The Swimmeret System of Crayfish: A Practical Guide for the Dissection of the Nerve Cord and Extracellular Recordings of the Motor Pattern
Summary
Here we describe the dissection of the crayfish abdominal nerve cord. We also demonstrate an electrophysiological technique to record fictive locomotion from swimmeret motor neurons.
Abstract
Here we demonstrate the dissection of the crayfish abdominal nerve cord. The preparation comprises the last two thoracic ganglia (T4, T5) and the chain of abdominal ganglia (A1 to A6). This chain of ganglia includes the part of the central nervous system (CNS) that drives coordinated locomotion of the pleopods (swimmerets): the swimmeret system. It is known for over five decades that in crayfish each swimmeret is driven by its own independent pattern generating kernel that generates rhythmic alternating activity 1-3. The motor neurons innervating the musculature of each swimmeret comprise two anatomically and functionally distinct populations 4. One is responsible for the retraction (power stroke, PS) of the swimmeret. The other drives the protraction (return stroke, RS) of the swimmeret. Motor neurons of the swimmeret system are able to produce spontaneously a fictive motor pattern, which is identical to the pattern recorded in vivo 1.
The aim of this report is to introduce an interesting and convenient model system for studying rhythm generating networks and coordination of independent microcircuits for students’ practical laboratory courses. The protocol provided includes step-by-step instructions for the dissection of the crayfish’s abdominal nerve cord, pinning of the isolated chain of ganglia, desheathing the ganglia and recording the swimmerets fictive motor pattern extracellularly from the isolated nervous system.
Additionally, we can monitor the activity of swimmeret neurons recorded intracellularly from dendrites. Here we also describe briefly these techniques and provide some examples. Furthermore, the morphology of swimmeret neurons can be assessed using various staining techniques. Here we provide examples of intracellular (by iontophoresis) dye filled neurons and backfills of pools of swimmeret motor neurons. In our lab we use this preparation to study basic functions of fictive locomotion, the effect of sensory feedback on the activity of the CNS, and coordination between microcircuits on a cellular level.
Introduction
The swimmerets of crayfish serve a function in posture control and beat rhythmically when the animals swim forward, ventilate their burrows or females aerate their eggs 5, 6. The swimmerets of the signal crayfish, Pacifastacus leniusculus, occur in pairs from the second to the fifth abdominal segment, with one limb on each side of the abdomen 7. The central nervous system produces on its own the rhythmic motor patter which drives the swimmeret movement in the intact animal as well as in the isolated nerve cord preparation. When there is no sensory feedback or descending input present the rhythmic motor pattern produced is called fictive locomotion 1, 2. In the swimmeret system this motor pattern does not differ in any parameter from the activity of the swimmerets measured in the intact animal.
The movement of each swimmeret is driven by a microcircuit that is located in and restricted to one corresponding hemiganglion 1 – 3. In each microcircuit there is a pattern generating kernel that comprises five identified non spiking interneurons. They can be functionally characterized as being either Inhibitor of Power Stroke (IPS) or Inhibitor of Return Stroke (IRS) 8. These IPS and IRS interneurons are not endogenous oscillators, rather their alternating activity is driven by reciprocal inhibition 9. Because these interneurons inhibit the swimmeret motor neurons directly, the alternating PS-RS movement is generated 10. Locomotion however, does not only require the generation of activity, but also coordination of the different independent microcircuits. In the swimmeret system such coordination is established by the coordinating microcircuit which ensures that limbs are active at correct times. This microcircuit is built by three identified neurons in each segment 11-15.
This protocol provides for the first time a step-by-step dissection guide to isolate the chain of ganglia (T4 to A6, Figure 1). We show how to pin the isolated abdominal nerve cord and desheathe each ganglion. In this isolated nervous system preparation, the neurons responsible for swimmeret movement are ready for use in electrophysiological and morphological experiments. The second part of this protocol demonstrates the main features of the swimmeret motor pattern. This includes a step-by-step guide to extracellularly record the activity of swimmeret motor neurons. Axons of RS motor neurons project through the anterior branch of nerve N1, while axons of PS motor neurons project through the posterior branch of the same nerve (Figure 1) 4. Therefore their activity can be recorded from these branches with differential pin electrodes.
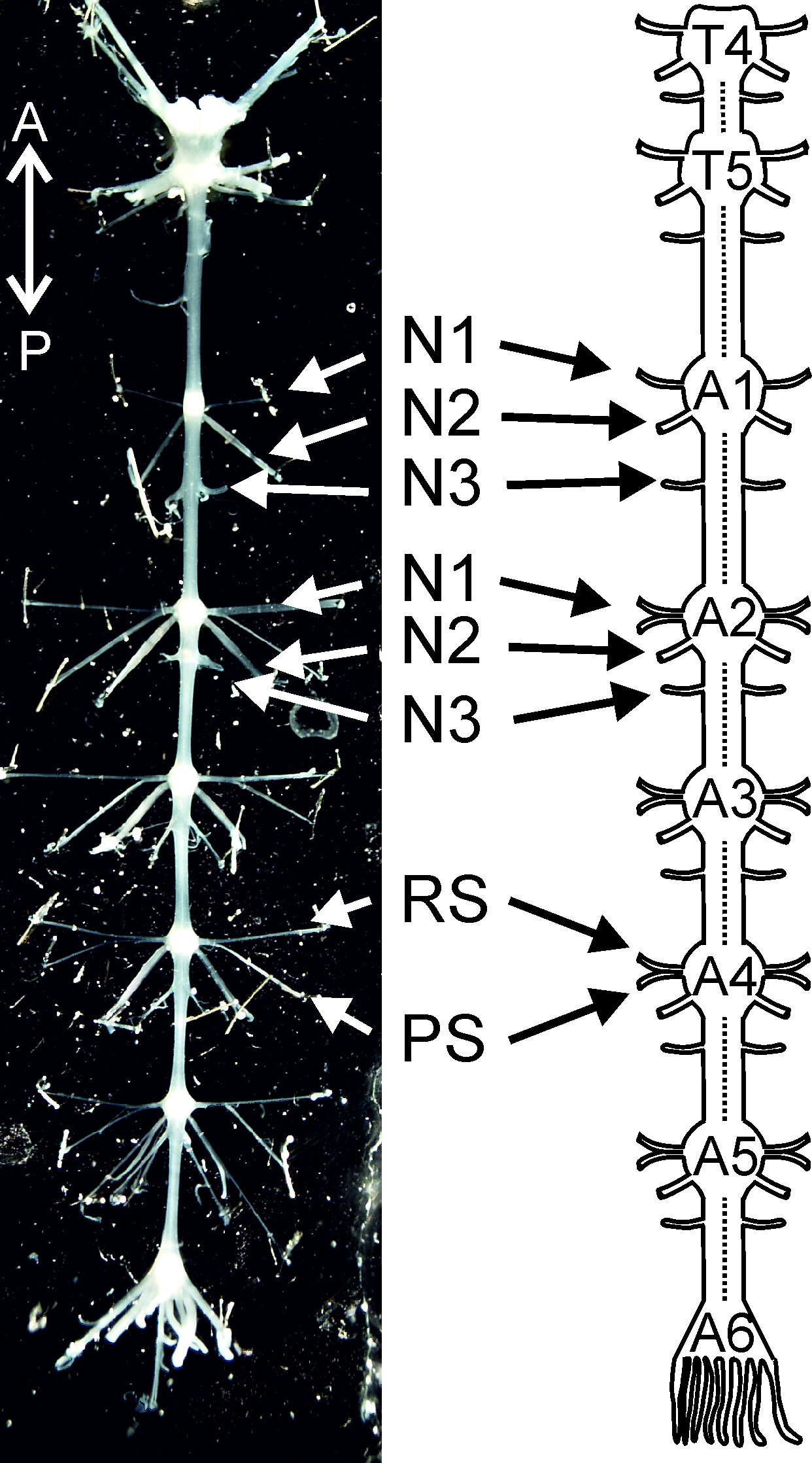
Figure 1: Isolated nervous system from thoracic ganglion 4 (T4) to abdominal ganglion 6 (A6) and a schematic diagram of it. T4: thoracic ganglion 4; T5: thoracic ganglion 5; A1, A2 … A6 abdominal ganglion 1, abdominal ganglion 2 … abdominal ganglion 6; N1: nerve N1; N2: nerve N2; N3: nerve N3; PS: power-stroke; RS: return-stroke. Directional abbreviations: A = anterior; P = posterior.
