Visualization of the Axonal Projection Pattern of Embryonic Motor Neurons in Drosophila
Summary
This work details a standard immunohistochemistry method to visualize motor neuron projections of late stage-16 Drosophila melanogaster embryos. The filleted preparation of fixed embryos stained with FasII antibody provides a powerful tool to characterize the genes required for motor axon pathfinding and target recognition during neural development.
Abstract
The establishment of functional neuromuscular circuits relies on precise connections between developing motor axons and target muscles. Motor neurons extend growth cones to navigate along specific pathways by responding to a large number of axon guidance cues that emanate from the surrounding extracellular environment. Growth cone target recognition also plays a critical role in neuromuscular specificity. This work presents a standard immunohistochemistry protocol to visualize motor neuron projections of late stage-16 Drosophila melanogaster embryos. This protocol includes a few key steps, including a genotyping procedure, to sort the desired mutant embryos; an immunostaining procedure, to tag embryos with fasciclin II (FasII) antibody; and a dissection procedure, to generate filleted preparations from fixed embryos. Motor axon projections and muscle patterns in the periphery are much better visualized in flat preparations of filleted embryos than in whole-mount embryos. Therefore, the filleted preparation of fixed embryos stained with FasII antibody provides a powerful tool to characterize the genes required for motor axon pathfinding and target recognition, and it can also be applied to both loss-of-function and gain-of-function genetic screens.
Introduction
Precise and selective connections between motor axons and target muscles during embryonic development are essential for normal locomotion in Drosophila larvae. The embryonic patterning of 30 muscle fibers in each of the abdominal hemisegments A2-A7 is established by stage 161. The 36 motor neurons that are generated in the ventral nerve cord extend their axons into the periphery to innervate specific target muscles2. Motor axon pathfinding and target recognition can be visualized by immunohistochemistry with an antibody (mouse monoclonal antibody 1D4)3,4. Multiple images of the motor axon projection patterns in wildtype embryos are available on the web5. The 1D4 antibody labels all motor axons and three longitudinal axon fascicles on each side of the midline of the embryonic central nervous system (CNS)4,6 (Figure 1C and Figure 2A). Therefore, immunohistochemistry with FasII antibody provides a powerful tool for identifying genes required for neuromuscular connectivity for demonstrating the molecular mechanisms underlying motor axon guidance and target recognition.
In each of the abdominal hemisegments A2-A7, motor axons project and selectively fasciculate into two principal nerve branches, the segmental nerve (SN) and the intersegmental nerve (ISN)2,4, and a minor nerve branch, the transverse nerve (TN)7. The SN selectively defasciculates to give rise to two nerve branches called the SNa and SNc, whereas the ISN splits into three nerve branches called the ISN, ISNb, and ISNd2,4. Among them, ISN, ISNb, and SNa motor axon projection patterns are most precisely visualized when late stage-16 embryos are stained with FasII antibody and are filleted (Figure 1C and Figure 2A). The ISN motor neurons extend their axons to innervate dorsal muscles 1, 2, 3, 4, 9, 10, 11, 18, 19, and 202,4 (Figure 2A). The ISNb motor neurons innervate ventrolateral muscles 6, 7, 12, 13, 14, 28, and 302,4 (Figure 2A and 2B). The SNa nerve branch projects to innervate lateral muscles 5, 8, 21, 22, 23, and 242,4 (Figure 2A). The TN, which consists of two motor axons, projects ipsilaterally along the segmental border to innervate muscle 25 and makes synapses with the lateral bipolar dendritic neuron (LBD) in the periphery7 (Figure 2A). These target muscle innervations require not only selective defasciculation of motor axons at specific choice points, but also target muscle recognition. In addition, some putative mesodermal guidepost cells that act as intermediate targets were found in both the ISN and SNa pathways, but not along the ISNb pathway4. This might suggest that ISNb motor axon pathfinding can be regulated in a distinct manner compared to ISN and SNa motor axon guidance, and it also indicates that peripheral motor axon guidance provides an attractive experimental model to study the differential or conserved roles of a single guidance cue molecule8.
