Understanding Early Organogenesis Using a Simplified In Situ Hybridization Protocol in Xenopus
Summary
The Xenopus laevis embryo continues to be exceptionally useful in the study of early development due to its large size and ease of manipulation. A simplified protocol for whole mount in situ hybridization protocol is provided that can be used in the identification of specific organs in this model system.
Abstract
Organogenesis is the study of how organs are specified and then acquire their specific shape and functions during development. The Xenopuslaevis embryo is very useful for studying organogenesis because their large size makes them very suitable for identifying organs at the earliest steps in organogenesis. At this time, the primary method used for identifying a specific organ or primordium is whole mount in situ hybridization with labeled antisense RNA probes specific to a gene that is expressed in the organ of interest. In addition, it is relatively easy to manipulate genes or signaling pathways in Xenopus and in situ hybridization allows one to then assay for changes in the presence or morphology of a target organ. Whole mount in situ hybridization is a multi-day protocol with many steps involved. Here we provide a simplified protocol with reduced numbers of steps and reagents used that works well for routine assays. In situ hybridization robots have greatly facilitated the process and we detail how and when we utilize that technology in the process. Once an in situ hybridization is complete, capturing the best image of the result can be frustrating. We provide advice on how to optimize imaging of in situ hybridization results. Although the protocol describes assessing organogenesis in Xenopus laevis, the same basic protocol can almost certainly be adapted to Xenopus tropicalis and other model systems.
Introduction
The expression pattern of a specific gene is an important piece of information in determining the potential role for that gene in the development of a specific organ or cell type. Simply put, if it is not expressed at the right time and place it is unlikely to play a key role. In Xenopus, as in most early embryos, the most commonly used assay for detecting the expression of a gene is whole mount in situ hybridization using labeled antisense RNA probes. The use of antibody staining to assess expression of a gene in Xenopus is becoming more common as researchers discover antibodies, usually raised against mammalian proteins, that cross react to the Xenopus homologue or generate their own 1-3. However, the vast majority of studies on Xenopus organogenesis still utilize antisense RNA probes. When antibodies are used, each individual antibody often requires optimization for the primary antibody concentration or fixation protocols. In contrast, the protocol for in situ hybridizations is essentially invariant for different probes. The basic concept is relatively simple and an excellent standard protocol has been well established 4. Our protocol is a streamlined version of the original protocol 4 that still provides excellent detection of gene expression patterns in the early embryo. The embryos are fixed and then prepared for hybridization by changing solutions and temperatures such that it allows for high stringency binding of the labeled antisense RNA probe to its target mRNA. The unbound probe is washed away and the embryos are then prepared for binding of an antibody against the label on the RNA probes. Excess antibody is then washed away and an enzymatic color reaction is used to localize where the RNA probe is bound in the embryo. There are now a number of Xenopus transgenic lines that drive expression of fluorescent proteins in specific tissues and these are available at the Xenopus stock centers such as the National Xenopus Resource in Woods Hole. While very useful for many experiments that require examining organogenesis in living embryos, this option requires separate housing for the transgenic lines.
In situ hybridization can clearly delineate where specific organs or cell types will form in the early embryo (Figure 1). The technique is remarkably sensitive given that one can detect gene expression in a small number of cells in a single embryo 5. However, in situ hybridization using the intensity of colorimetric staining is not considered quantifiable because the color reaction is not a linear one. Despite difficulty in quantifying staining intensity, changes in expression are often quite noticeable; particularly when the in situ hybridization shows quantifiable increases or decreases in the size of expression domains 6,7.
The clear advantages of whole mount in situ hybridization make it a critical assay in the study of early development. However, it is a time consuming one that requires many steps over several days. This protocol is a simplified version of the standard protocol that eliminates several steps without reducing the quality of the in situ result. The simplification also eliminates sources of variability, making trouble shooting easier if an in situ hybridization is not optimal. Specifically, we have eliminated the use of proteinase K and RNAse treatments of the embryo, two steps that can depend on reagent quality and can also reduce signal intensity if overdone. The protocol also provides some degree of cost saving due to eliminating the use of several reagents. Finally, this protocol also provides some simple guidelines for improved capturing of images of in situ hybridization results. Although this protocol is optimized for work in Xenopus embryos, it is likely that at least some of the simplifications will be applicable to in situ hybridization work in other embryo systems.
Protocol
1. Embryo Preparation
- If not done routinely as part of embryo culture, de-jelly the embryos using 2.5% cysteine, pH 8.0 prior to fixation 8. Although it is not absolutely necessary, it is useful to then manually remove the fertilization envelope prior to fixation using fine forceps.
- Use glass Pasteur pipettes to transfer the embryos. The pipette is not wide enough to transfer embryos so use a diamond pen to cut the glass pipette at a point wide enough to pick up an embryo. Eliminate the sharp edges of the pipettes after cutting by quickly passing the cut tip through the flame of a Bunsen burner to melt the sharp edges.
NOTE: Care needs to be taken, as the glass can still be hot enough to cause burns even though it appears to have cooled by visual inspection.
- Use glass Pasteur pipettes to transfer the embryos. The pipette is not wide enough to transfer embryos so use a diamond pen to cut the glass pipette at a point wide enough to pick up an embryo. Eliminate the sharp edges of the pipettes after cutting by quickly passing the cut tip through the flame of a Bunsen burner to melt the sharp edges.
- Perform the embryo fixation in stages. First, prepare glass vials for use in embryo fixation. Use these vials for all steps including final storage. Use vials that are clear with a good Teflon seal in the lid, allowing for monitoring the embryos during all steps of the process. Label the vials with appropriate experimental information using permanent marker and then cover the label with clear tape, as even permanent marker will be lost over the course of the procedure due to the use of alcohols and other solvents.
- Use Mempfa to fix the embryos. Make a stock of 8% paraformaldehyde in batches of 50-100 ml at a time. Use approximately 75% of the required H2O and heat to 50-60 °C, which is needed to get the paraformaldehyde into solution.
NOTE: This solution is slightly different than the Memfa used in older published protocol versions in that it utilizes paraformaldehyde rather than formaldehyde. - Add 2-3 drops of 10N NaOH or until pH is approximately 7.5 (use pH paper to check pH). Paraformaldehyde is very toxic, therefore generate the stock solution in a fume hood. Once the paraformaldehyde is in solution, filter the solution through Whatman paper into a fresh container and add H2O to the final volume. If required, store the paraformaldehyde solution at 4 °C for 1-2 weeks.