This dissection procedure and the electrophysiological technique demonstrated are convenient for undergraduate students and may complement student practical courses in physiology. The isolated chain of ganglia has been used in a number of experiments to study nervous system function, coordination, or modulation of swimmeret microcircuits 6 as well as neuronal control of adaptive behavior in locomotion 16, 17. The crayfish swimmeret system thus provides an enormous amount of interesting teaching or training opportunities which all begin with the dissection of the ventral nerve cord of crayfish and extracellular recording of the fictive motor pattern.
Protocol
This dissection procedure is in accordance with the European Communities Council Directive of 22nd September 2010 (2010/63/EU).
1. Preparation
- Obtain crayfish, Pacifastacus leniusculus (Dana), of both sexes ≥8 cm in size. Ensure that the animals are vital and the abdomen and abdominal limbs are intact.
- Take care to inspect the carapace and that this cuticle is hard and rigid. Pre- and postmolt animals have a soft carapace and are not suited for experiments because during the molting process many parameters change (e.g., decrease in locomotor activity).
- Assemble all tools and materials used during the dissection, pinning and desheathing of the nerve cord shown in Figure 2 and listed in the supplements provided.
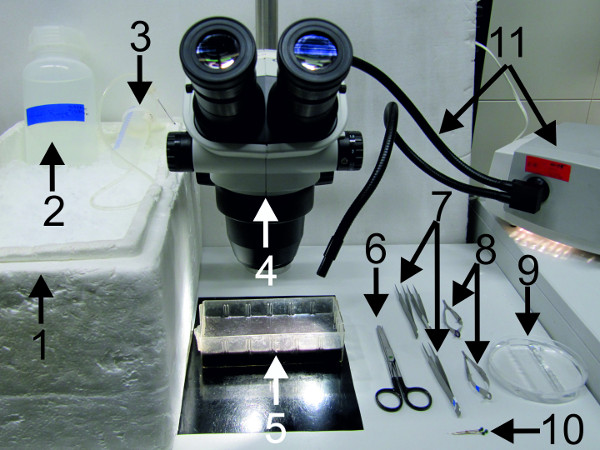
Figure 2: Materials and tools used for the dissection, pinning and desheathing of the nerve cord.
(1) big bucket filled with ice; (2) crayfish saline; (3) saline dispenser; (4) dissection microscope; (5) dissection dish; (6) strong scissors; (7) forceps (8) spring scissors; (9) Petri dish lined with clear sylgard; (10) fixing pins; (11) cold lamp source.
2. Gross Dissection
- Anesthetize animal on ice for 15 – 20 min. Carry out the first part of the gross dissection at a lab bench near the sink since it includes an exsanguination step and the specimen needs to be rinsed regularly with crayfish saline during the procedure.
- Hold animal ventral side up and use strong scissors to cut both claws at their bases near the thorax (Figure 3-1). Remove left and right uropod (Figure 3-2).
- Place the animal ventral side up in the dissection dish lined with black sylgard. Elevate cephalothorax by inserting ice underneath and pin the abdomen at the telson (Figure 4A).
- Fill the saline dispenser with ~60 ml chilled crayfish saline. Perfuse crayfish with cold saline through the claw opening (Figure 4B). Excess saline will drain off through the cuts at the uropods. Cover crayfish with ice during exsanguination
- Decapitate the animal with a single transverse cut just posterior to the animal’s eyes using strong scissors (Figure 5A). Remove all walking legs near the base joints as indicated in Figure 5B.
- Isolate the abdomen with the last thoracic segments from the rest of the cephalothorax. Make a first cut at the level of the second walking legs (thoracic segment 3) by inserting the tip of the scissors into the opening of the second walking leg and cutting to the opposite side. (Figure 6A-1).
- Extend this first cut to both sides through the cephalothorax (Figure 6A-2).
- Flip the animal open to make some of the internal organs visible. Push the prominent digestive gland (Figure 6B-3) to the anterior part of the specimen and use forceps to remove the reproductive organs from the abdominal cavity.
- Remove the anterior part of the cephalothorax (Figure 6C). Use lateral cuts to remove the lateral parts of the carapace, which cover the gills, on both sides of the remaining thorax (Figure 6D-4). Remove the gills and rinse the specimen with cold saline.
- Continue the dissection with a cut through the whole length of the sternal plate as indicated in Figure 6E-5. Make this cut at maximal lateral positions between pleuron and swimmerets (Figure 6E red markings). Proceed on the other side with a same cut. Rinse the specimen with cold saline.
- Carry out the remaining dissection under a dissection microscope. Place the crayfish’s abdomen ventral side up in the dissection dish lined with black sylgard and filled with crayfish saline so that it covers the specimen.
NOTE: The following steps (2.12–4.8) contain directional instructions that apply to right handed experimenters. In the following steps (2.12-6.8) it is important to replace crayfish saline in regular intervals, every 20-30 min with cold saline, to keep the nervous system healthy. - Fix the specimen with insect pins posteriorly at the telson and anteriorly at the remains of the carapace. Place the specimen so that the telson points to the left and is parallel to the table’s edge.
- Use coarse forceps in the left hand to grab through a walking leg opening (Figure 7A) and pull the specimen open (Figure 7B white arrow). Identify the large two dorsal flexor muscle strands (Figure 7B-1 and C-1) and cut their ventral basis as shown in Figure 7B.
- Identify the sternal artery (Figure 7C-2), that descends from dorsal (the heart) to ventral, at the 4th thoracic segment. This artery lies right above the nerve cord (Figure 7C-3), before it projects under the nerve cord, forming the ventral artery.
- Transect the sternal artery. As demonstrated in Figure 7C, lift the artery first, using one blade of the scissors, and only cut when the ventrally located nerve cord is visible.
- Fix the dorsal flexor muscles anteriorly (to the right side). This should be done in maximum stretched position with pins so they will not block vision and the specimen remains stretched. When the dorsal muscle strands are fixed (Figure 8-1), the first abdominal ganglia, A1 and A2, with associated nerves N1, N2, and N3 are visible (Figure 8).
- Use forceps in the left hand to grab the specimen at one opening of the walking legs. Throughout the following dissection steps pull gently to keep the specimen open.
- Transect the nerves N3 at the most distant position from the nerve cord. (Figure 8-3).
- Cut the flexor muscles close to the ventral apodeme as shown in Figure 8-4. Take care not to damage the nerve cord or the nerves N2.
- Repeat the steps 2.18 and 2.19 for the nerves N3 and the flexor muscles of the remaining abdominal ganglia A2 to A5.
- At the last abdominal ganglion, A6, cut the dorsal flexor muscles from the ventral apodeme and the specimen should look as shown in Figure 9.
- Cut the sternal plate posterior to the nerves of A6 (Figure 9-1) and keep the ventral part (Figure 9-2). Discard the dorsal part with flexor muscles (Figure 9-3). Fix the sternal plate anteriorly with pins through the openings of the walking legs, and posteriorly to A6.
