Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5
Summary
We describe here a protocol for the generation of iCMs using retrovirus-mediated delivery of Gata4, Tbx5 and Mef2c in a polycistronic construct. This protocol yields a relatively homogeneous population of reprogrammed cells with improved efficiency and quality and is valuable for future studies of iCM reprogramming.
Abstract
Direct conversion of cardiac fibroblasts (CFs) into induced cardiomyocytes (iCMs) holds great potential for regenerative medicine by offering alternative strategies for treatment of heart disease. This conversion has been achieved by forced expression of defined factors such as Gata4 (G), Mef2c (M) and Tbx5 (T). Traditionally, iCMs are generated by a cocktail of viruses expressing these individual factors. However, reprogramming efficiency is relatively low and most of the in vitro G,M,T-transduced fibroblasts do not become fully reprogrammed, making it difficult to study the reprogramming mechanisms. We recently have shown that the stoichiometry of G,M,T is crucial for efficient iCM reprogramming. An optimal stoichiometry of G,M,T with relative high level of M and low levels of G and T achieved by using our polycistronic MGT vector (hereafter referred to as MGT) significantly increased reprogramming efficiency and improved iCM quality in vitro. Here we provide a detailed description of the methodology used to generate iCMs with MGT construct from cardiac fibroblasts. Isolation of cardiac fibroblasts, generation of virus for reprogramming and evaluation of the reprogramming process are also included to provide a platform for efficient and reproducible generation of iCMs.
Introduction
Cardiovascular disease remains the leading cause of death worldwide, accounting for 17.3 million deaths per year1. Loss of cardiomyocytes resulting from myocardial infarction (MI) or progressive heart failure is a major cause of morbidity and mortality2. Due to limited regenerative capacity, adult mammalian hearts usually suffer from impaired pump function and heart failure following injury3-6. As such, efficient (re)generation of cardiomyocytes in vivo and in vitro for treatment of heart disease and for disease modeling is a critical issue needing to be addressed.
Recent development of direct reprogramming, which directly reprograms cells from one differentiated phenotype to another without transitioning through the pluripotent state, offers a promising alternative approach for regenerative medicine. The mammalian heart contains abundant cardiac fibroblasts (CFs), which account for approximately half of the cells in heart and massively proliferate upon injury7-9. Thus, the vast pool of CFs could serve as an endogenous source of new CMs for regenerative therapy if they could be directly reprogrammed into functional CMs. It has been shown that a combination of transcription factors, such as Gata4 (G), Mef2c (M) and Tbx5 (T), with or without microRNAs or small molecules can reprogram fibroblasts into iCMs10-26. Importantly, this conversion can also be induced in vivo, and results in an improvement in cardiac function and a reduction in scar size in an infarcted heart16,27-29. These studies indicate that direct cardiac reprogramming may be a potential avenue to heal an injured heart. However, the low efficiency of iCM reprogramming has become a major hurdle for further mechanistic studies. In addition, the reproducibility of cardiac reprogramming is another controversial issue of this technology11,30,31.
Very recently, we generated a complete set of polycistronic constructs encoding G,M,T in all possible splicing orders with identical 2A sequences in a single mRNA. These polycistronic constructs yielded varied G, M and T protein expression levels, which led to significantly different reprogramming efficiency25. The most efficient construct, named MGT, which showed a relatively high Mef2c and low Gata4 and Tbx5 expression, significantly improved reprogramming efficiency and produced large amounts of iCMs with CM markers expression, robust calcium oscillation and spontaneous beating25. Moreover, by using MGT polycistronic construct, our study avoided the use of multiple vectors and generated cells with homogenous expression ratio of G,M,T, thus providing an improved platform for cardiac reprogramming research. To increase experimental reproducibility, here we describe in detail how to isolate fibroblasts, produce retrovirus carrying MGT cassette, generate iCMs and evaluate the reprogramming efficiency.
Protocol
The protocol outlined here uses neonatal mice. Animal care and experiments are performed in accordance with the guidelines established by The Division of Laboratory Animal Medicine (DLAM) at University of North Carolina, Chapel Hill.
1. Preparation of Buffers and Media
- Prepare fibroblast (FB) medium: Supplement 500 ml IMDM media with 100 ml Fetal Bovine Serum (FBS) and 6 ml penicillin/streptomycin.
- Prepare iCM medium: Supplement 400 ml DMEM media with 100 ml M199 media and 50 ml FBS.
