Identification of Post-translational Modifications of Plant Protein Complexes
Summary
We describe here a protocol for the purification and characterization of plant protein complexes. We demonstrate that by immunoprecipitating a single protein within a complex, so we can identify its post-translational modifications and its interacting partners.
Abstract
Plants adapt quickly to changing environments due to elaborate perception and signaling systems. During pathogen attack, plants rapidly respond to infection via the recruitment and activation of immune complexes. Activation of immune complexes is associated with post-translational modifications (PTMs) of proteins, such as phosphorylation, glycosylation, or ubiquitination. Understanding how these PTMs are choreographed will lead to a better understanding of how resistance is achieved.
Here we describe a protein purification method for nucleotide-binding leucine-rich repeat (NB-LRR)-interacting proteins and the subsequent identification of their post-translational modifications (PTMs). With small modifications, the protocol can be applied for the purification of other plant protein complexes. The method is based on the expression of an epitope-tagged version of the protein of interest, which is subsequently partially purified by immunoprecipitation and subjected to mass spectrometry for identification of interacting proteins and PTMs.
This protocol demonstrates that: i). Dynamic changes in PTMs such as phosphorylation can be detected by mass spectrometry; ii). It is important to have sufficient quantities of the protein of interest, and this can compensate for the lack of purity of the immunoprecipitate; iii). In order to detect PTMs of a protein of interest, this protein has to be immunoprecipitated to get a sufficient quantity of protein.
Introduction
Immune pathways rely on various post-translational modifications (PTMs) of proteins to rapidly transduce signals and activate immune responses1. PTMs are rapid, reversible, and highly specific chemical alterations of protein structure that can affect protein conformation, activity, stability, localization, and protein-protein interactions2-4. In plants, more than 300 types of PTMs have been identified including ubiquitination, sumoylation, sulfation, glycosylation, and phosphorylation2,5. A growing body of evidence highlights the importance of PTMs in different aspects of plant immunity1,5,6. Protein phosphorylation, the reversible attachment of a phosphate group to a serine, threonine or tyrosine residue is a regulator of many cellular functions and not surprisingly the most highly studied PTM in plant defense signaling cascades5-11. Phosphorylation-dependent signaling is an integral part of plant defense activation initiated after extracellular perception of microbes by transmembrane receptors, or intracellular recognition by multidomain resistance proteins5,8.
Upon invading plants, microbes are detected by plasma membrane receptors called pattern recognition receptors (PRRs)12. Known PRRs are either receptor-like proteins (RLPs) or receptor-like kinases (RLKs), both carrying a ligand-binding ectodomain and a single-pass transmembrane domain. In contrast to RLPs, RLKs have an intracellular kinase domain13-15. The ectodomain of PRRs binds to conserved microbe elicitor molecules called pathogen-associated molecular patterns (PAMPs) leading, in the case of RLKs, to autophosphorylation and transphosphorylation within the intracellular domain6,16. Downstream of perception-induced phosphorylation of PRRs, subsequent phosphorylation of cytoplasmic proteins including kinases17,18, E3 ligases19, mitogen-activated protein kinases (MAPKs)20,21, calcium-dependent protein kinases (CDPK)22,23, and transcription factors24,25 leads to PAMP-triggered immunity (PTI).
In addition to the extracellular perception by PRRs, intracellular recognition of pathogens is achieved through cytoplasmic receptors. These multidomain resistance (R) proteins contain a variable N-terminus domain, a central nucleotide-binding (NB) motif, and C-terminal leucine-rich repeats (LRR) and therefore called NB-LRR proteins26,27. R proteins directly or indirectly recognize pathogen-derived virulence molecules called effectors, also known to target PRRs and other nodes of the immune system. Recognition induces strong defenses leading to effector-triggered immunity (ETI)28-31. NB-LRR proteins have a requisite ATP-binding motif within the NB domain30,32, but they lack a kinase domain. Phosphorylation of conserved domains of NB-LRRs has been reported in a large-scale proteomic survey33, but its relevance for ETI is unclear. Similarly to PTI, activation of NB-LRR proteins leads to phosphorylation of cytoplasmic proteins including MAPKs34-37 and CDPKs38-40. Most importantly, effector recognition can lead to phosphorylation of accessory proteins that interact directly with the NB-LRR proteins41. Some examples include RIN4 (Arabidopsis thaliana) and Pto (tomato, Solanum lycopersicum) proteins that interact with NB-LRR proteins, RPM142,43 and Prf44, respectively. During infection by Pseudomonas syringae bacteria, the effector protein AvrB induces RIN4 phosphorylation, most likely by the receptor-like kinase RIPK45,46. Similarly, in tomato the P. syringae effectors AvrPto and AvrPtoB induce phosphorylation of Pto47. In contrast to RIN4 which lacks a kinase domain and must be trans-phosphorylated, Pto is an active kinase capable of auto-phosphorylation48 and trans-phosphorylation of substrates49,50. While Pto requires its kinase activity for effector-dependent initiation of signaling, kinase-dead Pto mutants are still able to signal in an effector-independent way51. We recently explained these observations by demonstrating that the Prf/Pto complex is oligomeric, containing multiple Prf and Pto molecules52 and that Pto molecules within the same complex can trans-phosphorylate each other47. We proposed a model in which one molecule of Pto (sensor) interacts with the effector protein, causing a conformation change to the NB-LRR protein (Prf) that in turn activates a second Pto molecule (helper) within the complex. Subsequently, the helper Pto molecule trans-phosphorylates the sensor Pto leading to full activation of the resistance complex47.