3. Fine Dissection
- Place the specimen under the microscope with the anterior part directed away and the posterior part towards the tables’ edge.
- Use forceps as shown in Figure 10A to remove the most anterior parts of the cephalothoracic sterna.
NOTE: The thoracic ganglia and associated nerves are partly covered by the leg musculature and cephalothoracic sterna. The cephalothoracic sterna form a skeleton that separates the laterally located cavities of the walking legs from each other and from the medial cavity in which the ventral nerve cord resides. - Cut the muscles between the remaining exoskeletal structures as indicated in Figure 10B-1 and B2. Use forceps to grab and lift the anterior end of the ventral nerve cord (Figure 10C).
NOTE: The nerve cord will be damaged in the process so avoid picking up the nerve cord multiple times. - Cut the thoracic nerves laterally while lifting the nerve cord (Figure 10C-3). Keep these nerves at suitable length for pinning. Remove the squeezed part of the chain of ganglia, which was picked up with forceps, by cutting away all the tissue anterior to T4 (Figure 10C-4).
- Place the specimen with the anterior part to the left and focus on A1. Cut the nerves N1 and N2 of A1 at a suitable length (max. 1 cm) for pinning them out.
- Focus on A2 and identify the nerves N1, N2, and N3 of this segment (Figure 11). The nerves N1 of abdominal ganglia A2-A5 reside between two sternal cuticular infoldings in each segment (Figure 11A-1) and are covered by musculature. Make one cut along the posterior sternal cuticular infolding. Start at the lateral rim of the abdomen and proceed towards the midline as shown in Figure 11A.
- If the target N1 is still covered with tissue as shown in Figure 11B (red arrow), cut across the muscle bundle, but this time anterior to both sternal cuticular infoldings and the nerve N1 (Figure 11B-2).
- Cut nerve N1 as distally as possible (Figure 11C-3). Nerve N1 is fully visible and the anterior and posterior branch can be identified (Figure 11C).
- Proceed to the contralateral nerve N1 and first cut the muscles along the posterior sternal cuticular infolding, starting medially, near the ganglion (Figure 11D). If the nerve is still covered by tissue, cut across the muscle bundle, but this time anterior to both sternal cuticular infoldings and the nerve N1, similar to Figure 11B-2. Cut the nerve N1 as distally as possible.
- Cut the nerves N2 of this ganglion to a suitable length (approx. 0.5 cm) for pinning.
- Repeat steps 3.7-3.11 for the nerves of A3-A5.
- Cut the nerves of ganglion A6 as distally as possible (Figure 12A). Use forceps to grab multiple nerves of A6 to lift this ganglion and start isolating the chain of ganglia from the sternal plate.
- While lifting the nerve cord, pull it gently in the anterior direction as demonstrated in Figure 12B (white arrow). As the individual ganglia are lifted, remove the ventral artery that may be attached to the ventral side of the nerve cord (Figure 12C). Continue this (gently) pull-cut sequence until the nerve cord is completely isolated.
- Transfer the isolated chain of ganglia to a Petri dish lined with clear sylgard and filled with crayfish saline (Figure 12D).
4. Pinning the Nerve Cord into the Petri Dish
NOTE: Use small pins cut from stainless steel wire (see supplements) to pin the nerve cord. Touch only the nerve ending with the forceps and do not squeeze the connectives or ganglia.
- Pin the chain of ganglia in a straight line, while applying gentle stretch.
- Arrange the nerve cord in the Petri dish with the dorsal side facing upwards (Figure 13, black line). The ventral side of the ganglia can be identified by its convexity; the dorsal side is flat. Pin the thoracic nerves to the sides. Continue with the nerves of A6, stretching the nerve cord along its longitudinal axis.
- Pin out the nerves of A1 at a 90° angle relative to the nerve cord.
- Proceed to A2 and pin the nerves N2 at an angle of 35–45° relative to the nerve cord (Figure 1A).
- Separate the nerves N1 in their anterior and posterior branches before pinning as demonstrated in Figure 14. Use two pairs of fine forceps to pick up with one pair of forceps the anterior and with the other the posterior branch of the nerve N1. Take care to pick only the most distal ends of the nerve branches. Now pull them carefully apart.
- Pin the anterior branch of the nerve N1 at a 90° angle relative to the nerve cord (Figure 1A). Pin the posterior branch of nerve N1 between the anterior N1 branch and the nerve N2.
- Repeat the steps 4.4–4.6 for the nerves of ganglia A3–A5. While fixating the nerve cord stretch it in longitudinal as well as transversal directions.
5. Desheathing the Ganglia
- Place the preparation in such a manner that the experimenter’s hands are always resting on a stable plane to avoid shaking. In order to desheath the ganglia illuminate the nerve cord from below.
- Focus on any abdominal ganglion A1 to A5. Use fine spring scissors to make a small lateral cut through the ganglion sheath, posterior to the ganglion and between the nerves N2 and N3 (Figure 15A red arrow).
- Pick up the ganglion sheath using very fine forceps and cut transversely across the sheath above the connectives, as indicated in Figure 15A-1. Take care not to squeeze or cut the nerve cord with the scissors.
- Still holding and lifting the ganglion sheath with forceps continue to cut it along the lateral borders of the ganglion (Figure 15B-2 and -3). Remove the sheath. Alternatively pin it to both sides of the connectives in such a way that it is fixed but the nerve cord is not squeezed.
- Repeat steps 5.2-5.4 for all abdominal ganglia A1 to A5.
- Desheathe the thoracic ganglia and the ganglion A6 in a similar fashion. In order to desheathe A6 start anterior to the ganglion and proceed in posterior direction. Pin the ganglion sheath of A6 to the posterior end of the chain of ganglia.
6. Extracellular Recordings from Motor Neurons
- Assemble all tools and materials used for extracellular recordings shown in Figure 16B and listed in the supplements. An overview of the recording setup is shown in Figure 16A. Start all electronic equipment used in this experiment (Figure 16C), so that the amplifiers can warm up for at least 30 min prior to recording. Turn on the computer and start the recording software.
- Place the chain of ganglia on the microscope table and illuminate from below. Insert the recording electrode into the sylgard close to the target nerve and the reference electrode at a position nearby, but lateral to the ganglia (Figure 17A and B). Bend the target nerve around the recording electrode (Figure 17C).
- Stretch the nerve slightly, to ensure contact between the electrode and the nerve and pin it to the side (Figure 17D). Fix the electrode cables to the microscope table using modeling clay, so they stay in the desired position.
- Use a syringe filled with petroleum jelly and a 20 gauge needle (with rounded tip) (Figure 17E-3) to isolate the target nerve from the bathing solution. First dab some petroleum jelly on the sylgard around the recording electrode. The result is a layer of petroleum jelly covering the sylgard in the proximity of the recording electrode (Figure 17E-4). Take care not to directly dab onto the nerve and avoid air bubbles in this layer.
- Seal the recording electrode with petroleum jelly from all sides up to the surface level of the saline (Figure 17F).
- Repeat this procedure for all target nerves whose activity should be monitored.
- Start recording. Use a continuous or gap free acquisition mode and a sampling rate of 5 kHz. Set the extracellular amplifier to the following parameters; gain to 1,000 (amplifies the signal 1,000 times, take care to include this amplification parameter in the acquisition software settings) and a bandpass filter range of 300 Hz (low cut) to 2,000 Hz (high cut).
Representative Results
With the simultaneous extracellular recordings from RS and PS, motor neurons of one ganglion, the alternating activity of these motor neuron pools, can be monitored (Figure 18), representing the fictive locomotion pattern.