- Prepare B27 medium: Supplement 490 ml RPMI1640 media with 10 ml B27 media.
- Prepare Plat-E culture medium: Supplement 500 ml DMEM media with 50 ml FBS, 5 ml penicillin/streptomycin and 5 ml Non-essential amino acids.
- Prepare Plat-E transfection medium: Supplement 500 ml DMEM media with 50 ml FBS and 5 ml Non-essential amino acids.
- Prepare Gelatin coated 24 well plate: Add 0.5 ml 0.1% gelatin solution to a well of 24 well plate, incubate at 37 °C for at least 10 min and aspirate right before use.
- Prepare FACS labeling buffer: Supplement 500 ml DPBS with 10 ml FBS and 2 ml EDTA (0.5 M) to reach a final concentration of 2 mM. Pass through a 0.22 µm filter and store at 4 °C.
- Prepare MACS sorting buffer: Supplement 500 ml DPBS with 2.5 g BSA and 2 ml EDTA (0.5 M) to reach a final concentration of 2 mM. Pass through a 0.22 µm filter and store at 4 °C.
- Prepare 2x HBS buffer for transfection: supplement 500 ml HEPES buffer (50 mM) with 280 mM NaCl (8.1816 g), 10 mM KCl (0.37275 g), 1.5 mM Na2HPO4 (0.10647 g), 12 mM Glucose (1.08096 g). Add around 650 µl 10 N NaOH to adjust pH to 7.05-7.12. Filter through a 0.22 µm pore filter and store at 4 °C.
- Prepare 2.5 M CaCl2 solution for transfection: Add 18.376 g CaCl2 to 50 ml ddH2O. Filter through a 0.22 µm pore filter and store at 4 °C.
- Prepare ICC (immunocytochemistry) fixation buffer: Add 5 ml paraformaldehyde (PFA, 32%) to 35 ml DPBS to reach a final concentration of 4%.
- Prepare ICC permeabilization buffer: Add 0.1 ml Triton to 100 ml DPBS to reach a final concentration of 0.1%.
- Prepare ICC blocking buffer: Add 5 g Bovine serum albumin (BSA) to 100 ml DPBS to reach a final concentration of 5%.
- Prepare ICC staining buffer: Add 1 g BSA to 100 ml DPBS to reach a final concentration of 1%.
2. Generation of Neonatal Mouse Cardiac Fibroblasts
- Generate cardiac fibroblasts from explant culture method
- Clean neonatal αMHC-GFP transgenic mouse (P1 to P2) with 75% ethanol. Cut the head off with sterile scissors. Then make a horizontal incision from under one armpit to the other. Use a sterile blunt and bent forceps to dissect out the heart and place it in one well of the 24 well plate containing ice-cold PBS buffer.
- Alternatively, squeeze out the heart after the horizontal incision.
- Check GFP expression in heart by fluorescent microscope. Since the GFP expression is driven by αMHC promoter that is a cardiac specific promoter, the heart from transgenic mouse should be in green15, while the negative heart is dim in any fluorescence.
- Take 3-4 GFP positive hearts into a 60 mm center well culture dish and mince them into small pieces less than 1 mm3 in size by sterile scissors and forceps.
- Place 3-4 minced hearts into one 10 cm dish with 2 ml FB media. Let the tissues settle down for 3 hr.
- Slowly add 8 ml pre-warmed FB media to the dishes containing heart tissues. Do not disturb the tissues for three days.
- Replace the media every three days.
- On day 7, aspirate the culture medium and wash cells with DPBS. Add 3 ml 0.05% trypsin to each plate and digest at 37 °C for 5 min.
- Add 5 ml FB media to quench trypsin. Gently detach the cells with a cell scraper. Pipette the media up and down to further mechanically dissociate the tissue.
- Collect cells and filter through 40 µm cell strainers to avoid contamination of heart tissue fragments, and then pellet cells by spinning at 200 x g for 5 min.
- Wash cells once with MACS buffer and cells are ready for sorting.
- Clean neonatal αMHC-GFP transgenic mouse (P1 to P2) with 75% ethanol. Cut the head off with sterile scissors. Then make a horizontal incision from under one armpit to the other. Use a sterile blunt and bent forceps to dissect out the heart and place it in one well of the 24 well plate containing ice-cold PBS buffer.
- Generate cardiac fibroblasts from enzyme digestion method
- Harvest GFP positive hearts as 2.1.1. Transfer all the hearts into a 10 cm dish with 10 ml ice-cold DPBS.