These examples demonstrate that the identification of immune complex components and their potential PTMs upon effector recognition can lead to a better understanding of how signals are transduced from effector perception to downstream targets. Here we describe a protein purification method for NB-LRR-interacting proteins and the subsequent identification of their PTMs. We use Nicotiana benthamiana and the tomato Prf/Pto complex as a model, but the same protocol can easily be applied to RLKs from N. benthamiana and A. thaliana53 with small modifications as we describe within the protocol.
Protocol
Note: all steps are done at room temperature, unless stated otherwise.
1. Preparation of Plant Materials and Buffers
- For N. benthamiana
- Grow the Agrobacterium tumefaciens strains of interest for transient expression (in this example Prf-FLAG, Prf-3xHA, Pto-FLAG and empty vector as a control) by shaking (200 rpm) in liquid culture (L medium with appropriate antibiotics) at 28 °C until stationary phase.
- Collect and pellet agrobacteria by centrifugation (3,000 x g for 5 min). Discard supernatant and resuspend pelleted agrobacteria in infiltration buffer.
- Measure the amount of agrobacteria by obtaining the optical density (OD) value at an absorbance of 600 nm (Abs 600 nm). Adjust the OD of bacteria to 0.1-0.8.
- Infiltrate 4 week old N. benthamiana plants (22 °C, 16 hr light) with agrobacteria by hand (with a 1-ml needleless syringe) or by vacuum (add 0.02% v/v surfactant).
Note: Use the first nearly-fully expanded leaf, and the two immediately older leaves for most effective protein expression. For detection of ubiquitinated or acetylated proteins infiltrate leaves with 100 nM MG-132 or 100 ng/ml Trichostatin A, respectively, at 1 and 2 days post-infiltration. - Harvest the infiltrated leaves 2-5 days post-infiltration and freeze in liquid nitrogen. Store the samples in a -80 °C freezer.
Note: Determine the best level of expression of the protein of interest beforehand by taking samples over a time course of 5 days and detect protein levels by immunoblotting.
- For A. thaliana stable transgenic lines
- Grow A. thaliana seeds in 6-well plates supplemented with 5 ml of liquid MS medium (1% w/v sucrose) per well. Put three to five seeds (sterilized and stratified) of the transgenic line of interest or the untransformed control line per well. Shake at 200 rpm for two days before transferring the plate to a growth room and leave for 2 weeks (22 °C, 10 hr light).
- Harvest samples and freeze in liquid nitrogen.
Note: You can use as a control a transgenic line expressing an unrelated protein with the same tag as the protein of interest.
- Buffers
- Prepare Buffer A. De-gas the buffer for 1-2 hr (for RLKs, other membrane bound proteins and nuclear proteins, add 1% v/v IGEPAL CA-63053).
Note: You can keep Buffer A at 4 °C indefinitely. You can use 1% v/v Triton instead of IGEPAL CA-360. - Prepare 80 ml of cold Buffer B at least 1-2 hr before extraction.
Note: The ideal ratio of tissue vs. extraction buffer is 1:4 (w/v).
- Prepare Buffer A. De-gas the buffer for 1-2 hr (for RLKs, other membrane bound proteins and nuclear proteins, add 1% v/v IGEPAL CA-63053).
2. Protein Extraction
- Just prior to extraction prepare 80 ml of cold Buffer C.
Note: Add 10 μM MG-132 for the detection of ubiquitinated proteins. - Grind 20 g of N. benthamiana tissue (approximately 15 leaves) or A. thaliana seedlings (2-4 g per 6-well plate) in liquid nitrogen using a mortar and pestle.
- Add 80 ml of cold Buffer C to the 20 g of ground tissue, mix well, and thaw on ice.
Note: Grind all samples at the same time (protein of interest and control). - Homogenize with 3 bursts of 10 sec each at full speed with a tissue homogenizer (prechilled at 4 °C). Filter through a 22-25 μm tissue and centrifuge at 30,000 x g for 30 min at 4 °C.
Note: Alternatively, homogenize with a fresh prechilled mortar and pestle. - For the blocking and washing steps of the affinity matrix, prepare 20 ml of Buffer D.
- Block 100 ml of suitable affinity matrix using 500 ml of cold Buffer D containing 1 % BSA for 5 min at 4 °C.
Note: Preferred affinity matrices include anti-FLAG M2 agarose, Streptavidin agarose, GFP-Trap_A and anti-HA agarose. - Wash 3x with 1 ml cold Buffer D.
- Remove supernatant of leaf extract and filter through a 0.45 μm syringe filter into a new tube on ice.
Note: The extract color should be green or yellow; if the sample has turned brown, discard and start over. - Take an aliquot of the crude extract for immunoblot, add 5x SDS-PAGE loading buffer, vortex and denature by boiling samples for 10 min at 80-100 °C in a heat block.
3. Protein Immunoprecipitation
- Split the protein extract into two 50 ml tubes and add 50 μl of the affinity matrix suspended in Buffer D to each tube. Incubate with gentle rotation for 2 hr at 4 °C.
Note: Longer incubations have not been found to increase yield. - Prepare 600 μl (6x bead volume of affinity matrix) of elution buffer by adding to Buffer D (final concentrations): 0.5% BSA, 0.25 mg/ml FLAG peptide (for anti-FLAG M2 agarose) or 10 mM D-biotin (for Streptavidin agarose).