- Assemble the remaining components of the Mempfa fixation solution (Table 1). Ensure that the final working concentration of paraformaldehyde is 4%. Store all components of the fixation solution at 4 °C as stocks.
- Using a cut glass pipette, fix the embryos by adding the embryos to the labeled glass vials that have been filled with the approximately 3-4 ml of Mempfa solution (Table 1). Avoid fixing more than 20-30 embryos per vial. Add the embryos with a minimum of liquid transfer from the embryo medium. Fix embryos in Mempfa solution for 2 hr at room temperature or overnight at 4 °C.
- For late endoderm structures perform the in situs on manually isolated gut and endoderm derivatives 9,10, which allows for good penetration of the probe and also avoids cavity staining.
- Store a solution of 100% methanol at -20 °C. After the paraformaldehyde fixation, replace the Mempfa solution with approximately 4 ml of the -20 °C, 100% methanol for storage of the embryos.
- Swirl the vials after addition of the methanol to prevent the embryos from sticking to the glass or other embryos. Also, make sure the vials are tightly sealed because the methanol can evaporate in the freezer over time if loose.
NOTE: Embryos can be stored for at least a year, and likely longer, in the methanol prior to staining with no loss of in situ hybridization quality.
- Use Mempfa to fix the embryos. Make a stock of 8% paraformaldehyde in batches of 50-100 ml at a time. Use approximately 75% of the required H2O and heat to 50-60 °C, which is needed to get the paraformaldehyde into solution.
2. Probe Preparation
- Use 1-2 µg of template DNA to make the digoxigenin-labelled probes. Cut plasmid containing the appropriate DNA sequence at the 5’ end of the gene of interest with a restriction enzyme suitable for that vector to generate antisense probes.
NOTE: The plasmid should have an appropriate RNA polymerase binding site (e.g. T7, T3, or SP6). - Allow the DNA, water, NTP mix and polymerase buffer to warm to room temperature before assembling the probe synthesis reaction. Add the components of the RNA synthesis reaction to a 1.5 ml microcentrifuge tube in the order as listed in Table 2. Adjust the volume of water added to bring the total volume of the probe synthesis reaction to 20 µl. Assemble the reaction at room temperature because components of the polymerase buffer can precipitate the template DNA when in high concentrations and cold.
- Incubate the transcription reaction for 2 hr at 37 °C.
NOTE: Incubations of slightly longer than two hours leads to no adverse effects, but also show little increased yield. If incubated for one hour, the yield will be reduced but still sufficient to make a good probe.- While waiting for the transcription reaction to finish, make an agarose gel that will be used to test the quality of the probe. Melt 1 µg of agarose in 100 ml of 1x TAE buffer (see Table 1) by heating the solution to the boiling point. Remove the solution from heat when the agarose powder has completely dissolved.
- Add 2 µl ethidium bromide stock solution (10 mg/ml) to about 100 ml of agarose gel when the agarose has cooled to about 60 °C to allow visualization of RNA under ultraviolet (UV) light.
NOTE: This agarose gel solution can be kept in a 60 °C incubator so that future gels can be poured without repeated melting. Take care in handling ethidium bromide due to potential toxicity.
- Add 1 µl DNAseI (RNAse free grade) to the transcription reaction after the 2 hr incubation and incubate for a further 10 min at 37 °C to eliminate template DNA.
- Remove 1 µl of the reaction mix to check on the 1% agarose-TAE gel and to the rest (20 µl) add 80 µl of 1% SDS in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA), 10 µl of 5M NH4 Acetate, and 220 µl of cold ethanol. Vortex the mix vigorously and set aside on ice until the results of the RNA quality check are known.
- Run the 1 µl of RNA removed before precipitation on the 1% agarose-TAE gel. To ease loading on the gel, add 4-5 µl of water and 1 µl of standard loading dye to the RNA. View the RNA on the gel using a UV light transilluminator in order to check the quality of the probe (see discussion).
- Precipitate the remaining RNA that was set aside on ice in step 2.5 by spinning in a microcentrifuge at full speed for 10 to 15 min. Draw off the supernatant with a drawn out glass pipette and allow to dry briefly.
- Resuspend the probe with 1 ml of RNA hybridization buffer (Table 1) in the eppendorf tube. Vortex and briefly heat the tube to 37 °C and vortex again. Transfer the probe solution to a 15 ml screw cap polystyrene tube and fill to 7-10 ml with RNA hybridization buffer. Note: The probe can be diluted further (a 10 fold further dilution can still work) often resulting in low background but the staining reactions will also take longer.
3. In situ Hybridization
- Take the embryos that were stored in -20 °C methanol and allow to warm to room temperature. Perform the entire procedure in the glass vials. If different embryos from one group are going to be looked at by different probes, keep them in a single vial until just before the probes are added to reduce variability and labor on the first day.
- Rehydrate the embryos through a methanol series as outlined in Table 3 (see Table 1 for wash recipes) in preparation for addition of the probe. Gently swirl the embryos after each change to ensure they are not sticking to the sides or each other, and rock the embryos by mounting the vials on a nutator. When transferring the liquids, check the inside of the caps to ensure that embryos have not been trapped there.
- Once in the probe solution, hybridize the embryos overnight as described in Table 3.
- Remove the probe solution. Save the used probe by storing in a 15 ml screw cap polystyrene tube, marked with date and number of times the probe has been used, at -20 °C.
NOTE: The same probe can be reused many times for subsequent in situ hybridizations until the colorimetric reactions begin to take an abnormally long time to reach desired intensity.- Once the probe is removed, prepare the embryos for antibody staining against the probe through the series of washes outlined in Table 4. Change the temperature as required (Table 4) by moving the nutator, with vials of embryos attached, directly into hybridization ovens that are set to the appropriate temperature.
- Make up the appropriate volume of the MAB+HTSS+BR+anti-Dig antibody (Table 4) at the time that the embryos are being incubated in the MAB+HTSS+BR blocking solution so that the antibody is blocked prior to adding to the embryos. Make the blocking solutions fresh on the day of use.
- Remove the antibody solution and begin washes as outlined in Table 5 following overnight incubation with antibody. Perform at least twelve 30 min washes in order to prepare for staining with the alkaline phosphatase substrate and reduce the background as much as possible. Use either MAB buffer or TBT solution (Table 1) for the washing steps.