- Squeeze ventricles with sterile forceps to remove blood and rinse once with ice-cold DPBS.
- Trim the hearts to be free of other tissues and fat.
- Cut each heart into roughly 4 pieces that are still loosely connected.
- If less than 20 hearts, transfer the hearts into a 15 ml conical tube with 8 ml warm 0.05% Trypsin-EDTA, and incubate at 37 °C for 15 min.
- If there are 20-30 hearts, transfer the hearts into a 50 ml conical tube with 10 ml warm 0.05% Trypsin-EDTA, and incubate at 37 °C for 15 min.
- Discard the trypsin and add 5 ml (for less than 20 hearts) or 10 ml (for 20-30 hearts) warm type II collagenase (0.5 mg/ml) in HBSS.
- Vortex the tube on vortexer for 1 min at a speed level around 4 to 6 (if the liquid does not get up to the lid, then the speed is fine).
- Incubate the tube in 37 °C water bath for 3-5 min.
- Vortex the tube for 1 min.
- Sediment for 1 min by gravity until the tissue pieces settle down, collect the liquid into a new tube containing 5 ml cold FB medium.
- Repeat steps 2.2.6-2.2.10 3-5 times.
- Combine all the collections by filtering through 40 µm cell strainer to make single cell suspension.
- Centrifuge at 200 x g for 5 min at 4 °C.
- Resuspend cells in 10 ml MACS buffer, and centrifuge at 200 x g for 5 min at 4 °C.
- Optional: If there are a lot of blood cells, resuspend cells with 1 ml RBC lysis buffer (150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA), keep on ice for 1 min, then dilute with 10 ml MACS buffer and centrifuge at 200 x g for 5 min. Wash one more time with MACS buffer.
- Isolation of Thy1.2+ fibroblasts by MACS (magnetic-activated cell sorting)
- Determine viable cell number through trypan blue staining.
- Take 10 µl cells out from 10 ml cell suspension in step 2.2.14. Mix it with 10 µl 0.4% trypan blue solution.
- Add the mixture to a hemocytometer. Allow it to stand for 3-5 min and then examine immediately under a microscope. The dead cells are stained in blue and viable cells are unstained.
- Count all the viable cells in four 1 mm corner squares of the hemocytometer. Determine the viable cell number: the average count per square x 2 (dilution factor) x 104 x 10 (total volume of cell suspension).
- For less than 1 x 107 cells, resuspend cells with 10 µl Thy1.2 microbeads in 90 µl chilled MACS buffer. Add more beads proportionally if there are more than 1 x 107 cells. Mix well and incubate in a refrigerator (2-8 °C) for 30 min.
- Add 10 ml MACS buffer and spin samples at 200 x g for 5 min; wash once with 10 ml MACS buffer and centrifuge again.
- Bring volume to 2 ml in MACS buffer.
- Set up a MACS Separator in hood. Insert an LS column to the separator. Apply 3 ml MACS buffer to the column to equilibrate it.
- Pass the cell suspension through the equilibrated LS column.
- Wash 3 times with 2 ml MACS buffer each time.
- Take the LS column off the Separator and elute 3 times with 2 ml MACS buffer into a new 50 ml tube.
- Spin at 200 x g for 5 min.
- Resuspend cells in 10 ml FB media, count the cells and seed into plates at proper density. For fibroblasts generated from explant culture method, the seeding cells density could be around 2-3 x 104 cells/well of 24 well plate. For fibroblasts generated from enzyme digestion method, the cells density could be around 4-5 x 104 cells/well of 24 well plate.
- Determine viable cell number through trypan blue staining.
3. Generation of Retrovirus for iCM Reprogramming
Note: Perform the following steps in a BSL2 Biological Safety Cabinet under sterile conditions. The proper disposal of transfected cells, pipette tips and tubes is recommended to avoid risk of environmental and health hazards.
- Maintain Plat-E cells in Plat-E media supplemented with 1 µg/ml puromycin and 10 µg/ml blasticidin at 37 °C with 5% CO2.
- On day 2, replace the medium by Plat-E culture medium lacking puromycin and blasticidin.
- On day 0, split Plat-E cells into 10 cm dishes the day before transfection at approximately 4-5 x 106 cells per plate. Cells should be ~80-90% confluent on the day of transfection.
