMe (http://prime.psc.riken.jp/?action=coexpression_index) based on the data set of ATTEDII version 3 8,2 with the Pajek program (http://vlado.fmf.uni-lj.si/pub/networks/pajek/). Positive correlations (r<0.5) are used to make network connections. Red node: twelve anthocyanin enzymatic genes (At5g13930, CHS, TT4, chalcone synthase; At3g55120, CHI, TT5, chalcone isomerise; At3g51240, F3H, TT6, flavanone 3-hydroxylase; At5g07990, F3’H, TT7, flavonoid 3′-hydroxylase; At5g17050, Fd3GT, UGT78D2, flavonoid 3-O-glucosyltransferase; At5g17220, AtGSTF12, TT19; At5g42800, DFR, TT3, dihydroflavonol reductase; At4g22880, ANS/LDOX, TT18, anthocyanidin synthese; At4g14090, A5GT, anthocyanin 5-O-glucosyltransferase; At5g54060, A3G2″XT, putative anthocyanin 3-O-glucoside 2″-O-xylosyltransferase; At3g29590, A5GMaT, anthocyanin 5-O-glucoside 6”’-O-malonyltransferase; At1g03940, A3GCouT, anthocyanin 3-O-glucoside 6″-O–p-coumaroyltransferase) and two transcription factors for anthocyanin production (At1g56650, PAP1; At1g66390, PAP2) was used for searching candidate genes. Candidate genes were found by an “intersection of sets” search with a threshold value with a coefficient of r>0.50 queried by intersection of sets by all genes queried (Fourteen anthocyanin biosynthetic genes). A co-expression network, including correlated candidate genes (68 genes) and queried genes (14 genes), was re-constructed by an “interconnection of sets” search with r>0.50 using the PRIMe database. The output files that were formatted with a ‘.net’ file from the PRIMe database and networks were drawn using Pajek software. Blue node indicates candidate genes which correlated with anthocyanin genes.
| species | Major secondary metabolite |
| Arabidopsis thaliana | Glucosinolate, flavonol, anthocyanin, sinapoyl derivative |
| Populus trichocarpa | Flavonol, anthocyanin, salicylate derivative |
| Vitis vinifera | Flavonol, anthocyanin, tannin, stilbene |
| Solanum lycopersicum | Flavonol, anthocyanin, glycoalkaloid, chrologenate related, |
| Nicotiana tabacum | Flavonol, anthocyanin, nicotianamide, chrologenate related, acylsugar |
| Oryza sativa | Glycoflavone, anthocyanin, sterol derivatives |
| Zea may | Glycoflavone, anthocyanin, benzoxazinone, sterol derivatives |
| Medicago truncatula | Isoflavone, anthocyanin, saponin, |
| Lotus japonica | Isoflavone, flavonol, anthocyanin, saponin, |
Table I. Major secondary metabolites in model plant species.
| Co-expression database | Address |
| Plant cross species | |
| COP | http://webs2.kazusa.or.jp/kagiana/cop0911/ |
| PlaNet | http://aranet.mpimp-golm.mpg.de/ |
| Plant species | |
| ATEED-II | http://atted.jp/ |
| BAR | http://142.150.214.117/welcome.htm |
| COP | http://webs2.kazusa.or.jp/kagiana/cop |
| GeneCAT | http://genecat.mpg.de/ |
| Arabidopsis | |
| ACT | http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser |
| AthCoR@CSB.DB | http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath.html |
| CressExpress | http://cressexpress.org/ |
| PRIMe | http://prime.psc.riken.jp/?action=coexpression_index |
| Oryza sativa | |
| RiceArrayNet | http://arraynet.mju.ac.kr/arraynet/ |
| Rice Array Database | http://www.ricearray.org/coexpression/coexpression.shtml |
Table II. Available gene expression database for in silico co-expression analysis.
Discussion
Given that transcriptomics and metabolomics technologies have been used for several years, the process of data integration for metabolomics assisted gene annotation generally begins with the identification of a novel peak representing an unknown metabolite. This fact leads to the next stage which is to evaluate quantitative variance in metabolite peaks or the novel candidate genes thought to be responsible for their biosynthesis. The strategy described in this protocol, however, has three major problems i) difficulty of peak annotation, ii) complexity of pathway prediction, iii) resolution of gene information and quality of gene expression data. To counter the first problem, peak annotation should be carried out with co-elution of standard compounds or combinatorial approach utilizing information from MSn analysis, reference extract, mutant analysis, metabolite database search and literature survey (Figure 2, 12). For the second problem, pathway prediction can only be obtained by correct peak annotation. However, metabolite profiling of tissue specificity also can be support peak annotation, because metabolite accumulation should be correlated with the gene expressions of related genes. Therefore combination profiles of different tissues and growth conditions can be helpful for this second problem. The third problem concerning the resolution of gene information depends on the progress of sequence data. In case of the model plant without completion of genome sequence, co-expression analysis using orthologous genes in other model plants is useful. Detailed alignment comparison and phylogenetic tree analysis of amino acid sequence can support to connect model organisms to other species.
This protocol is suitable for all metabolisms. It is most efficient in the analysis of intermediate and secondary metabolisms which are well characterised to be subject to strong transcriptional control 1,5,11,16. In some examples, co-expression analysis succeeded to be performed in sulfur assimilation, genes for β-oxidation, branched-chain amino acid degradation, chlorophyll breakdown, and the lysine catabolism 3, cell wall metabolism 10,7 and light signalling cascade 14. Annotation of gene function via combined genomics, metabolomics and informatics is not only for biosynthetic gene and direct regulator of transcription factor but also for understanding physiological process and response (see example Figure 3. 14).