Note: Elution is not recommended in the case of GFP-Trap_A and anti-HA matrices, so proceed directly to boiling in 1x SDS-PAGE loading buffer, step 3.11. - Precipitate the affinity matrix by centrifugation at 4 °C (5 min at 1,000 x g).
- Take an aliquot of the unbound extract for immunoblot, discard the rest of the supernatant, leaving approximately 500 μl in the bottom of the tube.
- Resuspend the slurry solution with a wide-bore tip.
- Transfer the mixed slurry solution from both tubes to a single 1.5 ml tube.
Note: From now on, use 1.5 ml low-protein binding microfuge tubes. - Pulse 3x for 5 sec using a benchtop centrifuge to precipitate the affinity matrix and remove the supernatant.
- Wash 3-5x with 1 ml cold buffer D. Between the washes precipitate the affinity matrix as previously described and discard the supernatant. In the last wash remove excess Buffer D with the needle of a syringe.
Note: A final wash using Buffer D with 0.1% IGEPAL can be used to further reduce nonspecific binding. - Elute with 200 μl of the appropriate elution buffer (for anti-FLAG M2 or Streptavidin agarose see step 3.2) 3x, 5 min each time with constant shaking.
- Pool the 3 eluates in a single tube. Concentrate the proteins by using 30 μl of absorption resin. Vortex, let stand for 5 min and precipitate the resin (2 min at 10,000 x g). Discard the supernatant.
Note: For GFP-Trap_A and anti-HA agarose, skip the steps 3.9 and 3.10. - Add 50 μl of 1x SDS-PAGE loading buffer to the precipitated affinity matrix or absorption resin. Vortex and boil for 10 min at 80-100 °C in a heat block.
- Centrifuge the boiled affinity matrix or resin for 2 min at 10,000 x g. Aliquot 5 μl of the supernatant for immunoblot, and run with other fractions onto a ~10 cm long SDS-PAGE gel (Figure 2).
- For separation and identification of phosphorylated proteins by mass spectrometry, load the remaining 45 µl onto a ~19-cm long SDS-PAGE gel if extra separation is required (Figure 2).
Note: A blank lane can be included between all samples to reduce contamination of samples.
4. Protein Digestion
- Stain the SDS-PAGE gel with colloidal Coomassie Brilliant Blue (cCBB) stain.
Note: Minimize handling of the gel to reduce contamination with keratins. Make sure that none of the equipment or tools have been used for immunoblotting or other activities that might have left residual protein contamination. - Destain the gel with copious washing in water, preferably O/N. Cut the band of interest from the gel with a clean razor blade.
Note: Align the gel with the immunoblot if necessary to locate the correct area. As phosphorylation can delay migration of proteins on SDS-PAGE cut the area immediately above and below the band of interest. - Dice the gel slice into cubes of 2-4 mm (this ensures that the gel will be covered by solutions in the tube without blocking pipette tips). Place in a tube.
- Estimate the volume required to keep the gel well submerged. Use this amount for derivatization, incubation with protease and peptide extraction, and then use double to triple volumes for washing.
Note: In a typical digest with approximately 100 μl of the cut gel pieces, use 500 μl solution for washing, 2 x 180 μl for dehydration, 150 μl for reduction, 120 μl for alkylation, and 120 μl for digestion. - Wash the gel pieces 2 x 20 min (or until destained) by adding 50% acetonitrile (in 50 mM ammonium bicarbonate, ABC). Pipette off the solution taking care not to remove gel pieces.
- Dehydrate with 100% acetonitrile for 5 min and remove free liquid.
Note: The gel should appear shrunken and white. If the blue color persists, incubate longer in acetonitrile/ABC and/or at elevated temperature (45-55 °C). - Reduce protein disulfide bonds by adding 10 mM DTT in water and incubate for 30-45 min at 56 ºC with shaking. Remove free liquid.
- Alkylate cysteine residues by addition of 55 mM chloroacetamide (in 50 mM ABC) for 20-30 min (room temperature, dark). Remove free liquid.
- Wash gel pieces 2 x 10 min with 50% acetonitrile (in 50 mM ABC). Remove free liquid.
- Dehydrate gel pieces with 100% acetonitrile for 5 min and remove free liquid.
- For the tryptic digest, add 40 μl of trypsin working solution (100 ng of trypsin in digestion buffer: 50 mM ABC, 5% acetonitrile for an average Coomassie-stained band). Allow gel pieces to rehydrate for 10 min. Add enough of digestion buffer to cover gel pieces if necessary. Incubate at 37 ºC O/N.
- To stop the digestion and extract the peptides from the gel pieces, add 5% formic acid (in 50% acetonitrile) (add the same volume as the digestion volume). Sonicate for 5-10 min. Transfer the supernatant to new tube. Repeat the extraction 3x.
- Dry the peptides by lyophilization (preferred) or a vacuum concentrator (such as Speed-Vac or Savant). Store at -20 ºC.
- Submit your sample for mass spectrometry.
Note: We submitted our sample to the mass spectrometry facility at The Sainsbury Laboratory (Norwich, UK). Protocols at mass spectrometry facilities vary considerably, but typically peptides will be dissolved in 0.2% trifluoroacetic acid with 2% acetonitrile before loading onto a liquid chromatography system coupled to electro-spray tandem mass spectrometry.