- On day 1, change culture media to transfection media on cells within 1 hr prior to transfection. Transfection procedure is as follows:
- Transfection using commercial kits
- Mix 20 µl transfection reagent (e.g. Lipofectamine) and 500 µl reduced serum media (e.g., Opti-MEM). Add 10 µg of the retroviral plasmid to another 500 µl reduced serum media. Incubate each mixture at room temperature (RT) for 5 min.
- Slowly add the plasmid mixture to transfection mixture. This step may take 15-30 sec. Incubate the mixture for 20 min at RT (solution may appear cloudy).
- Add mixture drop wise to the cells.
- Incubate overnight in a 37 °C incubator.
- Transfection using Calcium Phosphate.
- Mix 40 µl 2.5 M CaCl2 and 440 µL ddH2O, then add 20 µg of the retroviral plasmid (adjust the amount of ddH2O so that total volume is 500 µl).
- Add 500 µl 2x HBS to a 15 ml conical tube.
- Vortex the HBS and meanwhile add the Calcium-DNA mixture to the HBS drop wise. This step takes around 30 sec for each sample. So maximally do 3-4 samples simultaneously.
- Incubate at RT for 3 min Incubation for longer time leads to larger precipitate and less cells will take up the precipitate.
- Add the mixture drop wise to the cells and observe under 20X microscope. Fine precipitates (a lot of small ones are good but a few large ones are not good) should be seen.
- Incubate overnight at 37 °C incubator.
- Transfection using commercial kits
- On day 2, change media on cells with 8 ml prewarmed culture media. Incubate for 24 hr.
- On day 3 and day 4, collect culture supernatant from the dishes by using a 10 ml sterile disposable syringe, filtering it through a 0.45 µm pore size cellulose acetate filter, and transferring into a 50 ml tube. Add 2 ml of retrovirus precipitation solution to every 8 ml virus-containing supernatant. Gently mix it and incubate overnight at 4 °C.
- On day 5, spin the viral mixture at 1,500 × g at 4 °C for 30 min. The viral particles should appear in white pellet at the bottom of the tube.
- Discard the supernatant. Spin down residual solution by centrifugation at 1,500 × g for 5 min. Remove all traces of fluid by aspiration, taking great care not to disturb the precipitated retroviral particles in pellet.
- Resuspend retroviral pellets with 100 µl DPBS buffer for every 8 ml viral supernatant. Aliquot the virus on ice. Use immediately or store at -80 °C .
4. Reprogramming of Cardiac Fibroblasts
- Add 4 µl of 10 mg/ml polybrene solution into the 10 ml iCM medium, and mix gently by pipetting up and down. The final concentration of polybrene is 4 µg/ml.
- Add 500 µl iCM media containing polybrene to each well of 24 well plate seeded with fibroblasts.
- Add 10 µl retrovirus to each well containing either 2-3 x 104 cells from explant culture or 4-5 x 104 cells from enzyme digestion method. The amount of virus should be proportionally increased with increased cell number in different culture plates or dishes. Put the plate in a 37 °C incubator for 24-48 hr to allow the viral infection. The time line for cardiac reprogramming is shown in Figure 1.
- After viral transduction, replace virus containing media with 500 µl regular iCM media.
- Change media every 2-3 days. For positive selection of viral transduced cells, add iCM media supplemented with puromycin at 2 µg/ml to target cells. Keep it for three days. After that, maintain puromycin in the iCM medium at the concentration of 1 µg/ml.
- Three days after viral infection, take the plate out from the incubator and place it under an inverted fluorescent microscope (20X) to observe GFP expression.
Note: Ten days after viral infection, cells could be harvested for FACS analysis and ICC analysis to determine the reprogramming efficiency (in step 5 and 6). - Fourteen days after viral infection, replace iCM media with B27 media. Change media every three days. Spontaneous beating cell loci may appear from three (cells from enzyme digestion method) or four (cells from explant culture method) weeks after viral transduction.
5. Immunocytochemical Analysis of Reprogramming Efficiency
- Rinse cells with ice cold PBS three times; remove excess solution.
- Add 0.5 ml ICC fixation buffer to each well of 24 well plates. Fix the cells at RT for 15-20 min.
- Rinse cells with PBS three times.
- Add 0.5 ml ICC permeabilization buffer to each well of the 24 well plates. Permeabilize the cells at RT for 15 min.
- Rinse cells with PBS three times.
- Add 0.5 ml ICC blocking buffer to each well of 24 well plate, incubate at RT for 1 hr.