NOTE: TBT solution is TTw that has 2 mg/ml of BSA added.- Replace the last wash solution with the BM Purple alkaline phosphatase substrate. Perform the staining reaction at either room temperature or 37 °C.
NOTE: Staining is more rapid at 37 °C but if left overnight, unacceptable background staining can often be a result. The staining reactions will often require overnight staining and room temperature is safest for this. - If further staining is required and the BM purple solution is taking on a blue color, replace the staining solution with fresh BM Purple and put the tubes into 37 °C. If the target mRNA is very abundant, put the embryos in the staining solution at 4 °C overnight and then move the embryos to room temperature or 37 °C to allow better monitoring of the staining reaction.
- Monitor new reactions carefully, as the time to final result varies considerably between different target RNAs. For consistency, stop the staining reaction (see 3.4) when there are no new sites of expression arising with longer incubation. Importantly, if in situ hybridization is being used as an assay for experiments looking at different treatment groups, use the same time for color reactions between treatment and control embryos.
- Replace the last wash solution with the BM Purple alkaline phosphatase substrate. Perform the staining reaction at either room temperature or 37 °C.
- Stop the staining reaction and prepare the embryos for storage and image recording by changing the liquids as outlined in Table 6. Do not rock the embryos at this stage because the removal of stain can be visualized as purple coloring of the methanol around the embryos.
NOTE: Often embryos have a light blue staining from the staining solution and the cold methanol can at least partially remove that, although this is not sufficient to remove heavy background or cavity staining. Cold methanol refers to methanol that is maintained in a -20 °C freezer.- Rehydrate the embryos and fix the stain with Mempfa (Table 6). Once fixed, remove the Mempfa and wash the embryos with 25% methanol.
- Remove the 25% methanol and add the bleaching solution if removal of endogenous pigment is required. Take care in handling the bleaching solution as it can cause burns. Observe the bleaching closely as it happens relatively quickly and the degree of bleaching can be varied for different effects (Figure 2).
- Dehydrate embryos through a methanol series to 100% methanol for long term storage following bleaching, or transfer to PBS for short term storage and subsequent imaging (see Table 6).
4. Imaging Embryos
- Once an embryo has completed the staining process, image the embryo so that the information on where the gene of interest is expressed is captured for a wider audience. Use 1% agarose as a background to view uncleared embryos.
NOTE: The agarose gives a blue/grey background that contrasts well with the embryo and the blue color of the staining reaction. It also helps diffuse distracting shadows and reflections that divert attention from the embryo.- Add agarose to water and then bring to a boil until the agarose is in solution and then let cool to 50 before pouring into the petri dish. As with the TAE gel solution, store the agarose solution in 55 °C incubator for multiple uses. Pour the agarose to a depth of about 2 mm into the petri dish. If required, adjust the depth of agarose to yield a slightly differently shade of background.
- Following rehydration from storage in methanol to an aqueous solution (PBS or TTw), using a methanol series, place the embryos in a petri dish with the agarose base (see Table 6). Keep the solution clean and if required, use simple filtering to eliminate small particulate matter that can disrupt the clean background of good images.
- To image the embryos from alternate views, such as from the ventral side, cut thin channels in the agarose to fit the embryo using fine forceps and place the embryos in those channels for orientation (Figure 3). Take care in manipulating the embryos as they are easily damaged.
NOTE: Most stages have a characteristic position that they assume when placed in solution. For example, blastula-stage embryos tend to sit animal side up. Once the embryos begin to elongate, they lay on their side.
- Use the fiber optic light source to illuminate the embryo from a shallow angle, creating shadows on the embryo that provide depth to the image and help discern surface structures. Because bleaching can eliminate pigment that provides useful landmarks, use shadowing for strongly bleached embryos.
- Clear the embryos to image staining that is deep within the embryo, such as in the notochord, lung, or regions of the brain, (Figure 4). To accomplish this, put the embryos through a methanol series until in 100% methanol.
- After complete immersion in methanol, transfer the embryos to a solution of one part benzyl alcohol and two parts benzyl benzoate (BABB). The embryos will initially float on the surface but as the methanol mixes with the BABB, they will sink into the BABB. Perform every step dealing with BABB in glass vials or dishes; BABB will melt any plastic or paint.
- Once cleared, view the embryos with transmitted light coming from below the embryo. Adjust the intensity of the light as well as the angle from below to improve contrast and provide the best color.
- When viewing cleared embryos raise the glass petri dish off of the base. Do this by simply using two other petri dish lids to raise it so that the region of the dish with embryos is elevated.
NOTE: This has the advantage of taking the base out of focus and eliminating distracting effects from imperfections or stains on the microscope base that can interfere with the image.
5. Double In situ Hybridization
- In order to simultaneously view the expression pattern of two different genes in a single embryo, synthesize two probes, one for each of the different genes. Synthesize one probe using DIG-11-UTP as a label as described above. Dilute the product of the transcription reaction in RNA hybridization buffer to yield a 3x more concentrated probe than used for single in situ hybridizations.
- Synthesize the other probe of interest using the same protocol as for DIG labelled probed except that fluorescein-12-UTP must be substituted for DIG-11-UTP. Dilute the product of the transcription reaction in RNA hybridization buffer to yield a 3x more concentrated probe than used for single in situ hybridizations.
- Mix the two concentrated probes in a 1:1 ratio. For best results, use the fluorescein-labelled probe for the gene that shows the strongest expression in the single in situ hybridization protocol.
- Use the same in situ hybridization protocol described for single in situ hybridizations, except use the double probe (probe containing the 1.5x concentrated mixture of digoxigenin-labelled and fluorescein-labelled probes) at the end of the first day in place of a single in situ hybridization probe.
- Follow the single hybridization protocol on the second day of the double in situ hybridization protocol, except use anti-fluorescein-AP Fab fragments at a 1:4,000 dilution in place of anti-DIG-AP Fab fragments. Wash out the excess antibody from the embryo as in the single in situ protocol and carry out the first color reaction using the BM-Purple AP substrate.
- Following the first color reaction, inactivate the flourescein antibody in 0.1 M glycine pH 2.0 for 40 min followed by five ten min washes in MAB. Block the embryos in MAB+HTSS+BR for 90 min. Add the anti-DIG antibody at a 1:2,000 dilution in MAB+HTSS+BR and incubate at 4 °C overnight.
- The following day, wash the embryos thoroughly in MAB (12 washes of 30 min) to remove the excess antibody.
