In Vitro Transcription Assays and Their Application in Drug Discovery
Summary
In this manuscript, we describe a protocol to functionally examine transcription and the inhibitory activity of antibacterial agents targeting bacterial transcription.
Abstract
In vitro transcription assays have been developed and widely used for many years to study the molecular mechanisms involved in transcription. This process requires multi-subunit DNA-dependent RNA polymerase (RNAP) and a series of transcription factors that act to modulate the activity of RNAP during gene expression. Sequencing gel electrophoresis of radiolabeled transcripts is used to provide detailed mechanistic information on how transcription proceeds and what parameters can affect it. In this paper we describe the protocol to study how the essential elongation factor NusA regulates transcriptional pausing, as well as a method to identify an antibacterial agent targeting transcription initiation through inhibition of RNAP holoenzyme formation. These methods can be used a as platform for the development of additional approaches to explore the mechanism of action of the transcription factors which still remain unclear, as well as new antibacterial agents targeting transcription which is an underutilized drug target in antibiotic research and development.
Introduction
Transcription is the process in which RNA is synthesized from a specific DNA template. In eukaryotic cells there are three distinct RNAPs: RNAP I transcribes rRNA precursors, RNAP II is responsible for the synthesis of mRNA and certain small nuclear RNAs, and the synthesis of 5S rRNA and tRNA is performed by RNAP III. In bacteria, there is only one RNAP responsible for the transcription of all classes of RNA. There are three stages of transcription: initiation, elongation and termination. Transcription is one of the most highly regulated processes in the cell. Each stage in the transcription cycle represents a checkpoint for the regulation of gene expression1. For initiation, RNAP has to associate with a sigma factor to form holoenzyme, which is required to direct the enzyme to specific sites called promoters2 to form an open promoter complex. Subsequently, a large suite of transcription factors are responsible for the regulation of RNAP activities during the elongation and termination phases. The transcription factor examined here is the highly conserved and essential protein, NusA. It is involved in regulating transcription pausing and termination, as well as anti-termination during rRNA synthesis3-5.
In vitro transcription assays have been developed as powerful tools to study the complex regulatory steps during transcription6. In general, a linear fragment of DNA including a promoter region is required as the template for transcription. The DNA template is usually generated by PCR or by linearizing a plasmid. Purified proteins and NTPs (including one radiolabeled NTP for detection purposes) are then added and product analyzed following the required period of incubation. Using appropriate templates and reaction conditions, all stages of transcription have been examined using this approach which has enabled detailed molecular characterization of transcription over the last half century7. In combination with information on the 3-dimensional structure of RNAP it has also been possible to probe the molecular mechanism of transcription inhibition by antibiotics and antibiotic leads, and use this information in the development of new, improved drugs8-10.
In this work we provide examples of how transcription assays can be used to determine the mechanism of regulation by transcription elongation/termination factor NusA, and how the mechanism of action of a novel transcription initiation inhibitor lead can be determined.
Protocol
Caution: Experiments involve the use of the radioactive isotope 32P and no work should be undertaken until all appropriate safety conditions have been met. Generally personnel are required to attend a safety course and undergo supervised practice prior to experiments using radioactive reagents. Please wear personal protection equipment (thermoluminescent dosimeter, safety glasses, gloves, radiation room lab coats, full length pants, closed-toe shoes) when performing the reaction.
1. Preparation of Assay Materials
- Obtain the Proteins Required for Transcription assay.
- For the experiment described here, overproduce and purify E. coli RNAP in the laboratory as described11, or obtain from a commercial source (Materials List).
Note: Both provide similar results. Other methods for purification of native or affinity labeled RNAP can also be used12,13. Sigma/NusA factors can be obtained as described in previous work14,15.
- For the experiment described here, overproduce and purify E. coli RNAP in the laboratory as described11, or obtain from a commercial source (Materials List).
- Preparation of Diethyl Pyrocarbonate (DEPC) Treated Water
Note: Alternatively, RNase-free water can be obtained from commercial sources (Materials List).- Add 1 ml diethyl pyrocarbonate (DEPC) to 1 L of purified water (0.1% v/v) and mix overnight.