Representative Results
We describe here a protocol for purification of tagged proteins from stable transgenic A. thaliana lines or after transient expression of proteins in N. benthamiana. As illustrated in Figure 1 the purification of the targeted protein is coupled with mass spectrometry to allow identification of interacting proteins and the PTMs of the targeted protein. The protocol has been designed for purification of proteins involved in plant immunity and identification of their PTMs but can be applied to purification of any tagged plant protein.
As an example we use the purification of the Prf/Pto complex from N. benthamiana. The transgenic N. benthamiana line 38-1254, expressing 35S:Pto, was transiently transformed with 35S:Prf-FLAG and the complex was immunoprecipitated with anti-FLAG M2 agarose (Figure 2A). The immunoblots (IB) in Figure 2A illustrate that most of the targeted protein (Prf) was immunoprecipitated. Pto and the interacting effector protein AvrPtoB were copurified with Prf and detected using antibodies and mass spectrometry analysis (Figures 2A and 2B). We found that the Pto that copurified with Prf migrated slower on SDS-PAGE gel when coexpressed with the effector constructs 35S:AvrPto and 35S:AvrPtoB (Figure 2A). Both effector proteins induced slower migration of the Prf-interacting Pto (Figure 2A). This effector recognition-dependent slow migration of Pto was previously attributed to phosphorylation as it could be removed by treatment with phosphatase44. To detect the phosphorylation sites contributing to the slow migration of Pto, we subjected the Prf copurifying Pto to mass spectrometry analysis. Despite the high expression of Pto and its easy detection with immunoblots we could retrieve peptides covering only 57% of Pto sequence and we failed to identify any phosphorylation sites (Figure 2B).
Subsequently, we changed strategies to target the Pto protein with anti-FLAG. N. benthamiana WT plants were transiently transformed with 35S:Prf-3HA and 35S:Pto-FLAG, with or without 35S:AvrPto and 35S:AvrPtoB. The total amount of Pto protein, comprising both the Prf-complexed and the free forms, was immunoprecipitated with anti-FLAG M2 agarose and subjected to SDS-PAGE fractionation, in-gel tryptic digestion and mass spectrometric analysis (Figure 2C). With this approach we identified Pto peptides spanning approximately 80% of its sequence and an unknown 100 kDa interacting protein. We identified a set of single and double phosphorylation sites for Pto peptides 187-202 and 188-202 (Table 1). Ser-198 and Thr-199 were the predominant phosphorylation sites but other phosphorylation sites were also detected (Table 1). Most importantly double phosphorylation on Ser-198 and Thr-199 was identified only in the presence of either effector protein (Table 1). We recently identified a similar set of Pto phosphorylation sites and illustrated the significance of the double phosphorylation event for signaling47. Following the protocol described here we were able to identify dynamic changes in the phosphorylation state of the target protein.
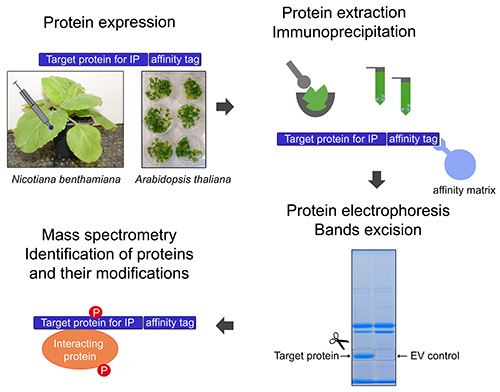
Figure 1. General experimental design. The target protein for immunoprecipitation (IP) is tagged and expressed in planta, transiently in Nicotiana benthamiana or stably in Arabidopsis thaliana. Following protein extraction the target protein is immunoprecipitated with an affinity matrix. The proteins are separated on an acrylamide gel by electrophoresis. Gel bands are excised. Proteins are extracted from the gel bands and subjected to mass spectrometry. Phosphorylation sites and interacting proteins are identified. Click here to view larger image.
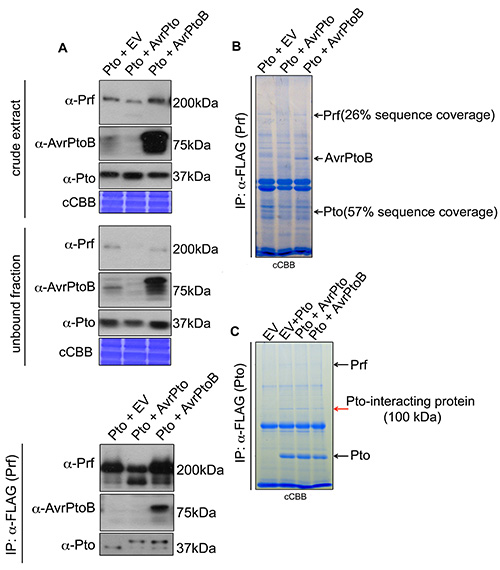
Figure 2. Prf and Pto immunoprecipitation. A). The construct 35S:Prf-FLAG was transiently expressed in 38-12 line (35S:Pto) along with empty vector (EV), 35S:AvrPto or 35S:AvrPtoB. Three days post-infiltration Prf was immunoprecipitated using anti-FLAG affinity matrix. Immunoblots (IB) for Prf, AvrPtoB and Pto were performed. As pointed by arrows at the bottom of Pto immunoblot panel, AvrPto and AvrPtoB induce a slower migration of Pto. B). The construct 35S:Prf-FLAG was transiently expressed in 38-12 line (35S:Pto) along with empty vector (EV), 35S:AvrPto or 35S:AvrPtoB. Three days post-infiltration Prf was immunoprecipitated using anti-FLAG agarose and separated on SDS-PAGE. The gel was stained with colloidal Coomassie Brilliant Blue (cCBB) and the visible Prf, Pto and AvrPtoB proteins bands were excised from the gel and analysed by mass spectrometry. The percentage peptide coverage of Pto and Prf sequences identified by mass spectrometry is indicated. C). The constructs 35S:Prf-3xHA and 35S:Pto-FLAG were transiently expressed along with 35S:AvrPtoB, 35S:AvrPto or empty vector (EV) in N. benthamiana. Three days post-infiltration the total amount of Pto-FLAG was immunoprecipitated using anti-FLAG agarose and resolved on SDS-PAGE. The visible Pto and Pto-interacting protein bands were excised from the gel and analysed by mass spectrometry. Pto and the 100 kDa Pto-interacting protein band are clearly visible on the colloidal Coomassie Brilliant Blue (cCBB) stained-gel, whereas Prf is slightly less visible (as indicated by arrows). IP: Immunoprecipitation; IB: Immunoblot; CBB: Coomassie Brilliant Blue staining; cCBB: colloidal Coomassie Brilliant Blue staining. Click here to view larger image.