To develop this approach from model plants to crop species, metabolic comparison of across plant species is powerful approach in some general metabolisms. For example, if same compound is detected in different plant species, and some orthologous genes are found in these plant species, cross species co-expression analysis using orthologous genes can provide strong support for your prediction. This approach can be performed in Arabidopsis, poplar, medicago, in addition important crops such as barley, rice, wheat and soybean, by co-expression analysis of plant species (6, PlaNet: http://aranet.mpimp-golm.mpg.de/,; 9, COP: http://webs2.kazusa.or.jp/kagiana/cop0911/; see an example, 15).
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Prof. Kazuki Saito in RIKEN PSC and Dr. Bjoern Usadel in MPIMP for useful discussions. TT is supported by a fellowship from the Alexander von Humboldt foundation.
Materials
| Name of the reagent | Company | Catalogue number |
| Distilled water ULC/MS grad | BIOSOLVE | 23214102 |
| Acetonitrile (ACN) ULC/MS grade | BIOSOLVE | 01204102 |
| Methanol (MeOH) ULC/MS grade | BIOSOLVE | 13684102 |
| Formic acid (HCOOH) ULC/MS grade for liquid chromatography | BIOSOLVE | 06914131 |
| Standard compounds | EXTRASYNTHESE | |
| Linear ion trap (IT) ESI-MS system FINNIGAN-LTQ | Thermo Finnigan | |
| HPLC system Surveyor | Thermo Finnigan | |
| Analytical column Luna C18(2), 2.0 mm diameter, 150 mm length, 100 Å pore size and spherical particles of 3 mm | Phenomenex | 00F-4251-B0 |
| Xcalibur software | Thermo Finnigan |
References
- Aharoni, A., Keizer, L. C. P., Bouwmeester, H. J., Sun, Z. K., Alvarez-Huerta, M., Verhoeven, H. A., Blaas, J., van Houwelingen, A., De Vos, R. C. H., van der Voet, H. SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 12, 647-661 (2000).
- Akiyama, K., Chikayama, E., Yuasa, H., Shimada, Y., Tohge, T., Shinozaki, K., Hirai, M. Y., Sakurai, T., Kikuchi, J., Saito, K. PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 8, 339-345 (2008).
- Araujo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., Witt, S., Obata, T., Schauer, N., Graham, I. A., Leaver, C. J., Fernie, A. R. Identification of the 2-Hydroxyglutarate and Isovaleryl-CoA Dehydrogenases as Alternative Electron Donors Linking Lysine Catabolism to the Electron Transport Chain of Arabidopsis Mitochondria. Plant Cell. 22, 1549-1563 (2010).
- De Vos, R. C. H., Moco, S., Lommen, A., Keurentjes, J. J. B., Bino, R. J., Hall, R. D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2, (2007).
- Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., Araki, R., Sakurai, N., Suzuki, H., Aoki, K., Goda, H., Nishizawa, O. I., Shibata, D., Saito, K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 104, 6478-6483 (2007).
- Mutwil, M., Klie, S., Tohge, T., Giorgi, F. M., Wilkins, O., Campbell, M. M., Fernie, A. R., Usadel, B., Nikoloski, Z., Persson, S. PlaNet: Combined Sequence and Expression Comparisons across Plant Networks Derived from Seven Species. Plant Cell. 23, 895-910 (2011).
- Mutwil, M., Ruprecht, C., Giorgi, F. M., Bringmann, M., Usadel, B., Persson, S. Transcriptional Wiring of Cell Wall-Related Genes in Arabidopsis. Molecular Plant. 2, 1015-1024 (2009).
- Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K., Ohta, H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Research. 35, D863-D869 (2007).
- Ogata, Y., Suzuki, H., Sakurai, N., Shibata, D. CoP: a database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics. 26, 1267-1268 (2010).
- Persson, S., Wei, H. R., Milne, J., Page, G. P., Somerville, C. R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proceedings of the National Academy of Sciences of the United States of America. 102, 8633-8638 (2005).
- Tohge, T., Yonekura-Sakakibara, K., Niida, R., Watanabe-Takahashi, A., Saito, K. Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthesis genes. Pure and Applied Chemistry. 79, 811-823 (2007).
- Tohge, T., Fernie, A. R. Web-based resources for mass-spectrometry-based metabolomics: A user’s guide. Phytochemistry. 70, 450-456 (2009).
- Tohge, T., Fernie, A. R. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nature Protocols. 5, 1210-1227 (2010).
- Tohge, T., Kusano, M., Fukushima, A., Saito, K., Fernie, A. R. Transcriptional and metabolic programs following exposure of plants to UV-B irradiation. Plant Signal Behav. 6, (2011).
- Tohge, T., Ramos, M. S., Nunes-Nesi, A., Mutwil, M., Giavalisco, P., Steinhauser, D., Schellenberg, M., Willmitzer, L., Persson, S., Martinoia, E., Fernie, A. R. Towards the storage metabolome: profiling the barley vacuole. Plant Physiol. , (2011).
- Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., Watanabe-Takahashi, A., Inoue, E., Saito, K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell. 20, 2160-2176 (2008).