- Autoclave DEPC water and tips twice.
- DNA Template Preparation
- Purify plasmid pSG115416 and/or pIA22617 using a plasmid purification kit (Materials List) according to manufacturer's instructions.
- To amplify the template for run-off transcription assays (a 360 bp fragment encompassing the Pxyl promoter region), assemble a PCR reaction on ice including 25 μl 2x PCR mix (Materials List), 2 μl each of the primers (5 µM): 5′-CAAAGCCTGTCGGAATTGG-3′ (forward) and 5′-CCCATTAACATCACCATC-3′ (reverse), 1 μl pSG1154 plasmid preparation (250 µM) and 20 μl purified water).
- Or alternatively, for single round transcription assays, amplify a 447 bp fragment encompassing the λ PR promoter region from pIA22617. Set up a PCR reaction on ice using 25 μl 2x PCR mix, 2 μl each of the primers (5 µM): 5′-GTGCTCATACGTTAAATCT-3′ (forward) and 5′-CAGTTCCCTACTCTCGCATG-3′ (reverse), 1 μl pIA22617 plasmid preparation (250 µM) and 20 μl purified water).
Note: This template lacks cytosine until the +26 nt position, which is the 26th nucleotide from the transcription start site. Therefore, transcription using this template will allow initiation, promoter clearance and stalling at the +26 nt to form a stable transcription elongation complex (EC) when only ATP, UTP and GTP are present in the reaction.
- Or alternatively, for single round transcription assays, amplify a 447 bp fragment encompassing the λ PR promoter region from pIA22617. Set up a PCR reaction on ice using 25 μl 2x PCR mix, 2 μl each of the primers (5 µM): 5′-GTGCTCATACGTTAAATCT-3′ (forward) and 5′-CAGTTCCCTACTCTCGCATG-3′ (reverse), 1 μl pIA22617 plasmid preparation (250 µM) and 20 μl purified water).
- Set up a PCR reaction with the following conditions: 95 °C for 30 sec, 50 °C for 30 sec, 72 °C for 30 sec, repeating for 30 cycles.
- Purify the product using a PCR clean-up kit (Materials List) according to manufacturer's instructions and elute in 100 μl transcription buffer for the transcription assay.
- Determine the template DNA concentration using a fluorospectrometer (Materials List).
- NTP Preparation
Note: NTPs can be obtained from different commercial sources (Materials List).- Dissolve NTPs (≥99% pure) in DEPC-treated water to make 1 M stock, aliquot 1 ml of the solution into 1.5 ml tubes (50 μl in each tube) and store at -20 °C.
- Order α-32P UTP (3,000 Ci/mmol) prior to the experiment. 32P has a short half-life of 14.29 days.
Note: Using alpha-labeled UTP avoids comparing the intensity of bands with different length.
- RNA Ladder Preparation
- Assemble the following reaction at room temperature: 0.5 μg RNA Template Mix (Materials List), 80 mM HEPES buffer pH7.5, 12 mM MgCl2, 10 mM NaCl, 10 mM DTT, 2 mM ATP/CTP/GTP, 0.5 mM UTP, 2 μl α-32P UTP, 0.5 μl T7 RNAP (Materials List), DEPC-treated water to 20 μl.
- Allow reaction to proceed at 37 °C for 1 hr. Use immediately or store at -80 °C.
2. Preparation of RNA Sequencing Gel
- Gel Preparation
- Clean the plates from a sequencing gel apparatus (Materials List) twice with 70% ethanol and purified water, and assemble the cell with a comb and two spacers.
Note: One plate can be coated with siliconizing reagent (Materials List) to facilitate gel removal in Step 4.2. - Dissolve 33.6 g urea in 22 ml water and 16.4 ml of 5x Tris/Borate/EDTA (TBE) buffer (54 g tris(hydroxymethyl)aminomethane; 27.5 g boric acid; 10 ml of 1 M ethylenediaminetetraacetic acid (EDTA), pH 7.4 in 1 L water) using a magnetic stirrer.
- Add 16 ml of 40% Acrylamide/bis-acrylamide solution (19:1) to the urea solution and filter through a 0.45 µm filter.