- Wash the embryos for 10 min in AP Buffer (Table 1) and then stain with BCIP (0.5mg/ml in AP Buffer).
NOTE: The in situ combination should give a dark blue-purple stain for the first color reaction and a light blue color reaction for the second (Figure 5). - Stop the final color reaction by removing the AP buffer and rinse three times with MAB. Fix the embryos with Mempfa for 10 min. Wash the embryos with 5 quick washes in MAB or TBT.
- With this color combination, the use of methanol in the post staining treatments is no longer possible, as it will eliminate the BCIP alone color. Store the embryos after staining and fixation in PBS with 0.02% sodium azide. Staining intensity can be weak with double in situs. If this is a problem, reduce the washes to four, each of 2 hr duration.
Representative Results
The use of tissue specific probes can provide outstanding information in regards to the state of development for specific organs. In the following examples, the stage of the embryo is based on the Nieuwkoop and Faber staging table 11. If one uses probes form genes expressed after differentiation, cardiac troponin I at stage 28-30, for example (Figure 1C), the presence or size of a differentiated organ can be assessed at any stage post differentiation. Years ago, embryologists were able to do such analysis based on remarkable knowledge of the histology of the early embryo 12,13 but that expertise has been largely lost to the current generation of embryologists. Although, the loss of this expertise can be considered regrettable, the reliability and ease of use of in situ hybridization techniques makes identification of specific tissues available to any researcher. Tissues that are relatively inconspicuous, the hatching gland at embryonic stage 25 for example (Figure 1A), can be vividly marked using in situs without the need for specific antibodies or histological techniques (Figure 1C). Also, the use of whole mount in situs allows one to view the entire organ in the context of the whole embryo rather than relying on inference from histological sections (Figure 2). Even tissues that are deep within the embryo including the optic stalk (Figure 4) can be viewed easily and the use of embryo clearing can provide sharp delineation of the organ boundaries.
Bleaching of embryos to remove endogenous pigment using peroxide solutions has largely eliminated the requirement for using albino embryos that lack pigment. Bleaching for different times can be useful. For example, if the stain is strong, a lighter bleaching that allows for some pigmentation to be seen can be useful because it allows for better staging and orientation of the embryo (Figure 2A). However, if the stain is an area where there are high levels of pigmentation, such as the kidney (Figure 2B), near complete elimination of the pigment by longer incubation in the bleaching solution gives a better result.
Embryos tend to take up particular positions when in solution. After neurulation, they tend to lay flat on their sides. This is fine for images of the flank (Figure 2B) but can be a problem with other areas. Use of an agarose base allows one to cut channels into the agarose that can be use to orient the embryos. For example, blood precursors are localized to the ventral side of the embryo (Figure 2A) and the full extent of the staining is hard to observe. Positioning of the embryo in a channel with the ventral side up, then allows for full viewing of that gene expression pattern (Figure 3A).
The use of double in situ hybridization can show the relationship between two gene expression patterns within a single organism (Figure 5) eliminating the necessity of comparing between different embryos that may have small differences in morphology. Perhaps most importantly, the use of in situ hybridization gives one the ability to clearly mark cells prior to clear histological differentiation based on the expression of genes that are expressed in early progenitors of a lineage, such as pax2 that is expressed in many tissues in the early embryo, prior to differentiation (Figure 4A). The ability to identify progenitors and tissues that are not clearly distinguished based on histology, such as myeloid cell precursors (Figure 1B), has allowed researchers to ask much more detailed questions about the state of an organ’s development and also to assess the results of experimental manipulation designed to cause ectopic differentiation of a tissue.
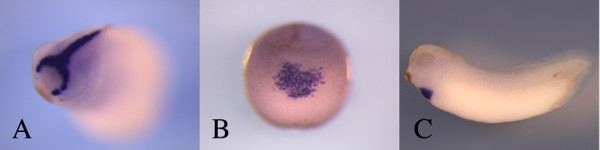
Figure 1: Examples of the whole mount in situ hybridization on Xenopus embryos. The blue staining from an in situ hybridization experiment can clearly delineate developing structures of the early Xenopus embryo. (A) An anterior view of a stage 26 embryo highlighting the hatching gland as demarcated by the expression of uvs.2 14. (B) A ventral view of an embryos showing the location of early myeloid cells at stage 20 using the expression of myeloperoxidase15 as a marker. (C) The early heart at stage 28 – 30 is visualized by the expression of cardiac troponin I 16. A clear advantage of using this method is that all of these gene expression patterns were visualized using different probes but the protocol used is identical in all cases. Please click here to view a larger version of this figure.
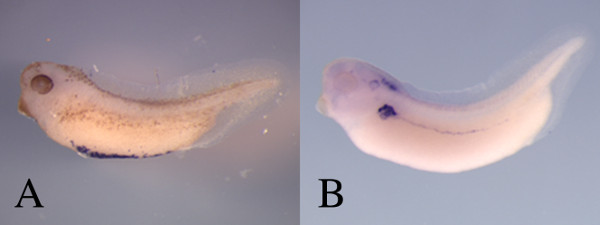
Figure 2: Different levels of bleaching can be used to visualize staining in the embryo. (A) This blue staining along the bottom of this embryo shows the expression of hemoglobin in the ventral blood islands at about stage 36. This is a region of the embryo that is only lightly pigmented and thus the embryo was not bleached for a long time as can be seen by the tan colored pigment in the eye and along the flank of the embryo. Being able to see the pigmentation allows for better visualization of the stage of the embryo. If the staining is in a region with greater natural pigmentation, such as the nervous system and the flank of the embryo, greater bleaching will help view the in situ as seen in B. (B) Here the expression of pax8 17 in the forming kidney, pronephric duct and hindbrain is best visualized after bleaching has removed almost all endogenous pigment. Please click here to view a larger version of this figure.
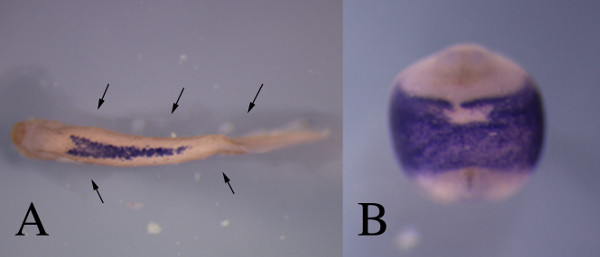
Figure 3: Manipulation of the agarose base can help with orientation of the embryos. Imaging specific regions of the embryo can be difficult because they tend to take up particular positions in the dish. (A) At tadpole stages the embryo will lie on its side. By cutting a fine channel in the agarose (black arrows) the embryo can be viewed from the ventral side, here showing hemoglobin expression at stage 36, with enough stability to capture a good image. (B) This ventral view of the hand1 expression at stage 20 outlines the lateral plate mesoderm 6. The embryo is placed in a small hole that stabilized its position with the ventral side up. Please click here to view a larger version of this figure.