| Prf | |||
| EV | AvrPto | AvrPtoB | |
| Pto peptide 188-202 | |||
| GTELDQ[pT195]HLSTVVK | 1 | 1 | 0 |
| GTELDQTHL[pS198]TVVK | 10 | 7 | 5 |
| GTELDQTHLS [pT199]VVK | 8 | 3 | 4 |
| G[pT190]ELDQTHLS[pT199]VVK | 0 | 1 | 2 |
| GTELDQTHL[pS198][pT199]VVK | 0 | 8 | 4 |
| GTELDQTHLSTVVK | 46 | 26 | 48 |
| Pto peptide 187-202 | |||
| KGTELDQ[pT195]HLS TVVK | 0 | 1 | 0 |
| KGTELDQTHL[pS198]TVVK | 17 | 17 | 12 |
| KGTELDQTHLS[pT199]VVK | 4 | 2 | 2 |
| KGTELDQ[pT195]HLS [pT199]VVK | 0 | 1 | 1 |
| KGTELDQTHL[pS198][pT199]VVK | 0 | 7 | 5 |
| KGTELDQTHLSTVVK | 21 | 37 | 18 |
| Peptides 187-202 and 188-202 | 83 | 88 | 50 |
| Percentage of peptides with two phosphorylation events | 0% | 26.9% | 11.2% |
| Pto Sequence Coverage | 78% | 79% | 82% |
Table 1. Double phosphorylation of a Pto peptide upon activation of signaling. 35S:Prf-3xHA and 35S:Pto-FLAG were transiently expressed along with empty vector (EV), 35S:AvrPto or 35S:AvrPtoB de N. benthamiana. Three days post-infiltration the total amount of Pto-FLAG protein was immunoprecipitated using anti-FLAG affinity matrix and resolved on SDS-PAGE. The visible bands of Pto on the colloidal Coomassie brilliant blue stained gel were excised and analysed with mass spectrometry. The number of Pto peptides identified with 0, 1, and 2 phosphorylation events is indicated.
| Table of buffers | ||
| L medium | Tryptone | 10 g/L |
| Yeast extract | 5 g/L | |
| NaCl | 5 g/L | |
| D-glucose | 1 g/L | |
| Agro-infiltration buffer | MgCl2 | 10 mM |
| MES | 10 mM | |
| Adjust to pH 5.5 | ||
| Acetosyringone (optional) | 150 μM | |
| Buffer A | Tris-HCl pH 7.5 | 150 mM |
| NaCl | 150 mM | |
| EDTA | 5 mM | |
| EGTA | 2 mM | |
| Glycerol | 5% (v/v) | |
| Buffer B | Buffer A | |
| PVPP | 2% (w/v) | |
| Buffer C | Buffer B | |
| Dithiothreitol (DTT) | 10 mM | |
| Plant Protease Inhibitor Cocktail | 1% (v/v) | |
| Phenylmethylsulfonyl fluoride (PMSF) | 0.5 mM | |
| Calyculin A | 50 nM | |
| Sodium fluoride (NaF) | 50 mM | |
| Sodium molybdate (Na2MoO4) | 10 mM | |
| Sodium Orthovanadate | 1 mM | |
| Okadaic acid | 100 nM | |
| Buffer D | Buffer C without PVPP | |
| 5x SDS-PAGE loading buffer | Tris-HCl pH 6.8 | 60 mM |
| SDS | 2% | |
| Glycerol | 0.15% | |
| Bromophenol blue | 0.10% | |
| DTT (add just before use) | 50 mM |
Table 2. Buffer and media recipes.
Discussion
Elucidating the mechanism of receptor activation by PAMPs and effectors can contribute greatly to our understanding of plant immunity. In the last 20 years, genetic and yeast two-hybrid screens have been instrumental for the discovery of PRRs and NB-LRR proteins. More recently, mass spectrometry-based protocols have been established for the identification of proteins regulated differentially during immune signaling55-58, their PTMs11,33,59,60, the composition of immune complexes61 and effector targets62. Here we describe a straightforward protocol for the identification of PTMs regulating the activation of immune complexes.