- Add 514 µl 10% (w/v) freshly prepared ammonium persulfate in water followed by 51.4 µl N,N,N′,N′-Tetramethylethylenediamine (TEMED) to the filtered solution.
- Gently invert the gel solution to avoid excessive aeration and inject immediately and smoothly into the horizontally positioned pre-packed sequencing cell from the bottom injection hole. Take care to avoid making bubbles during injection.
- Allow 1-1.5 hr until the gel is completely set before use in a transcription assay. The gel can be stored at room temperature for two or three days when wrapped with water-wet tissue on the top of the sequencing cell.
- Clean the plates from a sequencing gel apparatus (Materials List) twice with 70% ethanol and purified water, and assemble the cell with a comb and two spacers.
3. Transcription Reaction
- Transcription Elongation Pausing Assay
- Formation of a transcription elongation complex
- Add 1 µl of RNAP core enzyme (1 µM stock in transcription buffer) to a 1.5 ml tube.
- Add 1 µl of purified sigma-70 factor (σ70; 3 µM stock in transcription buffer) to the RNAP and incubate on ice for 10 min to form RNAP holoenzyme.
- Add 3 µl 500 nM purified template DNA (λ PR promoter from the pIA226 PCR product) in transcription buffer to the RNAP holoenzyme and incubate at 37 ºC for 5 min to form the open complex.
- Prepare a mixture of reaction components in another 1.5 ml tube using 5 µl of 10x transcription buffer (Materials List: 400 mM Tris-HCl, 1.6 M KCl, 100 mM MgCl2, 10 mM DTT, 50% glycerol, pH 7.9 in DEPC treated water), 2 µl of 250 µM each GTP and UTP, 1 µl of 1 mM ATP, and 1 µl of α-32P UTP (3,000 Ci/mmol). Add DEPC treated water to 41 µl.
Note: RNase inhibitor (Materials List) can be used to minimize the RNase contamination. - Transfer the mixture solution to the tube containing the open complex and incubate at 37 ºC for 15 min to form the transcription elongation complex stopped at +26 nt.
- Move the reaction to room temperature (~21 °C) and add 1 µl of 2.5 mg/ml rifampicin to a final concentration of 50 µg/ml.
- Add 1 µl of 0.1 mM purified NusA to the reaction mixture and incubate for 5 min at room temperature.
- Add 2 µl of 250 µM CTP into the reaction mixture to make a final volume of 50 µl and measure the reaction time using a timer.
- Transfer 5 µl of the reaction solution into 2 µl of RNA gel loading buffer (95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol) at 30, 60, 120, 240, 480, and 1,200 sec to quench the reaction.
- Dispose the 32P UTP tips and tubes in a radioactive waste container.
- Formation of a transcription elongation complex
- Transcription Initiation Assay
- Formation of the Open Complex
- Dilute purified σ70 to 0.5 µM.
- Repeat Steps 3.1.1.1 to 3.1.1.3 using 0.5 µM (instead of 3 µM) purified σ70, and the DNA template (the Pxyl promoter region from pSG1154 PCR product).
- Inhibition of transcription initiation assay
- Add the inhibitor designed to prevent RNAP holoenzyme formation to RNAP core enzyme prior to addition of σ70 (after step 3.1.1.1), followed by Steps 3.1.1.2 and 3.1.1.3 to examine whether the inhibitor is able to disrupt holoenzyme formation.
- Alternatively, after Step 3.1.1.2, add the inhibitor to examine whether it is able to competitively inhibit σ70 binding to RNAP.
- Use the same amount of DMSO, the solvent of the inhibitor, instead of the inhibitor stock in control reactions.
- Completion of the reaction
- Dissolve heparin in water to a stock concentration of 1 mg/ml and stock at -20 °C.
Note: Heparin is a competitor of DNA that can sequester RNAP that has not formed an open complex. Once the open complex is formed, heparin cannot enter the DNA binding site of RNAP and inhibit transcription from pre-formed open complex. Therefore, heparin is only required for single-round assays; it is not needed for multi-round assays. - Prepare the reaction mixture using 5 µl of 10x transcription buffer (Materials List), 1 µl of 1 mg/ml heparin solution, 1 µl of 10 mM each ATP, GTP, and CTP, 2.5 mM UTP, and 1 µl of α-32P UTP (3,000 Ci/mmol). Add DEPC treated water to 45 µl.