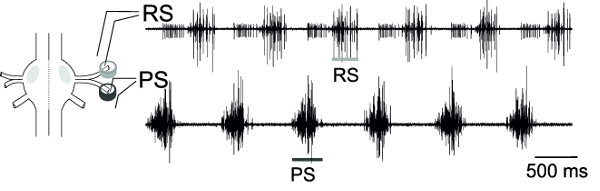
Figure 18: Schematic of one ganglion and placement of differential pin electrode. Extracellular recording of RS motor neurons (upper trace) and PS motor neurons (lower trace).
The extracellular PS activity of ipsilateral PS 2 to PS 5 motor neurons highlights interesting aspects of intersegmental coordination in the swimmeret system (Figure 19A).
First, the activity always starts with a PS burst in A5 (Figure 19A, 4th trace) and the excitation wave propagates in the anterior direction (Figure 19A, green line). Second, the period (which is the time measured from the onset of one burst to the onset of the next burst) in each segment remains constant (Figure 19A, cyan arrow). This is true, as long as the information flow from one ganglion to the next is intact and the experimental conditions do not change.
In different preparations or under different experimental conditions the periods and burst durations can vary substantially: from periods of 0.25 to 1 sec. In order to compare these parameters between different preparations, they need to be normalized and graphed in a phase histogram (Figure 19B). This reveals the third interesting aspect of the motor pattern: the phase-lag between segments, the intersegmental delay, remains constant and independent of the frequency of the rhythm.
To calculate a phase histogram, the following measurements and calculations are necessary:
First, a reference trace has to be chosen. Usually we use the PS trace of the most posterior ganglion (e.g., PS 5) as a reference (being time 0) for a phase histogram of the swimmeret rhythm recorded across several segments (Figure 19A).Then, measure the period (in ms) of the rhythm from the beginning of one PS 5 burst (time 0, Figure 19A, orange dotted line) to the beginning of the next PS 5 burst (Figure 19A, cyan arrow).Within this cycle, measure the latencies (in ms) between the start of the PS 5 burst (time 0) and the start (burst onset, Figure 19A and B, orange arrows) and end (offset Figure 19A and B, violet arrows) of the PS burst in each of the other ganglia. Therefore to calculate the phase onset and offset for each burst the latencies are divided by the concurrent period. These measurements will result in numeric values between 0 and 1. Finally, plot these values in a phase histogram as shown in Figure 19B.
The resulting phase histogram (Figure 19B) contains information about the duty cycle of the PS burst in each segment. And the intersegmental phase-lag of ~0.25 between neighboring segments is clearly visible.
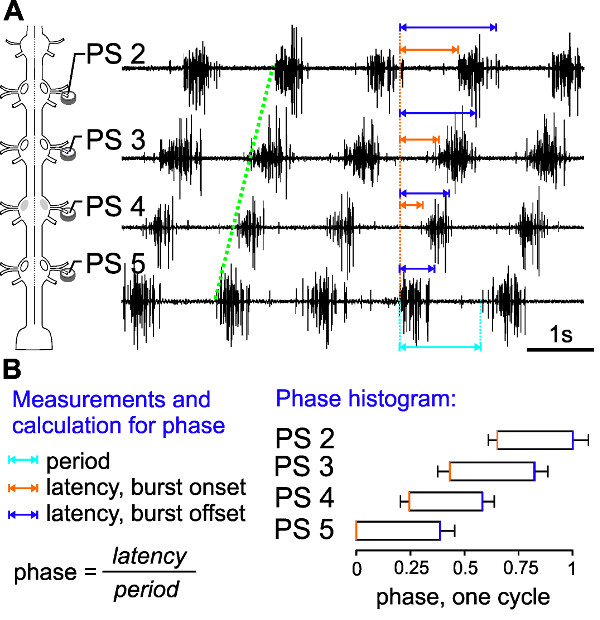
Figure 19: (A) Schematic of electrode placement and extracellular recording of ipsilateral PS 5 to PS 2. (B) Measurements and calculations needed for the phase histogram. The phase histogram calculated from ten consecutive bursts (n = 10) shows the intersegmental phase delay of ~0.25.
For more advanced experimenters, intracellular recordings with sharp microelectrodes from the dendrites of motor neurons can be useful. The membrane potentials of individual intracellularly recorded PS motor neurons show in-phase oscillations with the extracellularly recorded activity of all PS motor neurons of the same hemiganglion (Figure 20, gray panels). Alternatively, RS motor neurons or interneurons can be recorded intracellularly in the same way (data not shown).
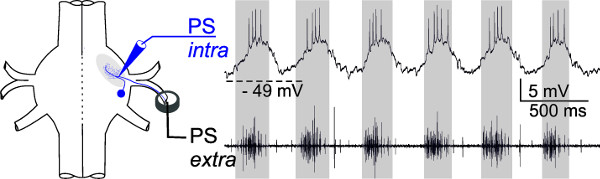
Figure 20: Schematic of electrode placement. The membrane potential of one intracellularly recorded PS motor neuron (upper trace) oscillates in phase (gray panels) with the extracellularly recorded PS activity (lower trace).
For identification of intracellularly recorded neurons (interneurons or motor neurons), cells can be dye filled using sharp microelectrodes and iontophoresis (see discussion). In Figure 21, two intracellularly dye filled PS motor neurons are shown. Their ventral somata are located posterior to the base of the nerve N1. The primary neurite projects anteriorly (Figure 21B-2) and branches within the Lateral Neuropil (Figure 21A and B-3); the axon projects through the posterior branch of the nerve N1 to the swimmeret musculature (Figure 21B-4).
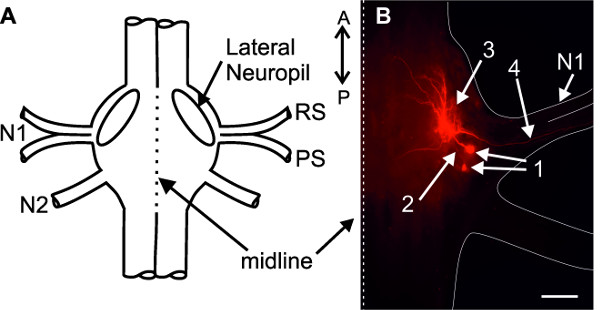
Figure 21: (A) Schematic of an abdominal ganglion. (B) Two intracellularly stained PS motor neurons. (1) somata; (2) primary neurites; (3) dendritic branching in the lateral neuropil; (4) axons project through the posterior branch of the nerve N1; Bar = 100 µm.
For less advanced experimenters, or those who want to study the morphology of the abdominal ganglion, backfills from the PS and RS motor neuron pools via the nerve N1, are an interesting exercise. In Figure 22, the RS and PS motor neurons of one hemiganglion were stained with Co2+ (RS) and Ni2+ions (PS), respectively. Somata of PS motor neurons are located posterior to the base of the nerve N1 (Figure 22-1). All somata of RS motor neurons are located anterior to the base of the nerve N1 (Figure 22-2).