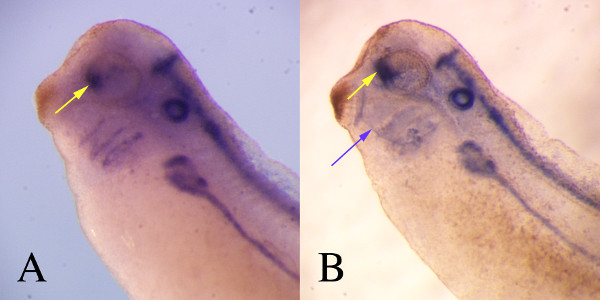
Figure 4: Internal organs can be viewed in both opaque, uncleared and in cleared embryos. In an uncleared embryo stained for pax2 at stage 34 (A), the optic stalk (yellow arrow) can be visualized relatively easily as can the staining down the neural tube. However, details are not sharp. By clearing the embryo (B) the boundaries of expression sites, including the optic stalk (yellow arrow) are sharper. The extent of clearing is shown by the ability to see both eyes in this cleared embryo. Also note that some staining has accumulated in the internal cavities (purple arrow), a common problem when viewing cleared embryos.
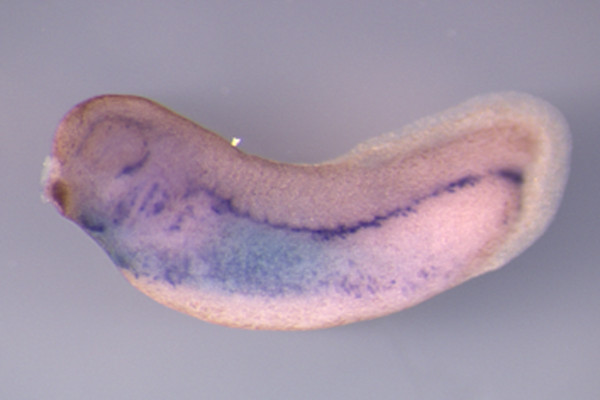
Figure 5: A representative double in situ hybridization on a Xenopus embryo. Expression of the lateral plate marker, hand1, is visualized by the light blue staining at stage 26. Expression of etv2 within the developing vasculature is visualized by dark blue purple stain. Expression of hand1 is usually very intense and thus it was utilized for the weaker fluorescein-labelled probe and BCIP combination. The utilized the digoxigenin-labelled probe and BM-purple combination for the weaker etv2 expression.
| Name | Final Concentration | Amount/100 ml |
| Mempfa | 1 mM MgSO4 | 100 µl of 1 M MgSO4 |
| (store at 4°C) | 2 mM EGTA, pH 8.0 | 10 ml of 20 mM EGTA, pH 8.0 |
| 0.1 M MOPS, pH 7.5 | 10 ml of 1 M MOPS, pH 7.5 | |
| 4% Paraformaldehyde, pH 7.5 | 50 ml of 8% Paraformaldehyde, pH 7.5 | |
| TTw | 50 mM Tris, pH 7.4 | 5 ml of 1 M Tris, pH 7.4 |
| 200 mM NaCl | 4 ml of 5 M NaCl | |
| 0.1% Tween 20 | 100 µl of Tween 20 | |
| 100X Denhardt's Solution | 2 % BSA | 2 g BSA |
| (filter through 0.2 µm filter, | 2% PVP-40 | 2 g PVP-40 |
| store at -20°C) | 2% Ficoll 400 | 2 g Ficoll 400 |
| 20X SSC | 3 M NaCl | 17.5 g NaCl |
| 300 mM Trisodium Citrate | 8.8 g Trisodium Citrate | |
| RNA Hybridization Buffer | 50% formamide | 50 ml of 100% formamide |
| (store at 4°C) | 5x SSC | 25 ml of 20x SSC |
| 1 mg/ml yeast RNA | 4 ml of 1mg/ml yeast RNA dissolved in 50% formamide | |
| 1X Denhardt's Solution | 1 ml of 100x Denhardt's Solution | |
| 0.1% Tween 20 | 100 µl of Tween 20 | |
| 5 mM EDTA, pH 8.0 | 1 ml of 0.5 M EDTA, pH 8.0 | |
| MAB | 100 mM Maleic Acid | 1.16 g of Maleic Acid |
| (pH 7.5, store at 4°C) | 150 mM NaCl | 0.88 g of NaCl |
| MAB+HTSS+BR | 100 mM Maleic Acid | 96 ml of MAB |
| 4% Heat Treated Sheep Serum | 4 ml of Heat Treated Sheep Serum (heat treated at 55°C for 30 min and aliquoted) | |
| 2% Blocking Reagent | 2 g of Roche Blocking Reagent | |
| MAB+HTSS+BR+anti-Dig antibody | 100 mM Maleic Acid | 96 ml of MAB |
| 4% Heat Treated Sheep Serum | 4 ml of Heat Treated Sheep Serum | |
| 2% Blocking Reagent | 2 g of Roche Blocking Reagent | |
| 1:10,000 Anti-Dig-AP, Fab Fragments Antibody | 1.5 µl of Anti-Digoxygenin-AP, Fab Fragments Antibody | |
| Alkaline Phosphatase (AP) Buffer | 100 mM Tris, pH 9.5 | 10 ml of 1 M Tris, pH 9.5 |
| 50 mM MgCl2 | 5 ml of 1 M MgCl2 | |
| 100 mM Nacl | 2.5 ml of 4 M NaCl | |
| 0.1% Tween 20 | 100 µl of Tween 20 | |
| Bleaching Solution | 0.3% H2O2 | 3.34 ml of 30% H2O2 |
| 5% Formamide | 5 ml of 100% Formamide | |
| 0.5% SSC | 2.5 ml of 20x SSC | |
| Clearing Solution | 1/3 Benzyl Alcohol | 33 ml |
| 2/3 Benzyl Benzoate | 67 ml |
Table 1: Solution Recipes
| Name | Amount |
| Dig-NTP Mix | 5 µl of 20 mM CTP |
| (40 µl reaction) | 5 µl of 20 mM GTP |
| 5 µl of 20 mM ATP | |
| 3.25 µl of 20mM UTP | |
| 3.5 µl of 10mM Dig-11-UTP | |
| 18.25 µl distilled, autoclaved water | |
| Probe Synthesis | x µl of template DNA (dependent on concentration) |
| (20 µl reaction) | x µl of distilled, autoclaved water |
| 4 µl of Dig-NTP mix | |
| 0.5 µl of RNAse inhibitor | |
| 2 µl of 10X RNA polymerase buffer | |
| 2 µl of RNA polymerase |
Table 2: Probe Synthesis Recipes
| Name of the Reagent | Approximate Volume | Duration | Temperature |
| 100% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 75% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 50% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| TTw | 2 ml | 10 min, rocking | Room Temperature |
| TTw | 2 ml | 10 min, rocking | Room Temperature |
| TTw | 2 ml | 10 min, rocking | Room Temperature |
| Pre-warm RNA Hybridization Buffer and Probe to 65°C | |||
| TTw | 4 ml | 5 min, rocking | Room Temperature |
| TTw | 4 ml | 5 min, rocking | Room Temperature |
| RNA Hybridization Buffer | 2 ml | 10 min, rocking | Room Temperature |
| Pre-warmed RNA Hybridization Buffer | 2 ml | 1 hr, rocking | 65°C |
| Probe Solution | 1 ml | Overnight, rocking | 65°C |
Table 3: Steps for First Day of In Situ Hybridization Protocol (about 3 hr total)
| Name of the Reagent | Approximate Volume | Duration | Temperature |
| Pre-warm RNA Hybridization Buffer and 02.x SSC to 65°C and 2x SSC to 37°C | |||
| Return probe solution to tube for repeat use | |||
| RNA Hybridization Buffer | 2 ml | 10 min, rocking | 65°C |
| 2X SSC | 2 ml | 20 min, rocking | 37°C |
| 2X SSC | 2 ml | 20 min, rocking | 37°C |
| 0.2x SSC | 4 ml | 1 hr, rocking | 65°C |
| 0.2x SSC | 4 ml | 1 hr, rocking | 65°C |