- Mix the reaction mixture with the transcription open complex, pulse spin and incubate at 37 °C for 20 min.
- Transfer 10 µl of the reaction solution into 5 µl of RNA gel loading buffer (95% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol) to quench the reaction.
- Dissolve heparin in water to a stock concentration of 1 mg/ml and stock at -20 °C.
- Formation of the Open Complex
4. RNA Gel Running and Development
- Running the Denaturing Polyacrylamide Gel
- Run the gel at 50 ºC and 50 W in 1x TBE buffer for approximately 1.5 hr until the temperature of the gel and the buffer is stable at 50 ºC.
- Heat samples at 90 ºC for 30 sec and move them onto ice prior loading onto the gel.
- Load samples into the wells on the top of the gel alongside one lane containing 2 µl RNA ladder (1:100 dilution).
- Run the gel at 50 W and 50 ºC in 1x TBE buffer until the bromophenol blue dye reaches ~2/3 down the gel.
- Gel Treatment
- Open slowly and carefully separate the upper plate from the sequencing cell. Take great care to avoid breaking the gel.
- Cut chromatography paper (Materials List) to match the gel size from the top to the bromophenol blue dye. Use a Geiger counter to confirm the approximate position of radioactively labelled RNA transcripts.
- Carefully cover the gel with the filter paper, and press firmly till the gel adheres to the filter paper. Cut off and discard the bottom of the gel to radioactive waste as the unincorporated NTPs run to the bottom of the gel, which can be very hot.
- Place a similar sized piece of filter paper on the drying bed of a vacuum-drying machine, and carefully lay the gel-coated paper onto it with the gel side up.
- Cover the gel with a plastic wrap and dry at 60 °C for 1.5 hr.
- Wrap the other side of the gel coated filter paper with plastic wrap, and place on a photostimulable phosphor plate in the imaging case, with the appropriate exposure time, which can vary from 1 hr to overnight depending on the reaction efficiency.
- Image the phosphor plate using an imager scanner according to manufacturer's instructions. Determine the relative band intensity using ImageJ software according to the software manual.
- Use a Geiger counter to check the radioactive contamination in the work area, wipe the suspected items and area with tissue and a cleaning agent and dispose the tissue in the disposal container.
Representative Results
Transcription efficiency can be determined by measuring the level of radiation in the bands at different time points. The pausing assay to test the function of NusA factor enabled visualization of the pause, termination, and run-off products (Figure 1A). In the presence of the N-terminal domain of NusA (NusA NTD; amino acid residues 1-137), appearance of the RNA products was significantly delayed compared to the control experiment lacking NusA. Deletion of amino acid residues 104-137 (Helix 3) of the NusA fragment, or alteration to alanine of a series of amino acids (residues K36, K37, R104, Q108, K111, Q112, Q116, and R119) resulted in the complete loss of NusA pause activity. Helix 3 residues R104 and K111 were shown to be essential for NusA NTD pause activity and with the R104A and K111A constructs giving similar pause half-life values to the control where NusA NTD was absent (Figure 1B). It should be noted that deletion of Helix 3 or alteration of these two amino acid residues has no effect on NusA binding to RNAP15. The result suggested that R104 and K111 were the key amino acids required for regulation of NusA pause activity.
Similarly, as shown in Figure 2A, the dose-response curve demonstrated the inhibition of in vitro transcription (multi-round) using different concentrations of a newly identified bacterial transcription inhibitor C514. C5 was identified following an in silico screen from project to rationally target the interaction of σ70 with its major binding site on RNAP called the Clamp-Helix region18. With addition of C5 into the transcription reaction, this compound can compete against σ70 for binding to RNAP, and thus inhibited transcription. Mechanistically, addition of C5 to RNAP followed by σ70 factor to the reaction mixture was significantly more efficient for transcription inhibition than the experiment using C5 after the formation of RNAP holoenzyme (Figure 2B). These results demonstrated that σ70 couldn't displace C5 bound to RNAP, or that C5 could not efficiently displace σ70 bound to RNAP in a single-round assay.
