Studying Microbial Communities In Vivo: A Model of Host-mediated Interaction Between Candida Albicans and Pseudomonas Aeruginosa in the Airways
Summary
While in vitro study of host-pathogen interactions allow the characterization of specific immune responses, in vivo models are required to observe the effects of complex responses. Using Candida albicans exposure followed by Pseudomonas aeruginosa-mediated lung infection, we established a murine model of microbial interactions involved in ventilator-associated pneumonia pathogenicity.
Abstract
Studying host-pathogen interaction enables us to understand the underlying mechanisms of the pathogenicity during microbial infection. The prognosis of the host depends on the involvement of an adapted immune response against the pathogen1. Immune response is complex and results from interaction of the pathogens and several immune or non-immune cellular types2. In vitro studies cannot characterise these interactions and focus on cell-pathogen interactions. Moreover, in the airway3, particularly in patients with suppurative chronic lung disease or in mechanically ventilated patients, polymicrobial communities are present and complicate host-pathogen interaction. Pseudomonas aeruginosa and Candida albicans are both problem pathogens4, frequently isolated from tracheobronchial samples, and associated to severe infections, especially in intensive care unit5. Microbial interactions have been reported between these pathogens in vitro but the clinical impact of these interactions remains unclear6. To study the interactions between C. albicans and P. aeruginosa, a murine model of C. albicans airways colonization, followed by a P. aeruginosa-mediated acute lung infection was performed.
Introduction
Animal models, especially mice, have been extensively used to explore immune responses against pathogens. Although innate and acquired immunity differ between rodents and humans7, the ease in breeding and the development of knockouts for numerous genes, make mice an excellent model to study immune responses8. The immune response is complex and results from the interaction of a pathogen, the resident microbial flora and several immune (lymphocytes, neutrophils, macrophages) and non-immune (epithelial cells, endothelial cells) cellular types2. In vitro studies do not allow observing these complex interactions and mainly focus on unique cell-pathogen interactions. While animal models must be used with caution and limited to very specific and relevant questions, mouse models provide a good insight into the mammal immune response in vivo and may address parts of important clinical questions7.
In the airways, the microbial community is complex associating a large number of different microorganisms6. While what constitutes a "normal" airway microbiome remains to be determined, resident communities are frequently polymicrobial, and originate from diverse ecological sources. Patients with suppurative chronic lung disease (cystic fibrosis, bronchectasis) or mechanically ventilated patients exhibit a particular flora due to colonization of the airways by environmentally-acquired microorganisms9. Pseudomonas aeruginosa and Candida albicans are both problem pathogens5, frequently isolated together from tracheobronchial samples, and responsible of severe opportunistic infection in these patients, especially in the intensive care unit (ICU)4.
Isolation of these microorganisms during acute pneumonia in ICU results in anti-microbial treatment against P. aeruginosa but yeast are usually not considered pathogenic at this site5. In vitro interactions between P. aeruginosa and C. albicans have been widely reported and showed that these microorganisms can affect the growth and the survival of each other but studies could not conclude if the presence of C. albicans is detrimental or beneficial for the host10. Mouse models were developed to address this relevance of P. aeruginosa and C. albicans in vivo, but the interaction between microorganisms was not the key point. Indeed, the model was established to evaluate the involvement of C. albicans in host immune response, and outcome.
A previous model established by Roux et al already used an initial colonization with C. albicans followed by an acute lung infection induced by P. aeruginosa. Using their model, the authors found a deleterious role of prior C. albicans colonization11. However Roux et al used a high load of C. albicans in their model with 2 x 106 CFU/mouse during 3 consecutive days. We established a 4-day model of C. albicans airway colonization, or at least persistence without lung injury, In this model C. albicans was retrieved up to 4 days after a single instillation of 105 CFU per mouse (Figure 2B) 12,13. After 4 days, no evidence of inflammatory cell recruitment, inflammatory cytokine production nor epithelial damage was observed. At 24 – 48 hr, at the peak presence of C. albicans, even though a cellular and cytokine innate immune response was observed, there was no evidence of lung injury. Surprisingly, mice thus colonized with C. albicans 48 hr prior to intranasal instillation of P. aeruginosa had attenuated infection compared to mice with P. aeruginosa infection alone. Indeed, mice exhibited lesser lung injury and decreased bacterial burden12,13.
Several hypotheses could explain this beneficial effect of prior colonization with C. albicans on P. aeruginosa-mediated acute lung infection. First, an interspecies cross-talk involving each microorganisms quorum-sensing systems, the homoserinelactone-based P. aeruginosa system and the farnesol-based C. albicans system, were evaluated. Second, C. albicans acting as a "decoy" target for P. aeruginosa diverting the pathogen from lung epithelial cells was studied. Both hypotheses were invalidated (unpublished data). The third hypothesis was that of a "priming" of the innate immune system by C. albicans responsible for an enhanced subsequent innate response against P. aeruginosa. This last hypothesis was confirmed. Indeed C. albicans colonization led to a priming of innate immunity through IL-22, mainly secreted by innate lymphoid cells, resulting in increased bacterial clearance and reduced lung injury12.
In conclusion, the host is a central actor in the interaction between microorganisms modulating the innate immune response and involving different inflammatory cell types. While these complex immune interactions can be dissected in vitro the initial hypotheses can only be provided by appropriate in vivo models. The following protocol provides an example of in vivo study of host-mediated pathogen interaction that may be adapted to others microorganisms.