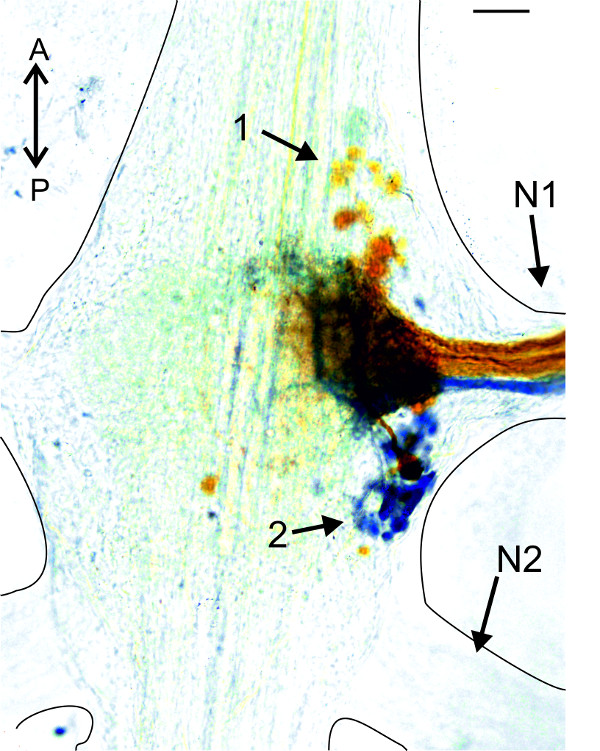
Figure 22: Backfills from the anterior (orange, Co2+) and posterior (blue, Ni2+) branch of nerve N1. (1) Somata of PS motor neurons; (2) somata of RS motor neurons. Bar = 100 µm.
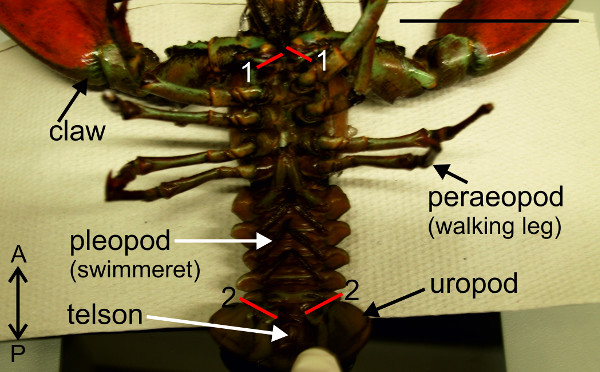
Figure 3: Removal the claws (1) and uropods (2). Red lines (1) and (2) indicate positions of cuts. Bar = 5 cm.
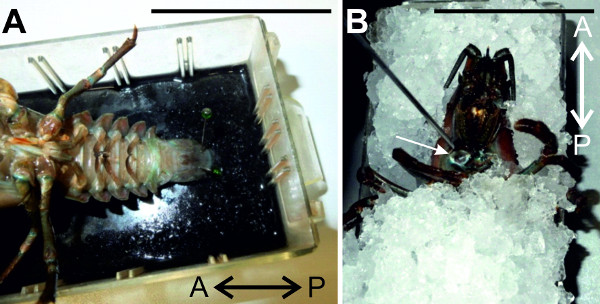
Figure 4: (A) The animal is pinned ventral side up and fixed at the telson. (B) The crayfish is exsanguinated through the claw opening with cold saline. Bars = 5 cm.
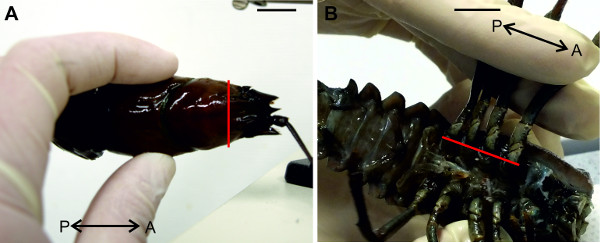
Figure 5: (A) Decapitation and (B) removal of walking legs. Red lines indicate positions of cuts. Bars = 1 cm.
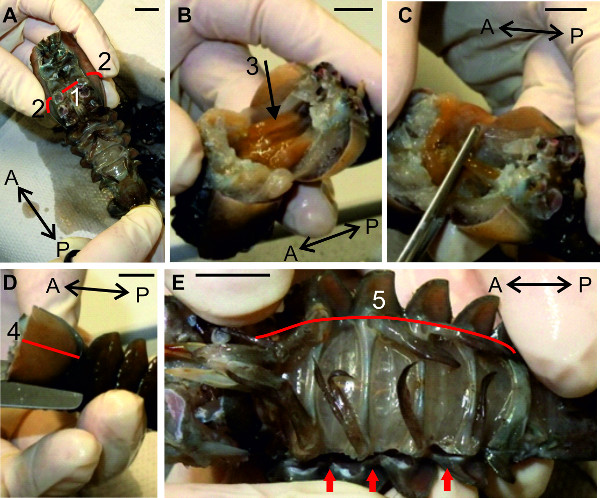
Figure 6: (A, B, C and D) Removal of the anterior and the lateral parts of the cephalothorax. E. Opening of the abdomen along the lateral side. Red lines (1), (2), (4), and (5) indicate positions of cuts. (3) Digestive gland. Red arrows point to the already opened abdominal cavity. Bars = 1 cm.
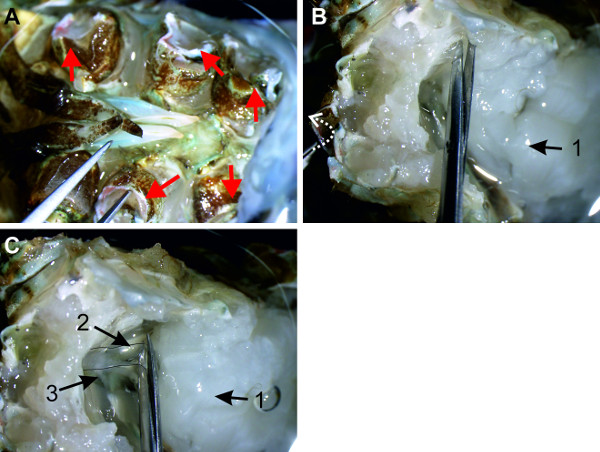
Figure 7: (A) The specimen is grabbed at an opening of the walking legs (red arrows). (B) The dorsal muscle strands (1) are transected while the specimen is opened. White arrow depicts the direction in which the sternal plate is pulled. (C) Overview of the abdominal cavity with the sternal artery (2) and nerve cord (3).
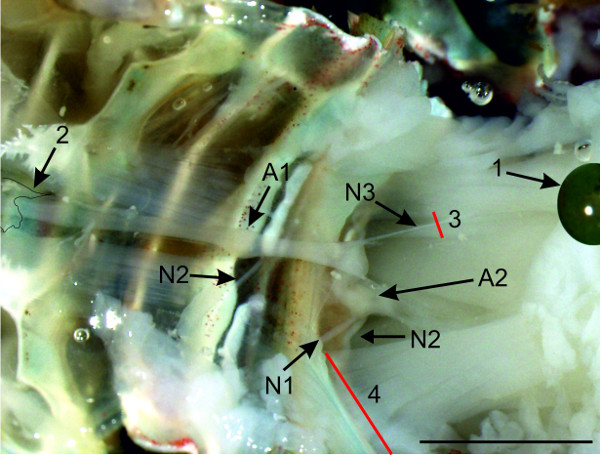
Figure 8: Transverse view of the crayfish abdomen with fixed dorsal muscle strands (1) and sternal artery (2); Red lines (3) and (4) indicate positions of cuts. Bars = 5 mm.
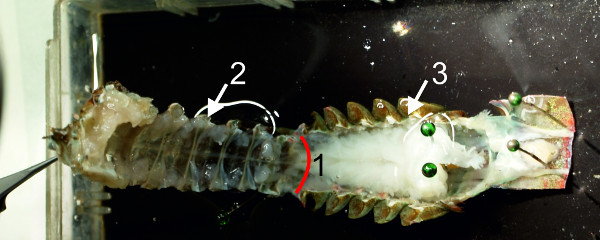
Figure 9: Dorsal and ventral parts of the abdomen are separated from each other. (1) Red line indicates position of cut. Sternal plate (2) with ventral nerve cord; dorsal part of the abdomen (3).