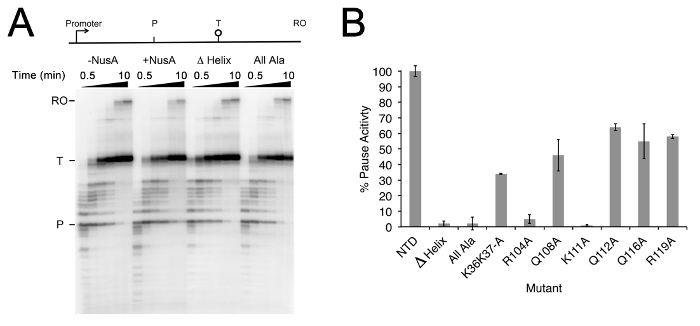
Figure 1: Effects of Mutant NusA NTD in transcription. (A) Transcription pause assays without NusA, using NusA NTD (+ NusA), NusA NTD with helix 3 deleted (Δ Helix), and NusA NTD with alanine substitutions at residues K36, K37, R104, Q108, K111, Q112, Q116, and R119 (All Ala). P, pause site; T, termination site; RO, run-off. Wedges above the gel demonstrate the time points sampled (0.5, 1, 1.5, 2, 5 and 10 min)15; (B) Pause activity of half-lives of mutant NusA NTD relative to wild-type protein. Average of 3 experiments with SD. This figure has been modified from Nucleic Acids Res. 43 (5), 2829-2840 (2015). Please click here to view a larger version of this figure.
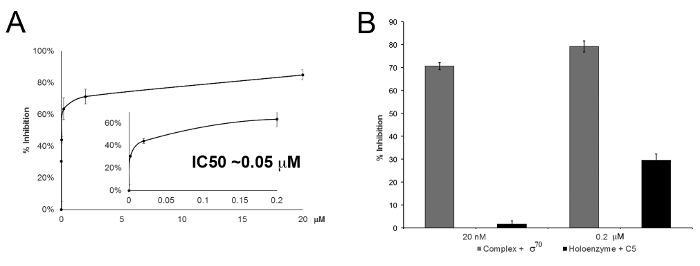
Figure 2: Transcription inhibition by compound C5. (A) Inhibition curve of E. coli RNAP transcription by compound C5 in a multi-round assay14. Percent inhibition of transcription was determined by the RNA product relative to a control (no C5 present) against the concentration of C5 used in the reaction. (B) Percent inhibitory activity of compound C5 in single-round transcription assays with addition of C5 before (complex + σ70; gray) and after (Holoenzyme + C5; black) holoenzyme formation. This figure has been modified from ACS Infect. Dis. 2, 39-46 (2016). Please click here to view a larger version of this figure.
Discussion
In all organisms, transcription is a tightly regulated process. In vitro transcription assays have been developed to provide a platform for testing the effects of transcription factors, small molecules and transcription inhibitors. In this method paper, an assay for general bacterial transcription was described. Transcription assays combined with sequencing gel electrophoresis of transcripts are very important for mechanistic studies as they allow visualization of all the transcription products along a timeline. As a result, this method is capable of identifying the influence of minor changes in reaction conditions and revealing a plausible mechanism of action. Furthermore, it becomes feasible to design specific transcription assays with regard to the function of transcription factors with unknown mechanisms of action.
In this paper we also described the method to confirm the mechanism of action of an inhibitor of RNAP holoenzyme formation. The method described here can display the change of each RNA transcript with time, and with appropriate modification can enable the testing of antibacterial agents against each step of transcription as appropriate. The multi-round assay can be used as a relative simple screening method to identify new transcription targeting inhibitors, while the single round assay can be used to study the mechanism of inhibitors, such as competitive/non-competitive properties. Note that small molecular aggregation can cause false positive hits as non-competitive inhibitors in the drug discovery process and the single round mechanistic study may provide a comprehensive approach for hit-to-lead evaluation. As a result, this controlled transcription process is capable of precisely revealing how an inhibitor affects transcription, which can help with rationalized de novo drug design. Together with high-resolution information on inhibitor binding sites by X-ray crystallographic studies, structure-based rational drug design will be highly amenable for application in drug discovery targeting transcription.