Protocol
The regional ethics regional committee for animal experiments has approved this method, in accordance with national and international animal care and use in investigational research guidelines.
1. Sample Collection
- Sample storage
- Collect all samples and immediately store at – 20 °C or on ice until freezer storage to avoid deterioration. Place sterile phosphate buffered saline (PBS) on ice to improve broncho-alveolar lavages (BAL) performance.
- Surgery
- Sterilize all surgical equipment using an autoclave.
NOTE: If possible, it is recommended to use two different sets of instruments for abdominal and thoracic steps to avoid cross-contamination. Required dissection equipment is detailed in Figure 4A
- Sterilize all surgical equipment using an autoclave.
2. Mice, Bacterial and Yeast Strains
- House mice in compliance with local use of animals in research committee guidelines in a ventilated rack without exceeding 5 mice per cage, with food and water ad lib, in a biosafety level 2 housing facility due to the use of biosafety level 2 micro-organisms : P. aeruginosa and C. albicans.
- Keep the bacterial strains at -80 °C in 40% glycerol medium.
- Add bacteria directly from frozen stock into culture tube containing 3 ml of sterile Luria-Bertani broth using a 10 µl inoculation loop. Leave O/N at 37 °C with orbital shaking (400 rpm) the day before instillation.
- Harvest bacteria by centrifugation at 2,000 x g for 5 min.
- Aspirate the supernatant into an appropriate closed biohazard waste disposal. Observe white adherent pellet at the bottom of culture tube.
- Wash and suspend the pellet using 5 ml of PBS.
- Repeat steps 2.2.2 to 2.2.3 a second time to perform a second wash.
- Resuspend the pelleted bacteria using 1 ml of PBS and brief vortexing.
- Determine the inoculum density using optical densitometer at 600 nm using an Optical Density Meter. A density of 0.9 corresponds to 109 CFU/ml for PAO1, dilute accordingly.
NOTE: This result has to be obtained for each used strain by determining density of successive dilutions of a calibrated inoculum. - Verify the inoculum by serial logarithmic dilutions and plate 100 µl of each dilution on bromocresol purple agar (BCP) plates and O/N culture. Administer each mouse intranasally 50 µl of the solution containing 1 x 108 to 2 x108 CFU per ml (5 x 106 to 1 x107 CFU per mouse).
- Use C. albicans SC5314 as a reference strain. Conserve the strain in 40% glycerol medium at -80°C.
- Supplement Yeast-Peptone-Dextrose broth with 0.015% amikacin to avoid bacterial contamination and facilitate further count.
- Add yeast using 10 µl inoculation loop into prepared YPD-broth supplemented with amikacin O/N at 37°C.
- Harvest yeast by centrifugation at 2,000 x g for 5 min.
- Remove supernatant into an appropriate bioharzard waste disposal. White adherent pellet should be observed in the bottom of culture tube.
- Wash and suspend the pelleted bacteria using 5 ml of PBS and brief vortexing.
- Repeat steps 2.2.2 to 2.2.3 a second time to perform a second wash.
- Resuspend the pellet using 1 ml of PBS and brief vortexing.
- Determine the size of the inoculum by counting on a Mallassez hematocytometer using a standard microscope at 40x magnification.
NOTE: Concentration (in CFU/ml) is obtained using the following formula: (number of yeast x 105)/(number of counted grid rectangles on the Mallassez hematocytometer). - Verify by serial logarithmic dilution to 10-5 and 10-6 to confirm that solution contains 2 x 106 CFU/ml.
- Plate on YPD agar plates supplemented with 0.015% amikacin.
3. Airways Colonization by C. albicans
NOTE: After environmental adaptation, mice are weighed twice a day.
- Under a fume hood, deposit 500 µl of Sevoflurane onto a 4 cm x 4 cm gauze (open-drop technique)14.
- Immediately place the gauze on the floor of an approximately 750 ml induction chamber. Immediately place a mesh-raised platform above the gauze to avoid direct contact between the animal and the gauze.
- Close airtight lid and wait 1 to 2 min to allow diffusion of Sevoflurane in the chamber.
- Transfer the mouse from cage to mesh platform and close lid. Light anesthesia with conserved spontaneous breathing should be achieved in 30 to 45 sec.
- Monitor for hypotonia by observing loss of righting reflex at which point the mouse can be removed from the box and instilled.
- Intranasal instillation (Can be performed in 10 sec by a trained operator).
- Hold the mouse one-handed nestled on its back held upright (Figure 4B).
- Using index finger, support the head and use the thumb to keep the jaw closed to avoid expectoration (Figure 4C).
- As described at step 2.3.8, ensure that the prepared C. albicans solution contains 2 x106 CFU per ml for an 50 µl instilled volume.
NOTE: The second critical point is the instilled volume. A volume lower than 50 µl could result in an insufficient colonization or inhomogeneous instillation of airway, a larger volume may induce drowning/suffocation and death. - Instil the mouse intra-nasally by approaching the pipette to the nostrils.
- Pipette a 50 µl drop forming a bubble containing the solution on the nostrils, subsequently inhaled by the spontaneously breathing mouse.
- Place mouse in a recovery area (e.g., a large well-aerated bare cage with an overhead heating lamp). Mice must be monitored until complete awakening. Do not leave an animal unattended until it has regained sufficient consciousness to maintain sternal recumbency and righting reflex. At this point, the mouse can be returned to a normal housing cage.