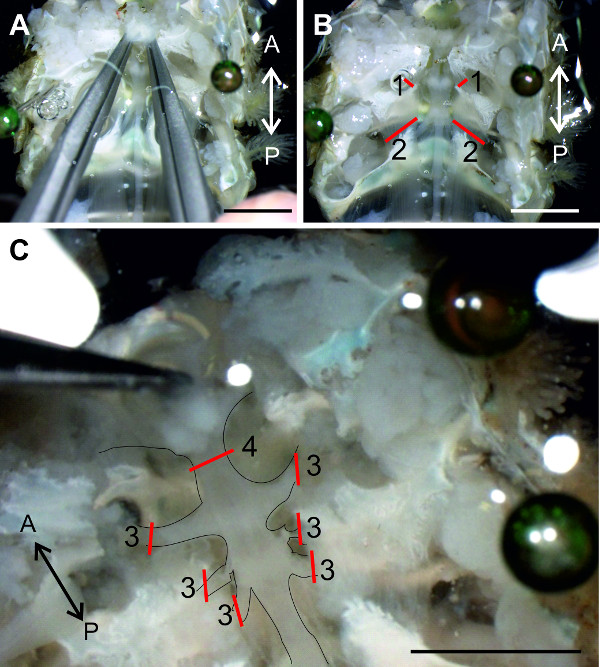
Figure 10: (A) Positions of the forceps to remove the anterior cephalothoracic sterna. (B) Red lines (1) and (2) indicate where to cut the muscles between the remaining cephalothoracic sterna. (C) The thoracic nerves are transected at the red lines (3) to isolate the thoracic ganglia from the surrounding tissue. Red line (4) indicates the cut between T3 and T4. Bars = 5 mm.
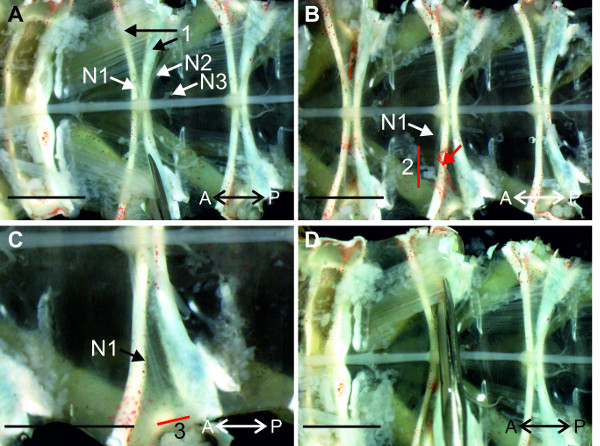
Figure 11: (A and B) View of the sternal plate at A2. The nerve N1 is isolated from the tissue with a cut along the posterior sternal cuticular infolding (1) and anterior to both sternal cuticular infoldings (B 2). (C) Higher magnification of the region around A2, with the isolated nerve N1. (D) Isolation of the nerve N1 on the contralateral side with similar cuts. Red lines (2) and (3) indicate positions of cuts; (1) anterior and posterior sternal cuticular infoldings; Bars = 5 mm.
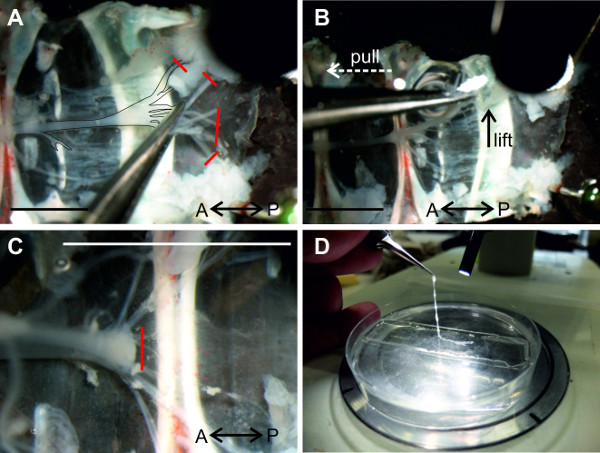
Figure 12: (A and B) A6 is isolated from the tissue and the nerve cord can be lifted at the nerves of A6. (C) All connections of the abdominal ganglia to the ventral abdominal artery are transected. (D) Isolated nerve cord is transferred into a Petri dish lined with clear sylgard and filled with saline. Red lines indicate positions of cuts. Bars = 5 mm.
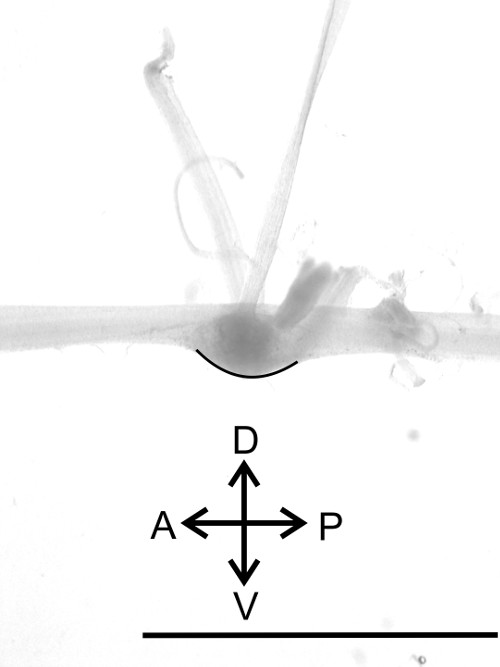
Figure 13: Lateral view of one ganglion. Identification of the dorsal and ventral side (black line) of the nerve cord. Bar = 5 mm
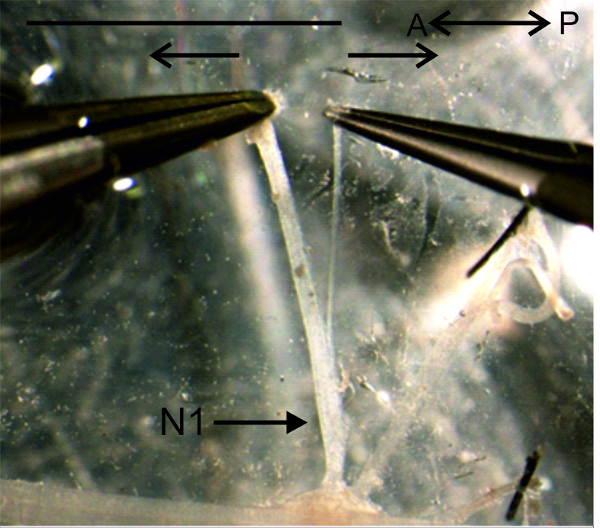
Figure 14: The anterior branch of the nerve N1 is separated from the posterior branch. Bar = 5 mm.
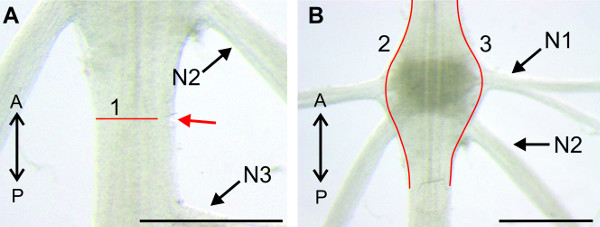
Figure 15: Directions for desheathing of an abdominal ganglion (A) Position of the first small cut through the sheath of the connectives (red arrow), placed lateral and posterior to the ganglion. The subsequent cut through the sheath above the connective is marked by the red line (1). (B) Red lines (1) and (2) mark the cuts along the lateral borders of the ganglion. The sheath can afterwards be pulled from the ganglion. Bars = 1 mm.