Although high-throughput results cannot be obtained with in vitro transcription assays, they are extremely useful in dissecting the precise mechanisms associated with the control of transcription. The components and assay conditions could be readily modified for individual needs. Various factors (e.g., NusA)4, 15, small molecules (e.g., ppGpp)19 and/or transcription inhibitors (e.g., rifampicin)20 of interest can be added to test their functions in transcription regulation. The DNA template should be individually designed, with careful choice of strong promoters, pause sequence and terminators. In order to achieve high reproducibility of the assay, proteins must be highly purified and free from residual RNase activity, and the transcription buffer, DNA template, and stock solution of NTP substrates must be made in DEPC treated water. The use of radioactive NTPs was described in this paper, however fluorescent nucleotides can also be employed21. Quality of the DNA template is critical for the success of transcription assay. Sufficient spacing between the promoter/pause/terminator must be included, allowing the various transcription products to be properly separated and examined. The sequencing gel must be properly poured and relatively fresh TBE buffer must be used for electrophoresis to achieve highest possible resolution of RNA transcripts. If degradation of transcription product is noticed, high quality RNase inhibitor must be incorporated in the transcription reaction mixture. Tubes and pipette tips must be double autoclaved and gloves must be worn at all times when performing the assay, in order to minimize introduction of RNase.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work acknowledges a Faculty Early Career Grant from the University of Newcastle (CM).
Materials
| Obtain the proteins required for transcription assay | |||
| E. coli RNAP | Epicentre | S90250 | |
| Preparation of DEPC-treated water | |||
| diethyl pyrocarbonate (DEPC) | Sigma-Aldrich | D5758 | |
| RNase-free water | |||
| Ambion Nuclease-Free Water | ThermoFisher | AM9937 | |
| DNA template preparation | |||
| Wizard Plus SV Minipreps DNA Purification System | Promega | A1330 | |
| ACCUZYME Mix | Bioline | BIO-25028 | |
| PCR primers | |||
| Wizard SV Gel and PCR Clean-Up System | Promega | A9281 | |
| NanoDrop 3300 fluorospectrometer | Thermo Scientific | ND-3300 | |
| NTP Preparation | |||
| ATP | Sigma-Aldrich | A6559 | |
| UTP | Sigma-Aldrich | U1006 | |
| GTP | Sigma-Aldrich | G3776 | |
| CTP | Sigma-Aldrich | C9274 | |
| High Purity rNTPs | GE Healthcare | 27-2025-01 | |
| α-32P UTP | PerkinElmer | BLU007C001MC | Radioactive compound |
| RNA ladder preparation | |||
| Novagen Perfect RNA Marker Template Mix 0.1–1 kb | Millipore | 69003 | |
| HEPES | Sigma-Aldrich | H7006 | |
| Sodium chloride | Sigma-Aldrich | S7653 | |
| Magnesium chloride | Sigma-Aldrich | M8266 | |
| DTT | Sigma-Aldrich | DTT-RO | |
| T7 RNAP | Promega | P2075 | |
| Gel preparation | |||
| Sequi-Gen GT nucleic acid sequencing cell | Bio-Rad | 165-3804 | |
| Sigmacote | Sigma-Aldrich | SL2 | |
| urea | Sigma-Aldrich | U6504 | |
| tris(hydroxymethyl)aminomethane | Sigma-Aldrich | 154563 | |
| boric acid | Sigma-Aldrich | B7901 | |
| ethylenediaminetetraacetic acid | Sigma-Aldrich | ED | |
| 40% Acrylamide/bis-acrylamide | Sigma-Aldrich | A9926 | |
| ammonium persulfate | Sigma-Aldrich | A3678 | |
| N,N,Nʹ′,Nʹ′-Tetramethylethylenediamine (TEMED) | Sigma-Aldrich | T9281 | |
| N,N,N”,N”-Tetramethylethylenediamine (TEMED) | Sigma-Aldrich | T9281 | |
| Transcription Assay | |||