| MAB+HTSS+BR | 1.5 ml | 30 min, rocking | Room Temperature |
| MAB+HTSS+BR+anti-DIG antibody | 1.5 ml | Overnight, rocking | 4°C |
Table 4: Steps for Second Day of In Situ Hybridization Protocol (about 4 hr total)
| Name of the Reagent | Approximate Volume | Duration | Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| MAB | 4 ml | 30 min, rocking | Room Temperature |
| BM Purple AP Substrate | 500-750 µl | Overnight, rocking (see text) | Room Temperature/37 °C (see text) |
Table 5: Steps for Third Day of In Situ Hybridization Protocol (about 7 hr)
| Name of the Reagent | Approximate Volume | Duration | Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 50% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 75% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 100% Methanol (cold) | 2 ml | 20 min, rocking | Room Temperature |
| 100% Methanol (cold) | 2 ml | Varies, no rocking | Room Temperature |
| 75% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 50% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| Mempfa | 2 ml | 30 min, rocking | Room Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| Bleaching Solution | 4 ml | 40 min-3 hr, rocking | Room Temperature/37 °C (see text) |
| Storing Embryos | |||
| 25% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 50% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 75% Methanol | 2 ml | 5 min, rocking | Room Temperature |
| 100% Methanol | 4 ml | Store at -20 °C | |
| Imaging Embryos | |||
| 1x PBS | 2 ml | 5 min, rocking | Room Temperature |
| 1x PBS | 2 ml | 5 min, rocking | Room Temperature |
| 1x PBS | 2 ml | 5 min, rocking | Room Temperature |
Table 6: Steps for Stopping In Situ Hybridization, Bleaching and Storage of Embryos
Discussion
The ability to use in situ hybridization to visualize the expression pattern of specific genes remains the most commonly used method to identify specific organs or cell types in the Xenopus embryo. This is because of several advantages offered by this technique. The expression of a gene can identify specific structures well before any histological sign of differentiation such as the case for nkx2.5 expression in the heart progenitors prior to any clear demarcation of those cells 18. Once all of the reagents are in hand, it is also very cost effective. Generation of many individual probes, that can be effectively reused, is possible with little extra time and cost investment other than obtaining the plasmids that encode the genes of interest. In our experience, probes can be reused for years with little loss in activity despite the inevitable small dilutions that come with each use. Finally, there is little optimization required when utilizing different probes.
The quality of the probe synthesis is a key factor in the success of the protocol. There are many ways to check quality of the RNA probe but simply running the RNA probe on a TAE gel is a rapid and easy method that is quite reliable. We routinely use ethidium bromide to stain the RNA and that requires special handling and disposal of the gels by most institutions. Non-toxic alternatives are commercially available although we have not specifically compared those for visualizing probes. The probe should run as a single band although it will appear somewhat fuzzy compared to DNA on a TAE gel. An estimate of the quantity of RNA can be obtained: a good reaction will have about 1 µg/µl of RNA. An easily visualized band will represent at least 0.5 µg of RNA and a bright band will represent over 1 µg. Although clearly not completely accurate and requiring some experience, quantification based on the gel is rapid and the robustness of the in situ procedure allows it to work well. If there is concern about repeatability, one can easily use UV absorbance to quantify a small amount of the transcription reaction, although we have not found this to be a major concern. Finally, the gel allows one to see if the template DNA has been eliminated. If the gel indicates that there is no RNA, the two likeliest explanations are a bad DNA template or that the transcription buffer is not working well. Heating the transcription buffer to 37 °C and thorough mixing before use, or addition of 1 µl of fresh 100 mM DTT to the transcription reaction can occasionally help with the latter. An important component of the transcription reaction buffer is spermidine and it can precipitate the DNA template at low temperatures making warming of the buffer important. DTT in the buffer can also precipitate significantly inhibiting the reaction. If a precipitate is seen in the buffer, warm the solution and vortex vigorously. There is no need for taking any unusual precautions, such as treating the water with DEPC or autoclaving tips and tubes, to prevent RNAse activity. The presence of the commercial ribonuclease inhibitor provides considerable protection against RNAses from the environment. Note that other UTP labels can be used, such as fluorescein, although in our experience, the digoxigenin-labeled UTP and anti-digoxigenin antibody combination provides the most consistent results.