4. P. aeruginosa-induced Acute Lung Infection
NOTE: Mice are weighed during the four following days. Normally, mice gain weight during C. albicans-mediated airway colonization (Figure 2A).
- Prepare the suspension containing P. aeruginosa the day of the instillation after O/N growth (section 2.2).
- Anesthetize briefly using inhaled Sevoflurane as described above (section 3.1).
NOTE: To perform acute lung infection, recommended bacterial burden are suggested in Table 1. - Instil the mouse as described above (section 3.2) with particular attention to post-instillation recovery.
5. Measure of Lung Injury Index
- Prepare a solution containing FITC-labeled albumin. Inject this solution 2 hr before animal euthanasia.
- Weigh 0.2 mg of albumin-FITC with the appropriate equipment.
- Add 0.2 mg into 1 ml PBS. Briefly vortex. If not used immediately, place the solution in foil to avoid exposure to ambient light.
- Inject intra-peritoneally 200 µl of FITC-labeled albumin solution to each mouse.
- Euthanasia
- Weigh the mice for the last weight data.
- Euthanize a single mouse in accordance with local use of animals in research committee guidelines using one intra-peritoneal injection of a lethal overdose of pentobarbital : 300 µl of 5.47% pentobarbital.
- Remove mouse from the cage and receives lethal injection by operator.
- Following injection, transfer the mouse alone to another cage, hidden from any other animals. Observe the mouse until absence of movement. Confirm death is by absence of movement, particularly respiratory movement, lack of pulse.
- Perform surgical collection of samples on dead animals, therefore without anesthetics nor analgesics.
- Surgical sample collection: Thoracic stage.
NOTE: To maintain sterile conditions, all surgery is performed using sterile equipment in a biosafety level 2 environment.- Apply ethanol to the skin. Perform a midline skin incision from sternum to mid abdomen with scissors. From midline incise along the ribcage on either side. Fold back the skin on either side of the thorax to visualize the rib cage.
- Perform a vertical incision of the ribcage on either side going up towards the clavicles in order to be able to recline the entire anterior chest wall with the sternum allowing the perfect visualization of heart and lungs (Figure 5A, 5B).
- Collect blood using a pre-heparined syringe by puncturing the heart next to the interventricular artery. Withdraw a minimum of 500 µl to obtain at least 100 µl of plasma. Place the blood sample on ice.
- Perform a midline cervical incision to visualize the trachea (Figure 5B and 5C). Carefully dissect the fascia around the trachea. Place a suture behind the trachea (Figure 5C and 5D). Subsequently the suture will be closed around the cannulating needle to ensure proper lavage.
- Catheterize the trachea using the 20-G modified gavage needle (Figure 5D and 4A). Tie a surgical knot around the cannulated trachea with the previously placed suture.
- To perform bronchoalveolar lavages (BAL), gently and progressively inject and draw 500 µl of ice-cold PBS into/from the lung. Place the sample on ice to avoid cellular lysis.
- Repeat step 5.3.6, 3 times to obtain a total of 1,500 µl BAL fluid and pool lavage samples into a 2 ml centrifuge tube (Figure 5E).
- Remove the lungs from the chest. Place a lung segment (size should correspond to the half of one lung or a lobe) into 1.5 ml centrifuge tube and store rapidly at – 80°C.
- Place a lung segment in a pre-weighed hemolysis tube containing PBS to determine bacterial burden and place it on ice.
- Surgical sample collection: abdominal stage.
- Perform another incision on the left-side of the abdomen. Observe the spleen through the peritoneum.
- Remove the spleen and place into a second hemolysis tube containing 1 ml PBS and place on ice.
- Lung injury index
NOTE: Alveolar-capillary membrane permeability is assessed by measuring FITC-labeled-albumin leakage from the vascular compartment to the alveolar-interstitial compartment.- Centrifuge blood sample and BAL fluid for 10 min at 1,500 x g. Collect the supernatants into new centrifuge tubes. The pellets correspond to recruited plasma or BAL cells and should be placed on ice.
- Add 100 µl of each blood supernatant (plasma, yellow) or BAL supernatant to a 96-well transparent plate (300 µl wells). Place a foil on the plate if not used immediately.
- Measure fluorescence levels in plasma and BAL supernatants using a fluorescence microplate reader (excitation, 487 nm; emission, 520 nm).
- Determine the lung injury index by calculating the fluorescence ratio [(BAL supernatant/blood supernatant) x 100].
- Bronchoalveolar lavage (BAL) differential cell count.
NOTE: Use the cell pellet obtained from centrifugation of BAL fluid at step 5.5.1.- If needed, use red-cell lysis buffer. Add 500 µl of red-cell lysis buffer into the centrifuge tube containing the cell pellet. Briefly vortex and leave 10 min on ice. Add 500 µl PBS to stop red-cell lysis.
- Harvest cells by centrifugation for 10 min at 1,500 x g. Remove supernatant and suspend the cell pellet into 1 ml of sterile PBS. Enumerate cells on a Mallassez hematocytometer. Using a Hemacytometer to Count Cells. Concentrate cells on a slide with a cytospin.
- Stain cells using coloration kit allowing cell identification and count (macrophages, lymphocytes, neutrophils).
- Lung bacterial burden and bacterial dissemination
NOTE: To assess lung bacterial burden and bacterial dissemination, lungs and spleen were respectively collected and stored into pre-weighed hemolysis tubes containing 1 ml of PBS (step 5.3.9).- Weigh hemolysis tubes containing 1 ml PBS and either lung or spleen. Homogenize the samples with a tissue homogenizer to obtain lung homogenates and spleen homogenates.