- Diluted primary antibodies with ICC staining buffer. Add 200 µl antibody solutions to each well and incubate overnight at 4 °C. The antibody dilution factors are listed in Materials.
- On the next day, wash cells with PBS three times.
- Add 200 µl secondary antibody solutions to the cells and incubate for 1 hr at RT in dark.
- Wash cells with PBS for three times.
- Add 150 µl mounting solution containing DAPI to each well. Allow to stain nuclei for 1 min. Then cover each well with round glass cover slip and gently press the slip against the bottom surface with a forceps to remove air bubbles.
- Aspirate the excess solution and the samples are ready for imaging. The representative images showing the reprogrammed cells that express cardiac marker cTnT and αActinin at d14 are shown in Figure 2B.
6. FACS Analysis of Reprogramming Efficiency
- Thoroughly wash cells once with DPBS. Add 0.3 ml trypsin (0.05%) to each well and incubate at 37 °C for 5 min.
- Gently tap the plate to facilitate lifting the cells. Check the cell detachment under microscope. Add 1 ml FACS buffer to each well when most cells are dissociated. Pipette up and down to further mechanically dissociate the cells.
- Transfer cells in the 24 well plate to a 96 well deep well plate well to well. Spin at 200 x g for 5 min at 4 °C to collect the cells.
- Wash cells once with FACS buffer.
- Add 100 µl fixation/permeabilization solutions to resuspend cells for 20 min at 4 °C.
- Wash cells twice with 1x wash solution, pellet, and remove supernatant.
- Prepare primary antibody solution against GFP and cTnT in 1x wash buffer. Thoroughly resuspend each sample with 50 µl antibody solution and incubate at 4 °C for 30 min.
- Wash cells once in 1x wash solution, pellet, and remove supernatant.
- Add 50 µl secondary antibody solution containing Alexa Fluor 488 conjugated donkey anti-rabbit IgG and Alexa Fluor 647 conjugated donkey anti-mouse IgG to each sample. Incubate at 4 °C for 30 min in dark.
- Wash cells once in 1x wash solution, pellet, and remove supernatant.
- Resuspend cells in 400 µl fixation buffer (DPBS with 1% PFA). Transfer cell into FACS tube with cell strainer cap. Samples are ready for FACS detection. The representative FACS analysis of reprogramming efficiency using pMxs-puromycin-MGT construct with or without puromycin selection is shown in Figure 2A.
Representative Results
The reprogramming steps are summarized by schematic in Figure 1. After MGT transduction, GFP expression in reprogramming cells could be detected as early as day 3. Puromycin selection of transduced cells starts from day 3 and is maintained during the first two weeks if pMx-puro-MGT construct is used. By day 10 to day 14, expression of cardiac markers like cTnT and αActinin could be detected by both ICC (Figure 2B, step 5) and FACS (Figure 2A, step 6), indicating that the starting fibroblasts are undergoing reprogramming toward a cardiac cell fate.
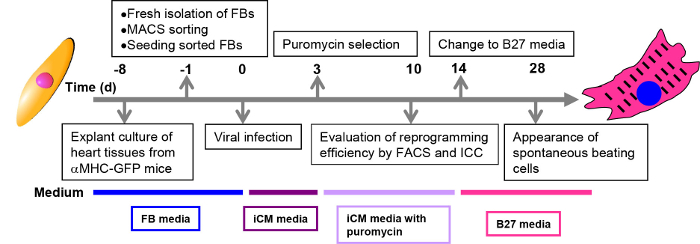
Figure 1: Schematic representation of the direct cardiac reprogramming process. Procedures at each time point are described in black boxes. Culture media for each stage are shown in colored boxes below the time line.