This work presents a standard method to visualize the axonal projection patterns of embryonic motor neurons in Drosophila. The described protocols include how to dissect fixed embryos stained with 1D4 antibody and processed in 3,3′-diaminobenzidine (DAB) for filleted preparations. One critical advantage of the flat preparations of fixed embryos is the better visualization of the axonal projections and muscle patterns in the periphery. Furthermore, this work also shows how to genotype fixed embryos to sort the desired mutant embryos using the LacZ staining method.
Protocol
1. Preparation
- Prepare 500 mL of phosphate-buffered saline (PBS) with t-Octylphenoxypolyethoxyethanol (PBT) solution by adding 0.5 g of bovine serum albumin (BSA) and 0.5 mL of t-Octylphenoxypolyethoxyethanol (see the Table of Materials) to 500 mL of 1X PBS and stirring for at least 30 min. Store at 4 °C. Use when relatively fresh and store the solution in a clean bottle.
- Make 10 mL of 4% paraformaldehyde by adding 2.5 mL of 16% stock paraformaldehyde solution and 1 mL of 10x PBS to 6.5 mL of deionized water. Store at 4 °C and use within one week.
CAUTION: Avoid skin contact with paraformaldehyde solution because it is highly toxic. - Make X-Gal substrate (10% w/v X-Gal in dimethyl sulfoxide (DMSO)) by dissolving 0.1 g of X-Gal substrate per 1 mL of DMSO. Store at -20 °C in 100 µL aliquots.
- Make X-Gal staining solution using 10 mM phosphate buffer (pH 7.2), 150 mM NaCl, 1 mM MgCl2, 3 mM K4[FeII(CN)6], 3 mM K3[FeIII(CN)6], and 0.3% t-Octylphenoxypolyethoxyethanol. Store at 4 °C in the dark.
- Make 3% hydrogen peroxide solution by diluting 30% stock hydrogen peroxide 1:10 with deionized water.
- Make 3,3′-diaminobenzidine (DAB) solution by completely dissolving 1 tablet (10 mg) of DAB in 35 mL of fresh PBT solution. Filter through a 0.2-µm syringe filter and store at -20 °C in 910 µL aliquots.
CAUTION: Dispose of all DAB waste in bleach to destroy the DAB. - Make egg plates with apple juice. Add 35 g of agar and 1 L of deionized water to a 2 L flask. Add 33 g of sucrose, 2 g of tegosept, and 375 mL of apple juice to a 1- L flask. Melt them by autoclaving for 30 min. Allow both solutions to cool to around 60 °C. Combine and mix these solutions well by stirring. Pour ~11 mL of combined solution per 60 mm culture dish.
- Make yeast paste by blending baker yeast into deionized water (1:0.8 ratio) and store at 4 °C.
2. Embryo Collection (Day 1)
- Collect young female and male flies (younger than 5 days), and for 1-2 days, maintain them in a plastic beaker cage with an egg plate (60 mm x 15 mm) that contains a small amount of yeast paste in the center.
- For the optimal collection of late stage-16 embryos, set up a bottle cage with flies (> 50) at around 5 PM and let them lay eggs at 25 °C for 3 h.
- Transfer the flies to a fresh food bottle.
- Close the egg plate with the lid and incubate it upside down at 25 °C in a humidified incubator (~70% humidity) overnight.
3. Preparation of Embryos for Immunostaining (Day 2)
- Add 1.8 mL of PBT solution to the egg plate around 10:20 AM and use a cotton swab to loosen the embryos from the plate while tilting it.
Note: By gently mashing the yeast paste using a cotton swab, dissolve it in case a large number of embryos are found on it. Begin collecting the embryos ~40 min before fixation (around 11:00 AM). - Transfer the embryos from the egg plate to a 1.5-mL microtube using a 1-mL pipette tip.