- Deposit 100 µl of tissue homogenates into centrifuge tubes containing 900 µl of sterile PBS to obtain serial logarithmic dilutions.
- Plate the two last appropriate diluted samples (10-3 and 10-2) on either BCP-agar for P. aeruginosa or YPD-amikacin-supplemented agar for C. albicans lung and spleen burden determination.
- Incubate the plates O/N at 37 °C. The following day, enumerate the colonies on plates.
- Index the result to lung weight to obtain a CFU per gram of lung. Lung sample sizes are not the same, the results should be expressed in CFU per gram of lung.
Formula for the index is: [CFU] x [weight of hemolysis tube and lung] – [weight of hemolysis tube].
Representative Results
As seen previously during the protocol description, the experiment needs 5 day to complete (Figure 1: experiment timeline). One operator is solicited during the entire run of the experiment and can handle the processes up to a maximum of 10 mice. If more animals are required, two persons are needed particularly for surgical sample collection. Indeed all samples must be collected in under 2 hr to avoid an increased passive alveolar-capillary leakage of FITC-labeled albumin in the last mice.
The first step is the preparation of C. albicans inoculum and intra-nasal instillation to obtain the airway colonization by C. albicans. A 4 days-persistence model is obtained by intranasal instillation of 5 x 105 CFU of C. albicans per mouse (Figure 2B). During these 4 days, mice gain weight (Figure 2A) and instillation of 5×105 CFU does not induce lung injury (Figure 2C). Although C. albicans may persist up to 4 days in this model, load decreases after 48 hr. Therefore, P. aeruginosa-induced acute lung infection is perfomed at 48 hr of C. albicans persistence.
P. aeruginosa strain PAO1 is a largely characterized laboratory strain comprising the major virulence factor, the type three-secretion system (T3SS), as in 75% of clinical lung isolates 15. For theses reasons, PAO1 is a relevant strain in animal models of acute lung infection. Lung injury is assessed through alveolo-capillar permeability measured by protein leak from the vascular compartment into the airway expressed as the lung injury index. Lung injury increases with the inoculum (Figure 3A). Here we report kinetics of the acute lung injury component of our model induced by PAO1 strain (5×106 CFU/mouse) (Figure 3B-3F) alone without prior C. albicans-mediated priming. Depending on strain and time course of the model, the choice of initial P. aeruginosa inoculum is discussed in the next section and is suggested in Table 1.
Five mice per group were used. Lung injury index (Figure 3B), bacterial burden in the lung (Figure 3C), bacterial burden in the spleen, reflecting bacterial dissemination (Figure 3D), BAL cellularity (Figure 3E) and differential cell count (Figure 3F) were determined every 12 hr. Lung injury was maximal between 24 hr and 36 hr after infection (Figure 3B). Bacterial burden showed a 1-log CFU/ml decrease every 24 hr (Figure 3C). Cumulative bacterial dissemination assessed by spleen homogenate cultures increased each day (Figure 3D). Finally, While BAL cellularity in uninfected mice is mainly composed (90%) of alveolar macrophages, in BAL from infected mice, neutrophils were widely recruited and differential cell count showed 90% neutrophils and 10% macrophages and lymphocytes (Figure 3E, 3F).
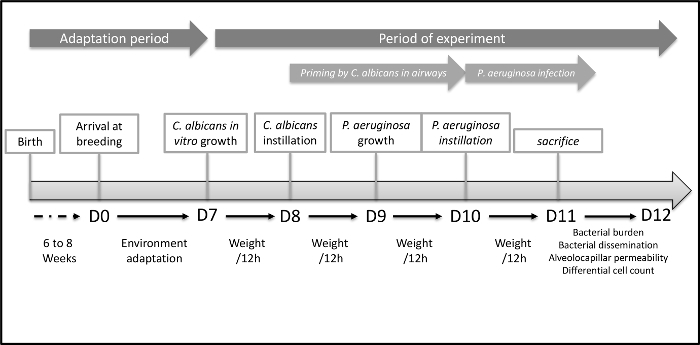
Figure 1. Timeline of the Acute Lung Injury Model to Explore Host-mediated Interaction between C. albicans and P. aeruginosa.
Graphic representation of the entire procedure. The first step is environmental adaptation of mice the housing facility. The Second step is C. albicans mediated airway colonization. Finally, the third step is the acute lung infection mediated by P. aeruginosa. Please click here to view a larger version of this figure.
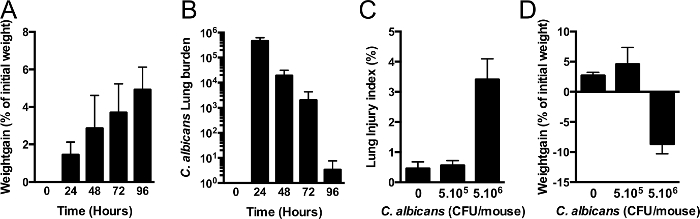
Figure 2. C. albicans airway Colonization.