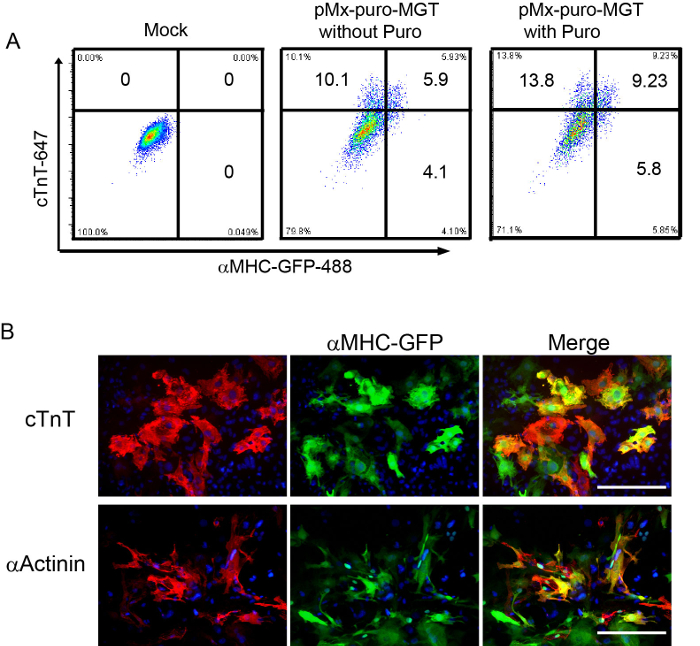
Figure 2: Reprogramming efficiency evaluated by FACS and ICC. (A) FACS analysis on iCMs generated from freshly isolated fibroblasts at day 10 after MGT transduction. (B) Staining of iCMs at 2 weeks with αMHC-GFP, cardiac cTnT, and αActinin with DAPI (4',6-diamidino-2-phenylindole). Scale bar: 200 µm
Discussion
For successful iCM generation when using this protocol, there are several important factors that have an impact on the overall efficiency. Particularly the conditions of starting fibroblasts and the quality of retrovirus encoding MGT can greatly affect the reprogramming efficiency.
It is important to generate fibroblasts as fresh and healthy as possible. For explant culture method, fibroblasts can be used before seven days after the explants were plated on dishes. For enzyme digestion method, fibroblasts can be used as early as the next day of isolation. Freezing or passaging the fibroblasts for reprogramming is not recommended. These additional treatments of fibroblasts will significantly decrease the reprogramming efficiency. The seeding density is another important factor that affects the reprogramming efficiency. If cells are sparsely seeded, they tend to become unhealthy with irregular cell morphology and in large size. Seldom could those cells be converted into iCMs. If cells are seeded too densely, the MOI (multiplicity of infection) for viral infection is decreased. Overgrowth of uninfected fibroblasts significantly dilutes the reprogramming events thus resulting in a low percentage of iCMs. We also notice the emergence of smaller cells in cobblestone-like morphology if more cells are seeded for reprogramming. They could hardly be reprogrammed and thus result in reduced reprogramming efficiency. When modifying the protocol for different size of tissue culture dishes, it is recommended that seeding cell numbers be adjusted proportionally to the surface area of the culture dish.
The purity of fibroblasts is critical for reprogramming. The high purity of fibroblasts could be achieved either by FACS sorting15 or MACS enrichment as we described here. Compared to FACS sorting, MACS sorting is easier, gentler, and more convenient. Cell purity after MACS could reach over 90% when strictly following the protocol. Low purity of fibroblasts may decrease reprogramming efficiency since the contaminated cells are harder to be reprogrammed than fibroblasts and that they proliferate much faster than reprogramming cells. It is also highly recommended to seed the cells right after MACS and perform viral infection overnight after the seeding. The sorted fibroblasts may partially become senescent after growing in dish for a long time, which decreases uptake of the retrovirus that only infects the proliferating cells. Tail-tip fibroblasts (TTFs) and embryonic fibroblasts (MEFs) have been reported to be converted into iCMs with some other transcription factors10,12,19,26. Our protocol here only focuses on cardiac fibroblasts. The application of this protocol to other fibroblast types may be further optimized due to the inherent variations of fibroblast signatures such as the proliferation status, genetic and epigenetic landmarks.
The quality of retrovirus for transduction is of utmost importance. Low titer viruses fail to convert fibroblasts into iCMs. Whereas viruses at excessive amount will cause cytotoxicity that directly leads to fibroblast cell death. It is necessary to determine the viral titer and optimize the viral dosage for reprogramming based on laboratory set-up and experimental conditions. The method for viral titration has been thoroughly described before23. In fact we found that the growth condition of Plat-E cells directly relates to viral quality. It is recommended to thaw Plat-E cells at lower passage (<30) each time. Cells obtained at passage three are best for packaging virus at a density around 4-5 x 106 cells per 10 cm culture dish. Additionally, the freshly harvested viruses yield higher reprogramming efficiency than frozen viruses. Please note that co-infection of MGT virus with another pMxs vector based retrovirus will decrease the reprogramming efficiency possibly due to the competition between the two viruses of the same type. Two different transfection methods are provided here. Both methods produce comparable retrovirus. While commercial Lipofectamine is costly but stable, transfection with calcium phosphate is cheaper. Preparation of HBS buffer needs to take great caution because the pH of HBS buffer significantly affects the transfection efficiency.