| Potassium chloride | Sigma-Aldrich | P9541 | |
| glycerol | Sigma-Aldrich | G5516 | |
| rifampicin | Sigma-Aldrich | R3501 | |
| formamide | Sigma-Aldrich | F9037 | |
| bromophenol blue | Sigma-Aldrich | B0126 | |
| xylene cyanol | Sigma-Aldrich | X4126 | |
| heparin | Sigma-Aldrich | 84020 | |
| RNasin Ribonuclease Inhibitor | Promega | N2511 | |
| Transcription buffer | |||
| Tris base | Sigma-Aldrich | T1503 | |
| Potassium chloride | Sigma-Aldrich | P9541 | |
| Magnesium chloride | Sigma-Aldrich | M2393 | |
| DTT | Sigma-Aldrich | DTT-RO | |
| glycerol | Sigma-Aldrich | G5516 | |
| Filter paper | |||
| Whatman 3MM Chr Chromatography Paper | Fisher Scientific | 05-714-5 | |
| Radioactive decontaminant | |||
| Decon 90 | decon | decon90 | |
| Gel Treatment | |||
| Typhoon Trio+ imager | GE Healthcare Life Sciences | 63-0055-89 |
References
- Mooney, R. A., Artsimovitch, I., Landick, R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 180 (13), 3265-3275 (1998).
- Burgess, R. R., Anthony, L. How sigma docks to RNA polymerase and what sigma does. Curr. Opin. Microbiol. 4 (2), 126-131 (2001).
- Gusarov, I., Nudler, E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 107 (4), 437-449 (2001).
- Ha, K. S., Toulokhonov, I., Vassylyev, D. G., Landick, R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 401 (5), 708-725 (2010).
- Yang, X., Lewis, P. J. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 7 (3), 272-275 (2010).
- Artsimovitch, I., Henkin, T. M. In vitro approaches to analysis of transcription termination. Methods. 47 (1), 37-43 (2009).
- Decker, K. B., Hinton, D. M. Transcription regulation at the core: Similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 67, 113-139 (2013).
- Artsimovitch, I., Vassylyev, D. G. Is it easy to stop RNA polymerase?. Cell Cycle. 5 (4), 399-404 (2006).
- Murakami, K. S. Structural biology of bacterial RNA polymerase. Biomolecules. 5 (2), 848-864 (2015).
- Ma, C., Yang, X., Lewis, P. J. Bacterial transcription as a target for antibacterial drug development. Microbiol. Mol. Biol. Rev. 80 (1), 139-160 (2016).
- Yang, X., Ma, C., Lewis, P. A vector system that allows simple generation of mutant Escherichia coli RNA polymerase. Plasmid. 75, 37-41 (2014).
- Burgess, R. R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J. Biol. Chem. 244, 6160-6167 (1969).
- Artsimovitch, I., Svetlov, V., Murakami, K. S., Landick, R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 278 (14), 12344-12355 (2003).
- Ma, C., Yang, X., Lewis, P. J. Bacterial transcription inhibitor of RNA polymerase holoenzyme formation by structure-based drug design: From in silico screening to validation. ACS Infect. Dis. 2, 39-46 (2016).
- Ma, C., Mobli, M., Yang, X., Keller, A. N., King, G. F., Lewis, P. J. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 43 (5), 2829-2840 (2015).
- Lewis, P. J., Marston, A. L. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene. 227 (1), 101-110 (1999).
- Artsimovitch, I., Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. 97 (13), 7090-7095 (2000).
- Vassylyev, D. G., et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 417, 712-719 (2002).
- Artsimovitch, I., et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 117 (3), 299-310 (2004).
- Bordier, C. Inhibition of rifampicin-resistant RNA synthesis by rifampicin-RNA polymerase complexes. FEBS lett. 45 (1-2), 259-262 (1974).
- Tang, G. Q., Anand, V. S., Patel, S. S. Fluorescence-based assay to measure the real-time kinetics of nucleotide incorporation during transcription elongation. J. Mol. Biol. 405 (3), 666-678 (2011).