(A,B) Mice are intranasally instilled with 105 CFU C. albicans (strain SC5314). Mice gain weight during C. albicans-mediated airway colonization (A). Colonization of the airway can be prolonged to 3-4 days with only one initial instillation. In a previous study, priming of innate immunity takes place between 24 and 48 hr. (n = 5 per group), error bars represent means ± SD. (C,D) mice are intranasally instilled 5 x 105 or 5 x 106 CFU of C. albicans. Lung injury index (C) assessed by alveolar capillary barrier permeability at 24 hr. Weight gain (D) expressed as percent of initial weight (n=5 per group). Error bars represent means ± SD. Please click here to view a larger version of this figure.
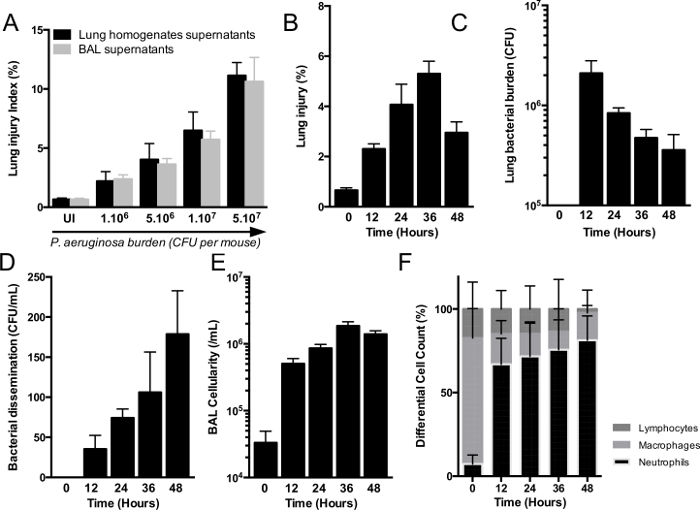
Figure 3. Model of Acute Lung Injury Induced by P aeruginosa.
(A) C57Bl/6J mice are intra-nasally infected with increasing loads of P. aeruginosa (from 1 x 106 to 5 x 107 CFU per mouse) (n=5 per group), error bars represent means ± SD. Mice are euthanized at 24 hr. Lung injury index is assessed by alveolar-capillary barrier permeability that increases proportionally with bacterial burden. Comparison of lung injury index obtained using old method with lung homogenates supernatants (black bars) and new combined-method using bronchoalveolar lavage supernatants (grey bars) (B-F) mice are intra-nasally infected with 5 x106 CFU per mouse. Mice are euthanized every 12 to 48 hr to acute injury model kinetics. Lung injury (B), lung bacterial burden (C), spleen bacterial burden (D), bronchoalveolar lavage (BAL) cellularity (E) and BAL differential cell count (F) are also assessed. (n=5) per group, error bars represent means ± SD. Please click here to view a larger version of this figure.
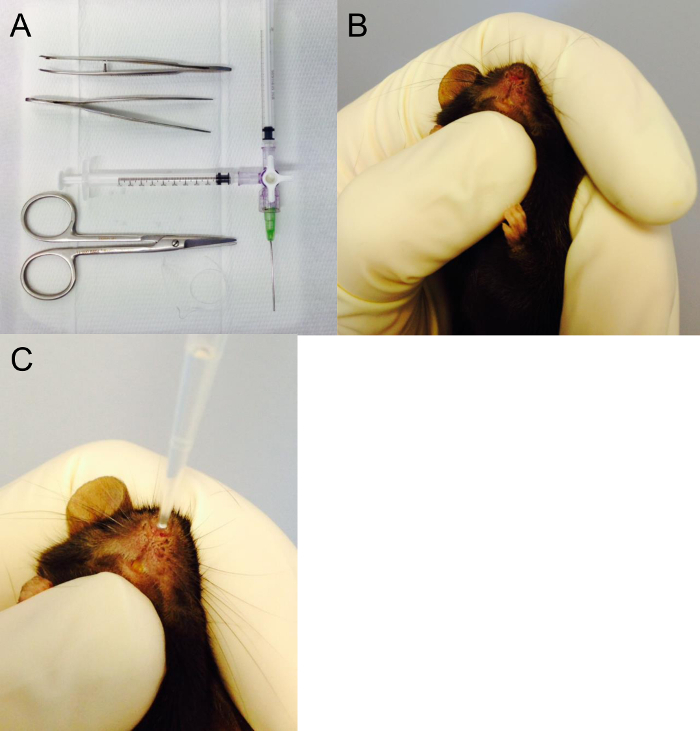
Figure 4. Surgical Equipment and Intranasal Instillation.
(A) Surgical equipment required to perform acute injury model and bronchoalveolar lavage. Here Tracheal cannula (20 G) and the two 1 ml syringes are connected to a Luer-lock 3-way valve. One syringe to inject water into the lungs, one to draw the bronchoalveolar fluid back out from the lungs. (B,C) Position of the mouse in the hand to perform the intra-nasal instillation. In this photo, the thumb under the jaw ensures a closed mouth during instillation. Please click here to view a larger version of this figure.
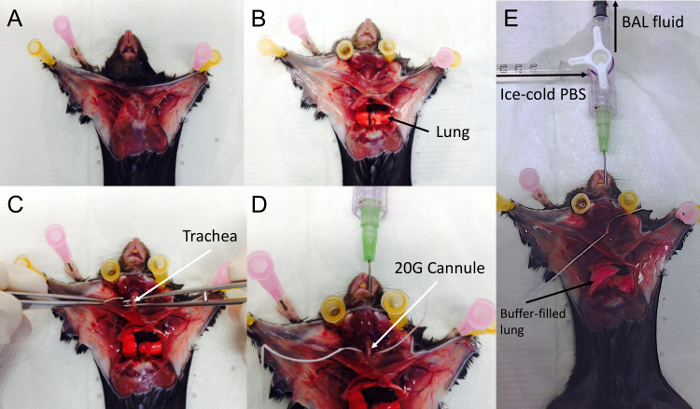
Figure 5. Surgery and Broncho-alveolar Lavage.