Early protocols for probe generation suggest that alkaline hydrolysis of the synthesized probes should be done 4 in order to allow for greater probe penetration. Hydrolysis of the probe does not appear to help with the in situ hybridization process; RNA probes of greater than 500 bp usually give an excellent signal and probes that are longer than 2 kb in length also work well. Shorter probes can work if the target RNA is abundant but longer probes will give better staining. There does not appear to be any clear advantage in using alkaline digestion to break up longer probes.
Care in the initial handling of the embryos is very important. Removal of the fertilization envelope is particularly helpful for embryos after neural fold closure and prior to natural hatching out of the fertilization envelope. During elongation of the embryo inside the fertilization envelope, the embryo becomes curled and if fixed in that position, the embryos are harder to image after the in situ hybridization. However, if the embryos are removed from the fertilization envelope before fixation, they rapidly straighten out. If embryos are damaged during membrane removal, allow them to heal the wound before fixation because damaged tissue can result in a false in situ hybridization signal at the wound site. If small, the wounds heal very rapidly, often within minutes 19. Later steps can also damage the embryos during liquid transfer and so care is needed when changing solutions. Damage in early steps usually means that the embryo will be badly damaged by the end of the procedure and damaged regions will cause false in situ signals at the sight of damage. Furthermore, up to 30 embryos can be probed in a single vial but with more embryos there are greater chances of high background developing during the colorimetric reaction, however increasing the washes on day 3 or washing overnight can compensate for this.
In this protocol, the number of steps has been reduced and the use of several commonly used reagents has been eliminated. Note that this protocol eliminates several steps that are used in many other protocols including a proteinase K digestion and use of acetic anhydride to block positively charged groups. Those steps may still be useful when looking at mammalian and avian embryos but in using Xenopus, eliminating those steps has little impact on the final results of the in situ hybridization. Our lab has not determined whether these same simplifications can be applied to in situ hybridization protocols for other species. Proteinase K digestion in particular may still be required in other embryos such as chick or mouse where probe penetration may have greater limitations.
The use of BM purple substrate is convenient and reliable. However, there are a number of other options available for the precipitating, color alkaline substrates that are required to localize the target RNA in the embryo. In particular, a combination of NBT/BCIP is commonly used 4 and works well. Some color reagents are soluble in methanol, in which case, the use of methanol after staining will eliminate the stain and must be avoided. Other color combinations also can be used when performing double in situs. The use of a similar combination (NBT/BCIP rather than BM purple) has also been shown to be very robust when using mouse embryos 20. The blue color of the staining and the endogenous pigment of the embryo are often difficult to clearly distinguish in pictures. Use of albino embryos will also eliminate pigment but the bleaching solution is almost as effective and is much more convenient as it does not require the maintenance of albino adults for breeding. If the staining is relatively weak, strong bleaching can emphasize that staining. If the staining is strong, leaving some light pigment can be an effective contrast and also help with orienting and staging the embryo.
After the overnight hybridization in RNA probe, the protocol can diverge from the strictly manual protocol outlined here to the use of an in situ hybridization robot. The use of the in situ hybridization robot saves almost a full day as the Day Two and Day Three of the manual protocol can be reduced to one day as the robot works through the night and can make all of the appropriate temperature changes. The robot also is very consistent in its results and allows for simultaneous probing of many samples efficiently. It remains advantageous to do the first day by hand as that allows for reuse of probes and use of smaller volumes of reagent. The only significant disadvantage of using a robot is the initial cost of the instrument.
This protocol discusses some of the ways to effectively image an in situ hybridization. It is important to do so as much of the information can be lost if the image is poor. In many cases, imaging of the in situ results of an experiment requires the same time as the initial experiment. Some elements of the procedure variables such as the degree of bleaching have an element of personal preference. Other imaging effects can be achieved by placing the dish on a colored base. Often a slight blue shade to the agar can emphasize the deep blue staining of the in situ hybridization. Simply put the petri dish containing the agar over a sheet of blue plastic to get the desired effect.
However, the methods outlined here provide a basis with which to explore imaging possibilities. Embryos can be viewed either cleared (made transparent to better view internal structures) or uncleared (viewed as they appear under normal lighting conditions). Some of the decision in regards to how to the image the embryo is dependent on the site of expression. If the expression is on or near the surface of the embryo, it is best to image the embryo without clearing. There are several advantages to using the uncleared embryos. The process of clearing the embryos requires many steps and the chemicals used to clear the embryos are difficult to handle. Also, several cavities in the embryo allow precipitation of alkaline phosphate substrates that can result in false cavity staining. In particular, the blastocoel of early embryos and the pharyngeal cavity often show staining (Figure 4). This false staining is not visible in embryos that are not cleared. For the most part, even moderately deep expression can be visualized in uncleared embryos, including mesodermal tissues such as heart, kidney, and somites, and endodermal stuctures such as thyroid and liver. Moving the embryos through a methanol series to either hydrate in PBS for viewing or put into 100% methanol for storage allows for multiple rounds of storage and imaging. The methanol storage allows one to try different modifications to the imaging over extended time.
There are some disadvantages to using the in situ hybridization technique to look at gene expression. Levels of expression are not strictly quantifiable although comparison within an embryo and gross changes in expression and changes in expression domain size are usually obvious. As with other RNA-based techniques, it does not provide any information as to the proteins encoded by the gene of interest, which can limit interpretation of results. Finally, it is often difficult to determine what might be background staining as compared to the true expression domain. This is particularly a problem with genes with an unknown expression pattern that can be widespread. Often, a probe generated from the sense strand of the same gene is used as a control for non-specific staining. Use of a sense strand control can provide some information regarding a problem with reagents but it does not provide definitive evidence that staining based on the antisense probe is completely accurate. Results from an in situ are usually remarkably consistent across embryos when multiple embryos are used and this can be used as a measure of confidence in a staining pattern. Also, different antisense probes, generated from different parts of the gene, particularly untranslated regions, can be used to see if they give the same staining pattern. If they do so, it also provides confidence in the observed staining pattern if it is identical. Diluted probes can also be used with prolonged exposure to the staining solution to see if the same staining pattern emerges. One factor that usually inspires confidence in a staining pattern is whether that pattern corresponds to a specific embryonic structure. Enough in situs have now been done with a large variety of genes that novel expression patterns are likely to relatively rare events. Large databases of in situ images have been established for different organisms that can be used to compare image results. For Xenopus embryos, Xenbase (www.xenbase.org) is an excellent example of a resource that can be used to understand expression patterns. Other model organisms also have similar, extensive databases of images.