There are some limitations of this protocol that need to be addressed. One of the limitations lies in the usage of retrovirus, which inevitably renders to biosafety issues. The heterogeneity of fibroblasts and the lack of specific cardiac fibroblast marker for isolation also affect the purity of cells that are reprogrammable. Variation of reprogramming efficiency may be observed due to batch variability inherent to fibroblast condition and virus production. In addition, although the reprogrammed cells express sarcomere structure proteins such as cardiac troponin T and αActinin, they are still not mature cardiomyocytes characterized by functional properties such as contractility and electrical propagation.
In summary, the methodology described here allows efficient generation of iCMs based on retroviral delivery of M,G,T transcription factors in a single construct. Our protocol provides a reproducible and valuable platform for iCM research, and will facilitate ongoing efforts on high-throughput screening and mechanistic studies of iCM reprogramming, and ultimately move iCM reprogramming field closer to clinical applications.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We are grateful for expert technical assistance from the UNC Flow Cytometry Core and UNC Microscopy Core. We thank members of the Qian lab and the Liu lab for helpful discussions and critical reviews of the manuscript. This study was supported by NIH/NHLBI R00 HL109079 grant to Dr. Liu and American Heart Association (AHA) Scientist Development Grant 13SDG17060010 and the Ellison Medical Foundation (EMF) New Scholar Grant AG-NS-1064-13 to Dr. Qian.
Materials
| anti-cardiac troponin T | Thermo Scientific | MS-295-PO | 1:200 for FACS and 1:400 for ICC |
| anti-GFP | Life Technologies | A11122 | 1:500 for both FACS and ICC |
| anti- aActinin | Sigma-Aldrich | A7811 | 1:500 for both FACS and ICC |
| anti-Connexin43 | Sigma-Aldrich | C6219 | 1:500 for ICC |
| anit-Mef2c | Abcam | ab64644 | 1:1000 for ICC |
| anti-Gata4 | Santa Cruz Biotechnology | sc-1237 | 1:200 for ICC |
| anti-Tbx5 | Santa Cruz Biotechnology | sc-17866 | 1:200 for ICC |
| Alexa Fluor 488–conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch Inc | 711-545-152 | 1:500 for both FACS and ICC |
| Alexa Fluor 647–conjugated donkey anti-mouse IgG | Jackson ImmunoResearch Inc | 715-605-150 | 1:500 for both FACS and ICC |
| Cytofix/Cytoperm kit for intracellular staining | BD Biosciences | 554722 | |
| Rhod-3 Calcium Imaging Kit | Life Technologies | R10145 | |
| Thy1.2 microbeads | Miltenyi Biotec | 130-049-101 | |
| Vectashield solution with DAPI | Vector labs | H-1500 | |
| FBS | Sigma-Aldrich | F-2442 | |
| Trypsin-EDTA (0.05%) | Corning | 25-052 | |
| PRMI1640 medium | Life Technologies | 11875-093 | |
| B27 supplement | Life Technologies | 17504-044 | |
| IMDM | Life Technologies | 12440-053 | |
| Opti-MEM Reduced Serum Medium | Life Technologies | 31985-070 | |
| M199 medium | Life Technologies | 10-060 | |
| DMEM, high glucose | Life Technologies | 10-013 | |
| Penicillin-streptomycin | Corning | 30-002 | |
| Non-essential amino acids | Life Technologies | 11130-050 | |
| Lipofectamine 2000 | Life Technologies | 11668500 | |
| blasticidin | Life Technologies | A11139-03 | |
| puromycin | Life Technologies | A11138-03 | |
| Collagenase II | Worthington | LS004176 | |
| polybrene | Millipore | TR-1003-G | |
| Triton X-100 | Fisher | BP151-100 | |
| CaCl2 | Sigma-Aldrich | C7902 | |
| HEPES | Sigma-Aldrich | H4034 | |
| NaCl | Sigma-Aldrich | BP358-212 | |
| KCl | Sigma-Aldrich | PX1405 | |
| Na2HPO4 | Sigma-Aldrich | S7907 | |
| Glucose | Sigma-Aldrich | G6152 | |
| Bovine serum albumin | Fisher | 9048-46-8 | |
| paraformaldehyde | EMS | 15714 | |
| Retrovirus Precipitation Solution | ALSTEM | VC-200 | |
| 0.4%Trypan blue solution | Sigma-Aldrich | T8154 | |
| gelatin | Sigma-Aldrich | G1393 | |
| Dulbecco's PBS without CaCl2 and MgCl2 (D-PBS, 1x) | Sigma-Aldrich | D8537 | |
| HBSS (Hanks Balanced Salt Solution) | Corning | 21022 | |