The chest is widely opened (A), and rib cage is opened laterally to avoid injury to the heart (B). Following blood collection, the cervical area is dissected to expose the trachea (C). Dental floss is used as a suture and is passed behind trachea (C,D). The trachea is then cannulated with the 20-G cannula combined (D) mounted on the syringe and 3-way valve. The trachea should be tightly secured around the cannula by tying a surgical knot using the suture in place behind the trachea. Finally 500 µl of PBS are gently instilled in the lungs and then the BAL is gently drawn out. (E) Fluid-instilled lungs. Please click here to view a larger version of this figure.
| Minimal Instilled Burden | Maximal Instilled Burden | |
| T3SS- | 5 x 107 | 1 x 108 |
| T3SS+ | 5 x 106 | 1 x 107 |
| T3SS+ exoU+ | 5 x 104 | 1 x 105 |
Table 1. P. aeruginosa Inocula Used in Acute Lung Infection Models.
Suggested optimal intranasal concentrations of inocula to induce acute lung injury according to strains.
Discussion
Animal models, particularly mammals, are useful to elucidate complex mechanisms of host-pathogen interaction in the fields of immunity. Of course, the need for information obtainable only from animal models must be essential; otherwise, use of animals must be replaced by in vitro models. This animal model illustrates the insight that can only be provided by an animal model since the interaction between pathogens is mediated by a multi-component host response. Mice currently used to study this host-pathogen interaction are young adults aged 6 to 10 weeks with a mature and unaltered immune response. When focusing on the innate immune response, C57Bl6/J background mice are preferred. To avoid an effect of sex and hormonal cycle (particularly estrogen) on the immune response, males are therefore the best choice. To achieve statistical significance, groups must have at least 5 individuals at the end of the experiment, but as suggested by all animal experimentation guidelines, the number of animals used should be reduced and refined to a strict minimum.
Transport from the breeder providing the animals to the research facility induces stress in mice. The consequence is an increased secretion of inflammatory cytokines that can alter subsequent experiments such as ours. Moreover, a new environment and new "cage mates" contribute to stress. Consequently, mice must be acclimated for at least seven days prior to study in their new housing environment. This housing environment has to be controlled, providing standard food and water ad-lib, a day/night cycle, and appropriate stable humidity and temperature.
Both lung colonization and lung infection models require practice and dexterity. Instillation can be performed intra-nasally or intra-tracheally. The latter is more difficult and requires greater expertise through training due to a high risk of anoxic cardiac arrest. Indeed, the procedure requires to successfully intubate a mouse in less than 15 sec and therefore required deeper anesthesia. Our choice of intra-nasal instillation is easier to perform since the route of administration is accessible, less risky requiring lighter anesthesia and is therefore more reproducible.
Boutoille et al already described our acute lung injury model, particularly the assessment of lung injury through measuring alveolar-capillary barrier permeability using FITC-labeled albumin16. To reduce the number of mice per experiment, this method was adapted and coupled with bronchoalveolar lavage (BAL). In the studies of Boutoille et al, we used the comparison between the fluorescence of FITC-labeled albumin in the lung homogenate supernatants and the blood supernatants17.
To fully perform this comparison, hemoglobin levels and hematocrit levels are also required. This method was adapted to allow the concomitant analysis of host response by dedicating one lung to homogenization and assessment of lung injury and the other to lavage and study of host response. Indeed, BAL fluid can be used to assess cytokine levels, protein secretion and cell recruitment. Our adaptation provides more results from a single animal experiment, reducing the cost and the required number of mice, particularly when using knock-out mice17 as recommended in animal experimentation guidelines. Moreover, a part of the lung is kept at -80 °C to perform total RNA extraction and quantitative polymerase chain reaction or histologic analysis. Comparison between the previous method and the new adapted method coupled with BAL shows comparable results (Figure 3A) to assess lung injury index.
In the study of Mear et al, using the same procedures, flow cytometer analysis was performed on BAL cells obtained by centrifugation of BAL fluid. Similar analyses were performed on total pulmonary cells from the lungs12. In this case, lung injury assessment with FITC-labeled albumin cannot be performed concomitantly, due to artefacts induced by FITC (same channel than green fluorescence protein). Therefore, if flow cytometry is required, experiments must be planned accordingly.
The timing of euthanasia is critical. A time course of the acute lung infection model over the first 72 hr is presented Figure 3. In this model, the acme of lung injury and host response occurs between 24 and 36 hr (Figure 3). Thus, in our design the 24 hr end-point was chosen as a readout for 3 reasons: first, it is easy to organize in the laboratory, second, to avoid loss of mice due to mortality between 24 and 36 hr, and finally, because host response was maximal at this time point.