Despite these potential caveats, in situ hybridization remains a powerful and reliable tool that is extremely useful for the study of organogenesis. It is likely to remain the method of choice for identifying cell types and examining change in gene expression domains for the foreseeable future.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the CIHR for fellowship support of Steve Deimling and the Department of Paediatrics, University of Western Ontario for support of Steve Deimling, Rami Halabi and Stephanie Grover. This work was supported by the NSERC grant R2654A11 and an NSERC Discovery Accelerator Supplement
Materials
| Name of the reagent or equipment | Company | Catalogue number |
| Labguake Tube Shakers | VWR | 17-08-2011 |
| VWR Vials | VWR | 10-07-2012 |
| L-Cysteine | BioShop | CYS342.500 |
| Ribonucleoside Triphosphate Set, 100mM | Roche | 11277057001 |
| Digoxigenin-11-UTP | Roche | 11209256910 |
| Rnase inhibator (Rnase OUT) | Invitrogen | 10777-019 |
| T7 RNA Polymerase | Fermentas | EPO111 |
| T3 RNA Polymerase | Fermentas | EPO101 |
| SP6 RNA Polymerase | Fermentas | EPO131 |
| Dnase 1 | Invitrogen | 18047-019 |
| Sheep Serum | Wisent | 31150 |
| Blocking reagent | Roche | 11096176001 |
| BM purple Ap Substrate | Roche | 11442094001 |
| Anti-Digoxigenin-Ap Feb fragments | Roche | 11093274910 |
| Methanol | VWR | CAMX0485-7 |
| NaCl | BioShop | SOD002.10 |
| SDS | EM | 7910 |
| EDTA | BioShop | EDT001.500 |
| Tris | BioShop | TRS003.5 |
| Tween-20 | EM | 9480 |
| MgSO4 | Sigma | M-2643 |
| Mops | BioShop | MOP001.250 |
| EGTA | Sigma | E-3889-25G |
| Paraformaldehyde | BioShop | PAR070.500 |
| Formamide | VWR | CAFX0420-4 |
| RNA | Roche | 10109223001 |
| Maleic Acid | VWR | CAMX0100-3 |
| tri-Sodium Citrate | BioShop | CIT001 |
| Hydrogen Peroxide (30% Solution) | EM | HX0635-2 |
| BSA | BioShop | ALB001.100 |
| PVP-40 | ICN | 195451 |
| Ficoll 400 | GE Healthcare | 17-0300-10 |
| Benzyl Alcohol | Sigma | B-1042 |
| Benzyl Benzoate | Sigma | B-6630 |
| UltraPure Agarose | Invitrogen | 16500-500 |
References
- Ninomiya, H., et al. Cadherin-dependent differential cell adhesion in Xenopus causes cell sorting in vitro but not in the embryo. J Cell Sci. 125, 1877-1883 (2012).
- Movassagh, M., Philpott, A. Cardiac differentiation in Xenopus requires the cyclin-dependent kinase inhibitor, p27Xic1. Cardiovasc Res. 79, 436-447 (2008).
- Zhao, Y., et al. The expression of alphaA- and betaB1-crystallin during normal development and regeneration, and proteomic analysis for the regenerating lens in Xenopus laevis. Mol Vis. 17, 768-778 (2011).
- Harland, R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685-695 (1991).
- Kirilenko, P., Weierud, F. K., Zorn, A. M., Woodland, H. R. The efficiency of Xenopus primordial germ cell migration depends on the germplasm mRNA encoding the PDZ domain protein Grip2. Differentiation. 76, 392-403 (2008).
- Deimling, S. J., Drysdale, T. A. Fgf is required to regulate anterior-posterior patterning in the Xenopus lateral plate mesoderm. Mech Dev. , (2011).
- Fletcher, R. B., Harland, R. M. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev Dyn. 237, 1243-1254 (2008).
- Sive, H. L., Grainger, R. M., Harland, R. M. . Early development of Xenopus laevis : a laboratory manual. , (2000).
- Park, E. C., Hayata, T., Cho, K. W., Han, J. K. Xenopus cDNA microarray identification of genes with endodermal organ expression. Dev Dyn. 236, 1633-1649 (2007).
- Horb, M. E., Slack, J. M. Endoderm specification and differentiation in Xenopus embryos. Dev Biol. 236, 330-343 (2001).
- Nieuwkoop, P. D., Faber, J. . Normal table of Xenopus laevis (Daudin) : a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. , (1994).
- Gebhardt, D. O., Nieuwkoop, P. D. The Influence of Lithium on the Competence of the Ectoderm in Ambystoma Mexicanum. J Embryol Exp Morphol. 12, 317-331 (1964).
- Nieuwkoop, P. D. Pattern formation in artificially activated ectoderm (Rana pipiens and Ambystoma punctatum). Dev Biol. 6, 255-279 (1963).
- Sato, S. M., Sargent, T. D. Molecular approach to dorsoanterior development in Xenopus laevis. Dev Biol. 137, 135-141 (1990).
- Smith, S. J., Kotecha, S., Towers, N., Latinkic, B. V., Mohun, T. J. XPOX2-peroxidase expression and the XLURP-1 promoter reveal the site of embryonic myeloid cell development in Xenopus. Mech Dev. 117, 173-186 (2002).
- Drysdale, T. A., Tonissen, K. F., Patterson, K. D., Crawford, M. J., Krieg, P. A. Cardiac troponin I is a heart-specific marker in the Xenopus embryo: expression during abnormal heart morphogenesis. Dev Biol. 165, 432-441 (1994).
- Carroll, T., Wallingford, J., Seufert, D., Vize, P. D. Molecular regulation of pronephric development. Curr Top Dev Biol. 44, 67-100 (1999).
- Tonissen, K. F., Drysdale, T. A., Lints, T. J., Harvey, R. P., Krieg, P. A. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 162, 325-328 (1994).
- Davidson, L. A., Ezin, A. M., Keller, R. Embryonic wound healing by apical contraction and ingression in Xenopus laevis. Cell Motil Cytoskeleton. 53, 163-176 (2002).
- Hurtado, R., Mikawa, T. Enhanced sensitivity and stability in two-color in situ hybridization by means of a novel chromagenic substrate combination. Dev Dyn. 235, 2811-2816 (2006).