In comparison with previously described protocols, this protocol allows the detailed identification of dynamic changes of PTMs. Protocols of large-scale proteomics approaches can identify PTMs of proteins but are not able to reveal the site plasticity of PTMs due to limited amounts of protein. In the case of protein phosphorylation, large-scale proteomics approaches typically only identify the predominant phosphorylation sites11,33,59. The detailed characterization of a protein phosphorylation status requires a substantial amount of protein that can be obtained only through partial purification of the target protein47. The protocol described here couples a protein purification approach that yields a substantial amount of the target protein, with mass spectrometry analysis of the purified protein. Following this protocol, the single phosphorylation event of Pto was attributed predominantly to Ser-198 as previously described52 but some mass spectrometry spectra also supported a single phosphorylation event on Thr-195 or Thr-199. When double phosphorylation events of Pto were observed, the predominant combination of phosphorylated amino acids was Ser-198 and Thr-199 although combinations on other sites were also observed (Table 1). These results clearly demonstrate the phosphorylation site plasticity of protein kinases and the ability of the described protocol to characterize in details all possible phosphorylation sites.
The most critical steps of this protocol are: 1) sufficient extraction of proteins; 2) protection of PTMs; and 3) sufficient amount of target protein. First, for sufficient extraction of proteins, it is important to grind the tissue in liquid nitrogen and subsequently to use a tissue homogenizer as described in the protocol. If a tissue homogenizer is not available, a mortar and pestle can be used. It is also important to use a ratio of one to three (or four) of grams of tissue to volume of extraction buffer. This tissue to buffer ratio and the high strength buffer that we suggest will ensure that the pH will remain neutral during the extraction process. We found that this is of particular importance for A. thaliana and tomato extractions52. Protection of PTMs can be achieved by including in all the buffers the appropriate inhibitors of enzymes that can remove PTMs. It is also critical to perform all steps at 4 °C and to prechill buffers and instruments. Higher yields of target protein can be achieved by using plants expressing the protein in sufficient amounts and by using high amounts of tissue (approximately 20 g). The most crucial step for obtaining sufficient amounts of target protein is concentrating the target protein by direct immunoprecipitation. The significance of this step is highlighted by our failure to identify PTMs of Pto after a Prf immunoprecipitation due to the limited amount of coimmunoprecipitated Pto (Figure 2B). In contrast, direct immunoprecipitation of Pto yielded substantially more measurable peptides (Figure 2C) leading to approximately 80% coverage of the protein sequence and identification of PTMs (Table 1). We also strongly recommend the use of epitope-tags for protein immunoprecipitation. In comparison to antibodies raised against native proteins epitope-tags affinity matrix can yield higher amounts of partially purified protein. By using an antibody raised against native Pto protein51 (Figure 2A) for immunoprecipitation, we were able to identify only single phosphorylation of the two predominant phosphorylation sites of Pto.
This protocol has primarily been developed for partial purification of N. benthamiana cytoplasmic protein and subsequent identification of their phosphorylation sites. However, using the modifications described herein, this protocol can be easily adapted for A. thaliana proteins and membrane bound proteins. The main objective of this protocol is to yield sufficient amounts of the target protein for the identification of PTMs and not to achieve the highest purity of the complex. If higher purity of the complex is required a second step of purification can be added to this protocol52. In that case, a first step will allow elution of the target protein from the affinity matrix without the use of high salts or acid and the subsequent step may entail a matrix, which requires harsh elution conditions. It is important to highlight that we have only tested this protocol for the identification of phosphorylation sites and that we are currently testing its ability for detailed identification of additional PTMs.
Our representative results clearly demonstrate the phosphorylation site plasticity of protein kinases and the ability of the described protocol to characterize phosphorylation sites. Most importantly, they highlight that identification of phosphorylation sites relying in low amount of proteins by mass spectrometry and by single point mutations of auto-phosphorylation sites can give confusing results. We show here and previously47 that double phosphorylation of Pto on Ser-198 and Thr-199 is associated with the activation of the Prf/Pto complex. In contrast, previously published results47,63 have shown that single point mutations of Ser-198 and Thr-199 to alanine, which prevent phosphorylation on these sites, are capable of signaling suggesting that phosphorylation of these sites is not a prerequisite for complex activation. These results can now be explained by phosphorylation of the secondary site Thr-195. Similar insight into the phosphorylation site plasticity of other protein kinase can be obtained following this protocol. Furthermore, a combined approach using phosphorylation sites point mutations, proteins that can inhibit specific kinases (effectors) coupled with protein immunoprecipitation and mass spectrometry analysis will lead to a better understanding of the evolutionary significance of the phosphorylation site plasticity of protein kinases.
Divulgations
The authors have nothing to disclose.
Acknowledgements
VN is supported by the Royal Society. JPR is an Australian Research Council Future Fellow (FT0992129). We thank Dr. Miriam Gifford for critically reading the manuscript.