The first step of our interaction study is the colonization of the airway with C. albicans. Using an instillation of 105 CFU per mouse, a 4-day persistence model is obtained without any lung injury. In fact, mice gained weight over the time course of this phase. Weight gain is a useful indicator of the absence of injury in line with colonization as opposed to infection. Indeed, an increased initial load (over 106 CFU per mouse) induced lung injury (Figure 2C) and a deleterious host-response far beyond priming, in which case mice lost weight (Figure 2D). On the contrary, using a smaller initial load (for example 104 CFU per mouse) an airway persistence model could not be obtained. Thus, to obtain the observed priming of host immunity described by Mear et al12, calibration of the instilled fungal burden is critical to the success and monitoring the weight curve is a key control.
The same precision is required for P. aeruginosa to induce an acute infection. An insufficient inoculum does not induce lung injury and P. aeruginosa is rapidly cleared from the airways by an appropriate host-response. Using an overly high initial P. aeruginosa surpasses the capacities of host defense or even induces an inappropriate and derleterious response resulting in load leads to massive acute injury and death erasing all differences between groups. The optimal inoculum to induce acute injury depends of the presence of a functional type-3 secretion system (T3SS) and the production of exotoxin U, a toxin translocated by the T3SS into the host cell cytoplasm. These two strain-specific attributes have to be considered in the choice of the initial bacterial burden. Initial bacterial burdens are suggested in this article as examples of lower and upper limits to induce acute lung injury with T3SS-negative or positive strain, and strains producing exotoxin U or not (Table 1). When studying involvement of T3SS, strain has to be grown O/N and revived with new LB medium 3 hr before the preparation of the inoculum to obtain optimal activity of T3SS.
The determination of bacterial and fungal burden 24 hr after P. aeruginosa-induced lung infection requires specific considerations discussed in this section. Indeed, as shown, when mice are infected with 5 x 106 CFU T3SS-positive strain, bacterial burden in the lungs at 24 hr will decreased to about 1 log (Figure 3C). Serial dilutions of samples must be performed up to 5-log dilution and plated on BCP-agar plates. Concerning C. albicans in the lungs, at 72 hr, fungal burden is decreased to about 2 log and samples must to be plated on YPD-agar supplemented with amikacin to facilitate colony identification. Finally, determination of bacterial dissemination can be assessed by plating 100 µl of blood sample on BCP-agar or by plating 100 µl of spleen homogenates on the same medium. The two methods were already been compared and spleen culture seems to be more accurate. Indeed, the spleen is an organ that "filters" whole blood and may "concentrate" and conserve bacteria having disseminated onto the blood. Thus, spleen homogenates reflect the systemic bacterial dissemination during acute lung infection with a higher sensitivity than blood. Blood samples represent bacterial dissemination at a very specific given time and may not truly reflect the phenomenon of bacterial dissemination as a whole. Therefore spleen homogenate cultures are preferred.
Lung injury index is a sensitive assessment of lung injury resulting from infection and/or inappropriate host response and the effect of potential therapeutics on these components. Additionally, this in vivo model allows the collection of several different samples to study host response. Lung can be used for RNA extraction and analysis of gene transcription. Lung samples can also be placed into paraformaldehyde (PFA) to further histological observations. BAL fluid can be used for assessment of protein secretion such as inflammatory cytokines. Finally, as discussed above the protocol can be adapted to provide samples for flow cytometry analysis.
Finally, this protocol can be adapted to modelize C. albicans-microbes interaction involved in ventilator-associated pneumonia such as Staphylococcus aureus18 or Enterobacteriae. Another adaptation could be airway colonization using bacteria instead of C. albicans to modelize bacterial interaction in chronic suppurative lung disease such as bronchiectasis. To this end, immunocompromised mice have to be used, because bacterial clearance in immune competent mice does not allow a persistent airway colonization.
For instillation, the position of the hand holding the animal is critical. As already underlined in the previous section, when intra-nasally instilling the mouse, the operator must ensure that mouth is perfectly closed to avoid expectoration of the solution. The thumb supports the jaw and maintains the mouth closed during the entire instillation procedure (Figure 4B). Then, with the other hand, the pipette is deposited on a nostril (Figure 4C) and solution progressively and gently instilled without air to prevent bubble formation. Obviously, instillation must be performed in a level 2-biosafety cabinet.
Collection of samples requires certifed small-animal surgical training of the operator. At each step, samples must be placed on ice to avoid cell lysis and preserve proteins from denaturation. Basic surgical instruments are required (Figure 4A). They must to be sterilized by autoclaving before use. It is recommended that different surgical instrument sets be used for abdominal and thoracic surgical steps, avoiding cross-contamination of lung samples. The different critical steps of surgical dissection are presented (Figure 5A-5E). Position of the mouse is critical; the animal should be immobilized flat on it's back head opposite from the operator and straight. Ethanol should be liberally used to prep before the first incision to avoid the spread of hair in the samples and potential bacterial contamination from the skin. Skin is well vascularized and must be retracted carefully to avoid bleeding (Figure 5A).
Then the ribcage is reclined following a lateral chest incision during which the ribcage should be constantly maintained in the tweezers grip to avoid injury to the heart and lungs during the incision. Lung and Heart are exposed (Figure 5B). Uninfected lung appear whitish (Figure 5B). Dissection of the cervical area exposes the trachea and a suture is carefully placed behind the upper trachea (Figure 5C). Cannulating the trachea must be performed with caution. Do not use an oversized cannula (maximum size is 20 G). A small anterior incision of the membraneous trachea between two cartilaginous rings allows insertion of the cannula. When the cannula is observed by transparency through the trachea above the carina (Figure 5D), the suture is tightly tied around the trachea securing it around the cannula (Figure 5E). The cannula is connected to a 3-way male Luer-lock valve with 2 syringes connected to the female ports. One syringe contains ice-cold PBS for bronchoalveolar lavage; the other is empty to draw back the BAL fluid. These syringes should be chaged between groups.