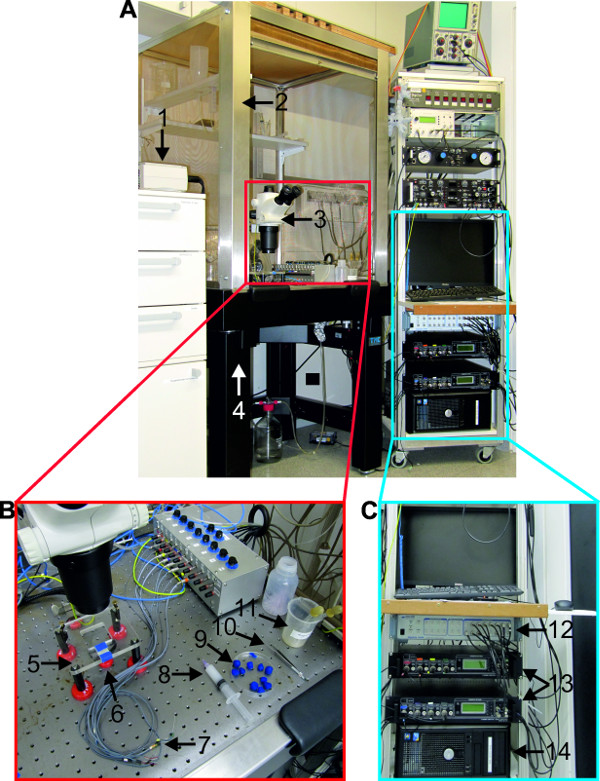
Figure 16: (A) Overview of the recording setup. (B) Materials and tools needed for extracellular recordings. C. Equipment necessary for electrophysiological recording. (1) cold lamp source; (2) faraday cage; (3) stereo microscope; (4) air-table; (5) microscope table; (6) mirror; (7) differential pin electrodes; (8) syringe filled with petroleum jelly; (9) modeling clay; (10) forceps; (11) liquid waste disposal; (12) digitizer; (13) extracellular amplifier; (14) computer, equipped with recording software and monitor.
Figure 17: (A and B) Position of recording (1) and reference (2) pin electrode. (C and D) The posterior branch of the nerve N1 is bend around the recording electrode. E. The base (4) of the recording electrode is isolated with petroleum jelly. F. The complete recording electrode is isolated with petroleum jelly from the bath solution. (3) Syringe filled with petroleum jelly. Bars = 2 mm.
Discussion
The anatomy of crayfish and their abdominal ganglia has been described previously 5, 18, 19, 20 and it is recommended to become familiar with them prior to the dissection in order to avoid cutting of important nerves.
It is critical to keep the preparation at temperatures below 23 °C to prevent degradation of the isolated nerve cord. This can be achieved easily by replacement of the bathing solution every 20–30 min with cold crayfish saline. Under these circumstances the chain of ganglia can be used for electrophysiological experiments for up to 12 hr.
Sometimes swimmeret motor neurons are inactive and show no spontaneous bursting activity. The cholinergic agonist carbachol is used to induce bursting activity in these preparations. Dissolved in cold crayfish saline, bath applications of 1-3 µM carbachol are sufficient to activate the swimmeret rhythm 21.
The fictive locomotion and especially the coordination of the limbs recorded in this system provide a lot of information about the cellular properties of rhythmic activity and coordination. In this system the neuronal components 8-10 of the local microcircuits are identified and additionally the coordinating network is known in cellular detail 11,14,15. The results drawn from the experiments performed with extracellular recordings, allow already predictions for mathematical models and for roboticists.
All swimmeret neurons show dendritic branching within the Lateral Neuropil and can be intracellularly recorded from these dendrites. In Figure 20, an intracellular recording of a PS motor neuron is shown. For this a sharp microelectrode (see supplements) filled with 1 M KAc + 0.1 M KCl (electrode resistance between 35 – 45 MΩ) is placed above the Lateral Neuropil. The oscillating membrane potentials as well spikes from the dendrites of PS and RS motor neurons can be recorded when the electrode is inserted near the base of nerve N1.
To stain individual neurons (e.g., motor neurons) intracellularly as shown in Figure 21, the sharp microelectrode must be additionally filled with fluorescent dye (e.g., 1% dextran Texas Red, see supplements) dissolved in 1 M KAc + 0.1 M KCl. To fill motor neurons intracellularly by iontophoresis, polarizing pulses of 1 nA with a duration of 250 ms at 2 Hz are applied for at least 10 min. For positively charged dyes use depolarizing pulses and for negatively charged dyes use hyperpolarizing pulses. Longer fills result in a stronger labeled neuron. For visualization the nerve cord must be fixed, dehydrated, and mounted 22.
One method to stain a pool of motor neurons is to use backfills from the nerve N1 (Figure 22) 4,23. Place a well of petroleum jelly around the posterior branch of the nerve N1 to backfill the pool of PS motor neurons. To stain the pool of RS motor neurons a well is placed on the anterior branch of the nerve N1. The crayfish saline in the well is replaced by ddH2O. Then cut this nerve inside the well and leave it for 15 min in ddH2O at room temperature (RT). Thereafter, replace the ddH2O in the wells with either 5% CoCl2– or 5% NiCl2– solution (dissolved in ddH2O) and close wells with petroleum jelly. The sample needs to be incubated for at least 15 hr at 4 °C. Open the wells, remove the dye and rinse the sample 3 times with saline. Precipitate the Ni2+or Co2+ ions with 10 drops of Dithiooxamid (saturated solution dissolved in 100% EtOH) for every 10 ml crayfish saline in the Petri-dish, under the hood for 20 min at RT. Ni2+– ions will precipitate to blue Ni(SCH) and Co2+-ions to orange CoS. The nerve cord must be fixed, dehydrated, and mounted in order to acquire images of the labeled cells under a light microscope equipped with a camera.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Jos Burgert for helping with some of the figures. We are grateful to Ingo Selbach (and the group “Edelkrebsprojekt NRW”) for his efforts to supply the lab with experimental animals. We thank Anna C. Schneider for proofreading first versions of the manuscript. This research was supported by an Emmy Noether DFG grant SM 206/3-1 and a startup grant of the University of Cologne for female faculty.