Materials
| MgCl2 | Sigma | M8266 | |
| MES | Sigma | M8250 | |
| Acetosyringone | Sigma | D134406 | toxic – 1 M stock in DMSO |
| Syringe 1mL sterile | Terumo | ||
| Syringe needle | Terumo | NN-2525R | |
| Silwet L-77 (surfactant) | Lehle Seeds | VIS-01 | toxic |
| MG-132 (Z-Leu-Leu-Leu-al) | Sigma | C2211 | 1 M stock in DMSO, store at -80 °C |
| MS salts | Sigma | M5524 | oxidizing, toxic |
| Sucrose | Sigma | 16104 | |
| Trizma base | Sigma | T1503 | adjust pH with 1 N HCl to make 1 M Tris-HCl buffer |
| NaCl | Sigma | S3014 | 5 M stock |
| EDTA | Sigma | E6758 | toxic – 0.5 M stock |
| EGTA | Sigma | E4378 | |
| Glycerol | National Diagnostics | EC-606 | |
| IGEPAL CA-630 | Sigma | I8896 | corrosive |
| PVPP (polyvinylpolypyrrolidone) | Sigma | P5288 | do not confuse with PVP |
| DTT (DL-dithiothreitol) | Sigma | 43815 | toxic – 1 M stock, store at -20 °C |
| Plant protease inhibitor cocktail | Sigma | P9599 | do not freeze/thaw too many times |
| PMSF (phenylmethylsulfonyl fluoride) | Sigma | P7626 | corrosive, toxic |
| Calyculin A | Cell Signaling Technology | 9902 | |
| Sodium Fluoride (NaF) | Sigma | S7920 | toxic – 1 M stock |
| Sodium Molybdate (Na2MoO4) | Sigma | S6646 | 0.5 M stock |
| Sodium orthovanadate (Na3VO4) | Sigma | 450243 | toxic – Prepare a 200 mM solution of sodium orthovanadate. Adjust the pH to 10.0 using either 1N NaOH or 1N HCl. The starting pH of the sodium orthovanadate solution may vary with lots of the chemical. At pH 10.0 the solution will be yellow. Boil the solution until it turns colorless (approximately 10 minutes). Cool to room temperature.Readjust the pH to 10.0 and repeat steps 3 and 4 until the solution remains colorless and the pH stabilizes at 10.0. |
| Okadaic acid | Santa Cruz Biotechnology | sc-3513 | |
| Kinematica Polytron tissue homogeniser | Fisher Scientific | 08-451-320 | |
| Miracloth | Merck Millipore | 475855-1R | |
| anti-FLAG M2 agarose | Sigma | A2220 | this matrix is recommended |
| Streptavidin-agarose | Thermo-Scientific | 20347 | this matrix is recommended |
| GFP-Trap_A | Chromotek | gta-20 | this matrix is recommended |
| Anti-HA-Agarose | Sigma | A2095 | this matrix is recommended |
| 0.45 μm filter | VWR | 513-1902 | |
| BSA | Sigma | A7906 | |
| FLAG peptide | Sigma | F3290 | |
| D-biotin | Sigma | 47868 | |
| StrataClean resin | Agilent | 400714 | this absorption resin is recommended for protein precipitation |
| Mini-PROTEAN Tetra Cell | Biorad | ||
| PROTEAN II XL Cell | Biorad | ||
| SimplyBlue SafeStain | Invitrogen | LC6060 | this stain is recommended |
| Protein low-binding tubes (LoBind) | Eppendorf | 0030 108.116 | these tubes are recommended |
| Ammonium bicarbonate (ABC) | Sigma | 9830 | toxic |
| Acetonitrile | VWR | 83639 | toxic, flammable |
| 2-chloroacetamide | Sigma | 22790 | toxic |
| Trypsin | Promega | V5280 | irritant, sensitizing |
| Formic acid | Sigma | 14265 | toxic, corrosive |
| Water-bath sonicator | Ultravawe | Ultra BT Ultrasonic Bath | |
| LTQ-Orbitrap XL | Thermo-Scientific | ||
| Trichostatin A | Sigma | T8552 | toxic |
References
- Jones, A. M., Monaghan, J., Ntoukakis, V. Editorial: Mechanisms regulating immunity in plants. Front. Plant Sci. 4, 64 (2013).
- Jensen, O. N. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33-41 (2004).
- Cohen, P. The regulation of protein function by multisite phosphorylation-a 25 year update. Trends Biochem. Sci. 25, 596-601 (2000).
- Narayanan, A., Jacobson, M. P. Computational studies of protein regulation by post-translational phosphorylation. Curr. Opin. Struct. Biol. 19, 156-163 (2009).
- Stulemeijer, I. J., Joosten, M. H. Post-translational modification of host proteins in pathogen-triggered defence signalling in plants. Mol. Plant Pathol. 9, 545-560 (2008).
- Park, C. J., Caddell, D. F., Ronald, P. C. Protein phosphorylation in plant immunity: insights into the regulation of pattern recognition receptor-mediated signaling. Front. Plant Sci. 3, 177 (2012).
- van Bentem, S. D., Hirt, H. Using phosphoproteomics to reveal signalling dynamics in plants. Trends Plant Sci. 12, 404-411 (2007).
- Peck, S. C. Early phosphorylation events in biotic stress. Curr. Opin. Plant Biol. 6, 334-338 (2003).
- Thurston, G., Regan, S., Rampitsch, C., Xing, T. Proteomic and phosphoproteomic approaches to understand plant-pathogen interactions. Physiol. Mol. Plant P. 66, 3-11 (2005).
- Xing, T., Ouellet, T., Miki, B. L. Towards genomic and proteomic studies of protein phosphorylation in plant-pathogen interactions. Trends Plant Sci. 7, 224-230 (2002).
- Nuhse, T. S., Bottrill, A. R., Jones, A. M., Peck, S. C. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931-940 (2007).
- Boller, T., Felix, G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann. Rev. Plant Biol. 60, 379-406 (2009).
- Wang, G., et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503-517 (2008).
- Wang, G. D., et al. The Diverse Roles of Extracellular Leucine-rich Repeat-containing Receptor-like Proteins in Plants. Crit. Rev. Plant Sci. 29, 285-299 (2010).