In conclusion, priming prior to acute lung injury model is a relevant and powerful model to explore in vivo host-mediated interactions between pathogens. The use of animals is the main limitation and should be carefully weighed against the information obtainable in vitro.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the University of Lille and the Pasteur Institute of Lille, especially Thierry Chassat and Jean-Pierre Decavel, responsible for animal housing breeding safety and husbandry. This work was supported by the “Société de Pathologies Infectieuses de Langue Française” (SPILF).
Materials
| Sevorane, Sevoflurane | Abott | 05458-02 | 250 mL plastic bottle |
| Fluorescence Reader Mithras LB940 | Berthold Technologies | reference in first column | no comment |
| Bromo-cresol purple agar | Biomerieux | 43021 | x20 per unit |
| Pentobarbital sodique 5,47% | CEVA | 6742145 | 100 mL plastic bottle |
| 2-headed valve | Distrimed | 92831 | no comment |
| Sterile inoculation loop 10 µL | Dutscher | 10175 | x1000 conditioning |
| Insuline syringes 1 mL | Dutscher | 30003 | per 100 conditioning |
| 2 positions Culture tube 8 mL | Dutscher | 64300 | no comment |
| Ultrospec 10 | General Electric life sciences | 80-2116-30 | no comment |
| Hemolysis tubes 13 x 75 mm | Gosselin | W1773X | per 100 |
| PBS – Phosphate-Buffered Saline | Life technologies | 10010023 | packaged in 500 mL |
| amikacin 1g | Mylan | 62516778 | per 10 |
| Heparin 10 000 UI in 2 mL | Pan pharma | 9128701 | x 10 per unit |
| RAL 555 coloration kit | RAL Diagnostics | 361550 | 3 flacons of 100 mL |
| 1,5 mL microcentrifuge tube | Sarstedt | 55.526.006 | x 1000 |
| Transparent 300 µL 96-well plate | Sarstedt | 82 1581500 | no comment |
| Yest-peptone-Dextrose Broth | Sigma | 95763 | in powder |
| FITC-albumin | Sigma | A9771 | in powder |
| Luria Bertani Broth | Sigma | L3022 | in powder |
| 25-gauge needle | Terumo or unisharp | A231 | x100 conditioning |
| Cytocentrifuge | Thermo Scientific | A78300003 | no comment |
References
- Casadevall, A., Pirofski, L. -. A. The damage-response framework of microbial pathogenesis. Nat. Rev. Micro. 1 (1), 17-24 (2003).
- Eddens, T., Kolls, J. K. Host defenses against bacterial lower respiratory tract infection. Curr. Opi. Immunol. , (2012).
- Beck, J. M., Young, V. B., Huffnagle, G. B. The microbiome of the lung. Translational research : J. Lab. Clin Med. 160 (4), 258-266 (2012).
- Hogan, D. A., Kolter, R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 296 (5576), 2229-2232 (2002).
- Nseir, S., Ader, F. Pseudomonas aeruginosa and Candida albicans: do they really need to stick together. Crit. Care Med. 37 (3), 1164-1166 (2009).
- Hibbing, M. E., Fuqua, C., Parsek, M. R., Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Micro. 8 (1), 15-25 (2010).
- Gibbons, D. L., Spencer, J. Mouse and human intestinal immunity: same ballpark, different players; different rules, same score. Mucosal Immunol. 4 (2), 148-157 (2011).
- Ariffin, J. K., Sweet, M. J. Differences in the repertoire, regulation and function of Toll-like Receptors and inflammasome-forming Nod-like Receptors between human and mouse. Curr. Opi. Micro.. , (2013).
- Slutsky, A. S., Ranieri, V. M. Ventilator-Induced Lung Injury. NEJM. 369 (22), 2126-2136 (2013).
- Peleg, A. Y., Hogan, D. A., Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Micro. 8 (5), 340-349 (2010).
- Roux, D., Gaudry, S., et al. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit. Care Med. 37 (3), 1062-1067 (2009).
- Mear, J. B., Gosset, P., et al. Candida albicans Airway Exposure Primes the Lung Innate Immune Response against Pseudomonas aeruginosa Infection through Innate Lymphoid Cell Recruitment and Interleukin-22-Associated Mucosal Response. Infect. Immun. 82 (1), 306-315 (2013).
- Ader, F. Short term Candida albicans colonization reduces Pseudomonas aeruginosa load and lung injury in a mouse model. Crit. care. , 1-33 (2009).
- Risling, T. E., Caulkett, N. A., Florence, D. Open-drop anesthesia for small laboratory animals. Can Vet J. 53 (3), 299-302 (2012).
- Stover, C. K., Pham, X. Q., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 406 (6799), 959-964 (2000).
- Boutoille, D., Marechal, X., Pichenot, M., Chemani, C., Guery, B. P., Faure, K. FITC-albumin as a marker for assessment of endothelial permeability in mice: comparison with 125I-albumin. Exp. Lung Res. 35 (4), 263-271 (2009).
- Faure, E., Mear, J. -. B., et al. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. American Journal of Respiratory and Critical Care Medicine. 189 (7), 799-811 (2014).
- Peleg, A. Y., Hogan, D. A., Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Micro. 8 (5), 340-349 (2010).