| LS column | Miltenyi Biotec | 130-042-401 | |
| 0.45 μm cellulose acetate filter | Thermo Scientific | 190-2545 | |
| 24-well plates | Corning | 3524 | |
| 10cm Tissue culture dishes | Thermo Scientific | 172958 | |
| 60mm center well culture dish | Corning | 3260 | |
| 96 Well Clear V-Bottom 2mL Polypropylene Deep Well Plate | Denville Scientific | P9639 | |
| Polystyrene round-bottom tubes with cell-strainer cap | BD Biosciences | 352235 | |
| Centrifuge | Eppendorf | 5810R | |
| Vortexer MINI | VWR | 58816-121 | |
| EVOS® FL Auto Cell Imaging System | Life Technologies | AMAFD1000 | |
| MACS MultiStand | Miltenyi Biotec | 130-042-303 | |
| MidiMACS Separator | Miltenyi Biotec | 130-042-302 | |
| Round glass cover slip | Electron Microscopy Sciences | 72195-15 |
References
- Mozaffarian, D., et al. Heart Disease and Stroke Statistics–2015 Update: A Report From the American Heart Association. Circulation. , (2014).
- Whelan, R. S., Kaplinskiy, V., Kitsis, R. N. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 72, (2010).
- Senyo, S. E., et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 493, 433-436 (2013).
- Soonpaa, M. H., Field, L. J. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol. 272, 220-226 (1997).
- Hosoda, T., et al. Clonality of mouse and human cardiomyogenesis in vivo. Proc Natl Acad Sci U S A. 106, 17169-17174 (2009).
- Choi, W. Y., Poss, K. D. Cardiac regeneration. Curr Top Dev Biol. 100, 319-344 (2012).
- Souders, C. A., Bowers, S. L., Baudino, T. A. Cardiac fibroblast: the renaissance cell. Circ Res. 105, 1164-1176 (2009).
- Ieda, M., et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 16, 233-244 (2009).
- Moore-Morris, T., et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 124, 2921-2934 (2014).
- Addis, R. C., et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 60, 97-106 (2013).
- Chen, J. X., et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 111, 50-55 (2012).
- Christoforou, N., et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 8, e63577 (2013).
- Fu, J. D., et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 1, 235-247 (2013).
- Hirai, H., Katoku-Kikyo, N., Keirstead, S. A., Kikyo, N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 100, 105-113 (2013).
- Ieda, M., et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 142, 375-386 (2010).
- Inagawa, K., et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res. 111, 1147-1156 (2012).
- Islas, J. F., et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U.S.A. 109, 13016-13021 (2012).
- Jayawardena, T. M., et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 110, 1465-1473 (2012).
- Muraoka, N., et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. Embo J. 33, 1565-1581 (2014).
- Nam, Y. J., et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 141, 4267-4278 (2014).
- Nam, Y. J., et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 110, 5588-5593 (2013).
- Protze, S., et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 53, 323-332 (2012).
- Qian, L., Berry, E. C., Fu, J. D., Ieda, M., Srivastava, D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 8, 1204-1215 (2013).
- Wang, H., et al. Small Molecules Enable Cardiac Reprogramming of Mouse Fibroblasts with a Single Factor, Oct4. Cell Rep. , (2014).
- Wang, L., et al. Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming. Circ Res. , (2014).
- Song, K., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 485, 599-604 (2012).
- Qian, L., et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 485, 593-598 (2012).
- Mathison, M., et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc. 1, e005652 (2012).
- Srivastava, D., Ieda, M., Fu, J., Qian, L. Cardiac repair with thymosin beta4 and cardiac reprogramming factors. Ann NY Acad Sci. 1270, 66-72 (2012).
- Muraoka, N., Ieda, M. Stoichiometry of transcription factors is critical for cardiac reprogramming. Circ Res. , 116-216 (2015).
- Srivastava, D., Ieda, M. Critical factors for cardiac reprogramming. Circ Res. 111, 5-8 (2012).