- Schwessinger, B., Ronald, P. C. Plant innate immunity: perception of conserved microbial signatures. Ann. Rev. Plant Biol. 63, 451-482 (2012).
- Schulze, B., et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444-9451 (2010).
- Lu, D., et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496-501 (2010).
- Zhang, J., et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 7, 290-301 (2010).
- Lu, D., et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 332, 1439-1442 (2011).
- Asai, T., et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 415, 977-983 (2002).
- Rasmussen, M. W., Roux, M., Petersen, M., Mundy, J. MAP Kinase Cascades in Arabidopsis Innate Immunity. Front. Plant Sci. 3, 169 (2012).
- Boudsocq, M., et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 464, 418-422 (2010).
- Dubiella, U., et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744-8749 (2013).
- Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., Yoshioka, H. Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell. 23, 1153-1170 (2011).
- Mao, G., et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 23, 1639-1653 (2011).
- Dangl, J. L., Jones, J. D. G. Plant pathogens and integrated defence responses to infection. Nature. 411, 826-833 (2001).
- Eitas, T. K., Dangl, J. L. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472-477 (2010).
- Jones, J. D., Dangl, J. L. The plant immune system. Nature. 444, 323-329 (2006).
- Boller, T., He, S. Y. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 324, 742-744 (2009).
- Bonardi, V., Dangl, J. L. How complex are intracellular immune receptor signaling complexes. Front. Plant Sci. 3, 237 (2012).
- Bonardi, V., Cherkis, K., Nishimura, M. T., Dangl, J. L. A new eye on NLR proteins: focused on clarity or diffused by complexity. Curr. Opin. Immunol. 24, 41-50 (2012).
- Takken, F. L., Albrecht, M., Tameling, W. I. Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383-390 (2006).
- Nakagami, H., et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161-1174 (2010).
- del Pozo, O., Pedley, K. F., Martin, G. B. MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072-3082 (2004).
- Ekengren, S. K., Liu, Y., Schiff, M., Dinesh-Kumar, S. P., Martin, G. B. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905-917 (2003).
- Romeis, T., et al. Rapid Avr9- and Cf-9 -dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell. 11, 273-287 (1999).
- Zhang, S., Klessig, D. F. Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 95, 7433-7438 (1998).
- Romeis, T., Piedras, P., Jones, J. D. G. Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 12, 803-815 (2000).
- Romeis, T., Ludwig, A. A., Martin, R., Jones, J. D. G. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20, 5556-5567 (2001).
- Romeis, T. Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 4, 407-414 (2001).
- Dodds, P. N., Rathjen, J. P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539-548 (2010).
- Grant, M. R., et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 269, 843-846 (1995).
- Mackey, D., Holt, B. F., Wiig, A., Dangl, J. L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 108, 743-754 (2002).
- Mucyn, T. S., et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 18, 2792-2806 (2006).
- Chung, E. H., et al. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 9, 125-136 (2011).
- Liu, J., Elmore, J. M., Lin, Z. J., Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 9, 137-146 (2011).
- Ntoukakis, V., et al. The Tomato Prf Complex Is a Molecular Trap for Bacterial Effectors Based on Pto Transphosphorylation. PLoS Pathog. 9, (2013).
- Sessa, G., D’Ascenzo, M., Martin, G. B. Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19, 3157-3157 (2000).
- Ntoukakis, V., et al. Host Inhibition of a Bacterial Virulence Effector Triggers Immunity to Infection. Science. 324, 784-787 (2009).
- Zhou, J. M., Loh, Y. T., Bressan, R. A., Martin, G. B. The Tomato Gene Pti1 Encodes a Serine/Threonine Kinase That Is Phosphorylated by Pto and Is Involved in the Hypersensitive Response. Cell. 83, 925-935 (1995).
- Wu, A. -. J., Andriotis, V. M. E., Durrant, M. C., Rathjen, J. P. A Patch of Surface-Exposed Residues Mediates Negative Regulation of Immune Signaling by Tomato Pto Kinase. Plant Cell. 16, 2809-2821 (2004).
- Gutierrez, J. R., et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 61, 507-518 (2010).
- Ntoukakis, V., Schwessinger, B., Segonzac, C., Zipfel, C. Cautionary notes on the use of C-terminal BAK1 fusion proteins for functional studies. Plant Cell. 23, 3871-3878 (2011).
- Rommens, C. M., Salmeron, J. M., Oldroyd, G. E., Staskawicz, B. J. Intergeneric transfer and functional expression of the tomato disease resistance gene Pto. Plant Cell. 7, 1537-1544 (1995).
- Jones, A. M., et al. Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochemistry. 65, 1805-1816 (2004).
- Widjaja, I., et al. Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics. 9, 138-147 (2009).
- Keinath, N. F., et al. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39140-39149 (2010).
- Elmore, J. M., Liu, J., Smith, B., Phinney, B., Coaker, G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteom. 11, (2012).
- Benschop, J. J., et al. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteom. 6, 1198-1214 (2007).
- Maor, R., et al. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell. Proteom. 6, 601-610 (2007).
- Elmore, J. M., Coaker, G. Biochemical purification of native immune protein complexes. Methods Mol. Biol. 712, 31-44 (2011).
- Win, J., Kamoun, S., Jones, A. M. Purification of effector-target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 712, 181-194 (2011).
- Sessa, G., D’Ascenzo, M., Martin, G. B. Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19, 2257-2269 (2000).