Materials
| Name of Material/ Equipment | Type | Company | Catalog Number | Comments/ Description |
| 4-channel extracellular amplifier: MA 102 | Amplifier | Elektroniklabor, Zoologie, Universität zu Köln, Germany | for extracellular recording | |
| air-table | Technical Manufacturing Corporation (TMC) a unit of AMETEK Ultra Precision Technologies, Peabody, MA, USA |
63-534 | for intracellular recording | |
| Axon Digidata 1440A | Digitizer | Axon Instruments, Molecular Devices Design, Union City, CA | DD1440A | digitizes recorded signals |
| big bucket | filled with ice | |||
| Clampex & Clampfit | pClamp 10, recording and analysis software | Molecular Devices Design, Union City, CA | pClamps 10 Standard | for extracellular recording |
| cold lamp source | with flexible light guide (fiber optic bundle) | Euromex microscopes holland, Arnhem, BD | LE.5211 & LE.5235 | |
| computer and monitor | equipped with recording software | for extracellular recording | ||
| container and pipette for liquid waste | ||||
| crayfish saline | contains (in mM): 5.4 KCl, 2.6 MgCl2, 13.5 CaCl2, and 195 NaCl, buffered with 10mM Tris base and 4.7mM maleic acid; aerated for 3 hours. Adjust at pH of 7.4. | always keep at temperatures ~ 4° C | ||
| dextran, Texas Red (3000MW, lysine fixable) | fluorescent dye, lysine fixable | Life Technologies GmbH, Darmstadt, Germany | D3328 | for intracellular dyefill of neurons |
| differential pin electrodes | made from stainless steel ɸ 0.2 mm | for extracellular recording | ||
| dissection dish | (l x w x h) 15x7x5 cm; linned with black silicone | used in the gross disection | ||
| faraday cage | for extracellular recording | |||
| fixing pins | for pinning the specimen | |||
| forceps (biology, Dumont #5) | Forceps: Biology, tip 0.05 x 0.02 mm, length 11cm, INOX | Fine Science Tools (FST), Germany | 11252-20 | fine forceps: used to pick nerves |
| forceps (biology, Dumont #55) | Forceps: Biology, tip 0.05 x 0.02 mm, length 11cm, INOX | Fine Science Tools (FST), Germany | 11255-20 | extra fine forceps: used for desheathing |
| forceps (electronic, Dumont #5) | Forceps: Standard, tip 0.1 x 0.06 mm, length 11cm, INOX | Fine Science Tools (FST), Germany | 11251-20 | coarse forceps: used to grab specimen and pins |
| intracellular electrode | Borosilicate glass capillaries (outer/inner diameter: 1mm/0.5mm), with filament | Sutter Instruments, Novato, CA | BF100-50-10 | for intracellular recording and dyefill of neurons |
| Leica S8 Apo StereoZoom | Dissection Microscope Zoom 1x – 8x | Leica, Germany | 10446298 | for extracellular recording |
| microscope table | for extracellular recording | |||
| mirror | to illuminate preparation from below | for extracellular recording | ||
| modeling clay | for extracellular recording | |||
| Olympus SZ61 | Dissection Microscope Zoom 0.67x – 4.5x | Olympus, Germany | for the dissection | |
| petri dish | 94 x 16 mm; lined with clear silicone | Greiner bio-one, Germany | 633180 | used to pin the isolated chain of ganglia |
| ring scissors | ThoughCut, cutting edge: sharp/blunt, straight: 13cm | Fine Science Tools (FST), Germany | 14054-13 | for gross dissection (steps 2.1 – 2.11) |
| saline dispenser with a 16 gauge needle (outer ɸ 1.6mm) attached via a flexible tube. | Volume ~ 60ml, | used for exsanguination | ||
| spring scissors or alternative: Vannas spring scissors | cutting edge: 8 mm, tip diameter: 0.2mm, straight: 10cm or cutting edge 2.5 mm, tip diameter 0.075 mm, straight: 8cm | Fine Science Tools (FST), Germany | 15024-10 or 15000-08 | for desheathing |
| stainless steel wire ɸ 0.125 mm | to cut pins of 4-7 mm length | Goodfellow GmbH, Bad Nauheim, Germany | for pinning of the nerve cord | |
| student Vannas spring scissors or alternative: Moria Spring Scissors | cutting edge: 5mm, tip diameter: 0.35mm, straight: 9cm or cutting edge: 5mm, tip diameter 0,1 mm, straight: 8 cm | Fine Science Tools (FST), Germany | 91500-09 or 15396-00 | for gross and fine disection (steps 2.11 – 3.14) |
| sylgard | 184 Silicone Elastomer Base and Curing Agent; for black sylgard add activated carbon | Dow Corning, Midland, MI, USA | ||
| syringe filled with petroleum jelly and equipped with a 20 gauche needle with rounded tip | for extracellular recording |
References
- Hughes, G. M., Wiersma, C. A. G. The Co-Ordination of Swimmeret Movements in the Crayfish, Procambarus-Clarkii (Girard). J Exp Biol. 37 (4), 657-670 (1960).
- Mulloney, B., Smarandache, C. Fifty Years of CPGs: Two Neuroethological Papers that Shaped the Course of Neuroscience. Front Behav Neurosci. 4, 45 (2010).
- Murchison, D., Chrachri, A., Mulloney, B. A Separate Local Pattern-Generating Circuit Controls the Movements of Each Swimmeret in Crayfish. J Neurophys. 70 (6), 2620-2631 (1993).
- Mulloney, B., Hall, W. M. Functional organization of crayfish abdominal ganglia. III. Swimmeret motor neurons. J Comp Neurol. 419 (2), 233-243 (2000).
- Davis, W. J. Lobster Righting Responses and Their Neural Control. Proc R Soc Ser B-Bio. 170 (1021), 435-456 (1968).
- Mulloney, B., Smarandache-Wellmann, C. Neurobiology of the crustacean swimmeret system. Prog Neurobiol. 96 (2), 242-267 (2012).
- Huxley, T. H. . The crayfish: An introduction to the study of zoology. , (1980).
- Smarandache-Wellmann, C., Weller, C., Wright, T. M., Mulloney, B. Five types of nonspiking interneurons in local pattern-generating circuits of the crayfish swimmeret system. J Neurophys. 110 (2), 344-357 (2013).
- Skinner, F. K., Mulloney, B. Intersegmental coordination of limb movements during locomotion: mathematical models predict circuits that drive swimmeret beating. J Neurosci. 18 (10), 3831-3842 (1998).
- Mulloney, B. During fictive locomotion, graded synaptic currents drive bursts of impulses in swimmeret motor neurons. J Neurosci. 23 (13), 5953-5962 (2003).
- Smarandache-Wellmann, C., Grätsch, S. Mechanisms of coordination in distributed neural circuits: Encoding coordinating information. J Neurosci. 34 (16), 5627-5639 (2014).
- Mulloney, B., Hall, W. M. Local commissural interneurons integrate information from intersegmental coordinating interneurons. J Comp Neurol. 466 (3), 366-376 (2003).
- Mulloney, B., Harness, P. I., Hall, W. M. Bursts of information: Coordinating interneurons encode multiple parameters of a periodic motor pattern. J Neurophys. 95 (2), 850-861 (2006).
- Smarandache, C., Hall, W. M., Mulloney, B. Coordination of Rhythmic Motor Activity by Gradients of Synaptic Strength in a Neural Circuit That Couples Modular Neural Oscillators. J Neurosci. 29 (29), 9351-9360 (2009).
- Smarandache-Wellmann, C., Weller, C., Mulloney, B. Mechanisms of Coordination in Distributed Neural Circuits: Decoding and Integration of Coordinating Information. J Neurosci. 34 (3), 793-803 (2014).
- Chrachri, A., Neil, D., Mulloney, B. State-Dependent Responses of 2 Motor Systems in the Crayfish, Pacifastacus leniusculus. J Comp Physiol A. 175 (3), 371-380 (1994).
- Chrachri, A., Neil, D. M. Interaction and Synchronization between 2 Abdominal Motor Systems in Crayfish. J Neurophys. 69 (5), 1373-1383 (1993).
- Skinner, K. The Structure of the 4th Abdominal-Ganglion of the Crayfish, Procambarus-Clarki (Girard) II. Synaptic Neuropils. J Comp Neurol. 234 (2), 182-191 (1985).
- Skinner, K. The Structure of the 4th Abdominal-Ganglion of the Crayfish, Procambarus-Clarki (Girard) I. Tracts in the Ganglionic Core. J Comp Neurol. 234 (2), 168-181 (1985).
- Mulloney, B., Tschuluun, N., Hall, W. M. Architectonics of crayfish ganglia. Microsc Res Techniq. 60 (3), 253-265 (2003).
- Braun, G., Mulloney, B. Cholinergic modulation of the swimmeret motor system in crayfish. J Neurophys. 70 (6), 2391-2398 (1993).
- Davis, W. J. Motoneuron Morphology and Synaptic Contacts – Determination by Intracellular Dye Injection. Science. 168 (3937), 1358-1360 (1970).
- Altman, J. S., Tyrer, N. M., Strausfeld, N. J., Miller, T. A. Filling Selected Neurons with Cobalt through Cut Axons. Neuroanatomical Techniques. , 373-402 (1980).