- Allow the embryos to settle down and aspirate the PBT solution as much as possible. Rinse the embryos twice with 1 mL of PBT solution. Aspirate the PBT solution as much as possible.
- Fill the tube with 1 mL of 50% bleach (i.e., diluted in deionized water) and dechorionate the embryos by incubating them on a nutator for 3 min at room temperature (RT).
Note: This step does not kill the dechorionated embryos. - Allow the dechorionated embryos to settle down, aspirate the 50% bleach as much as possible, and rinse the embryos 3 times with 1 mL of PBT solution.
- Add 0.5 mL of heptane and subsequently add 0.5 mL of 4% paraformaldehyde solution to the tube at RT at around 11:00 AM.
- Incubate the embryos on a nutator for 15 min at RT.
- Remove the 4% paraformaldehyde solution (the bottom layer).
- Add 0.5 mL of 100% methanol and devitellinize the embryos by shaking the tube vigorously for 30 s.
- Remove the heptane (i.e., the top layer).
- Add 0.5 mL of 100% methanol and shake the tube vigorously for 10 s.
- Allow the embryos to settle down by tapping the tube and aspirate the methanol as much as possible. Rinse the embryos with 1 mL of 100% methanol, shaking for less than 10 s.
Note: Extended exposure to methanol destroys β-Gal activity. - Remove the methanol and rinse the devitellinized embryos 3 times with 1 mL of PBT solution.
4. Genotyping the Embryos Using LacZ Staining (Day 2)
- Add 1 mL of PBT solution to the tube and incubate the tube in a 37 °C heat block or water bath for 15 min.
- During incubation, place X-Gal staining solution (1 mL per tube) at 65 °C in a water bath until it gets cloudy. Incubate it in a 37 °C water bath.
- Aspirate the PBT solution and add 1 mL of X-Gal staining solution and 20 µL of X-Gal substrate to the tube.
- Incubate the tube at 37 °C with nutation until a blue precipitate is evident.
Note: The incubation time for the 2nd and 3rd blue balancers in general ranges from 2-4 h. - Remove the X-Gal staining solution and rinse the embryos 3 times with 1 mL of RT PBT solution.
- Using a needle probe or forceps, hand-sort the embryos of the desired genotype (i.e., non-stained white embryos) with a light dissecting microscope (1.6X-2.5X objectives).
Note: Since some blue balancers, such as CyO, actin-LacZ and TM3, actin-LacZ, carry a transgenic construct expressing bacterial β-Gal (LacZ) under the control of the actin promoter, embryos stained blue in a ubiquitous manner contain one or two copies of blue balancer chromosomes. Therefore, non-stained white embryos are homozygous for the desired lethal allele.
5. Immunostaining of Embryos with Anti-Fasciclin II Antibodies (Day 2-3)
- Collect and transfer the embryos of the desired genotype (i.e., non-stained white embryos) to a 0.5 mL microtube using a 1 mL pipette tip.
Note: In this step, the embryos can be roughly staged according to morphological criteria, including the segmentation patterns of the cuticle and the structures of the head and tail9. During head involution, between 10 and 16 h after the eggs are laid, the embryo undergoes a pronounced reduction of the most anterior segment9. - Wash the embryos once with 0.4 mL of PBT solution and add 0.3 mL of blocking solution (5% normal goat serum and 5% DMSO in PBT solution).
- Block the embryos with nutation at RT for 15 min.
- Add 75 µL of anti-Fasciclin II antibody and incubate the tube with nutation at RT overnight.
- Wash the embryos 4 times with 0.4 mL of PBT solution for at least 20 min per wash.
- Add 0.3 mL of blocking solution (5% normal goat serum in PBT solution) containing goat anti-mouse-HRP antibody (2 µg/mL) and incubate the tube with nutation at RT overnight.
- Wash the embryos 6 times with 0.4 mL of PBT solution for at least 20 min per wash.
- Add 0.3 mL of DAB solution to the tube.
CAUTION: Wear gloves and place all DAB wastes/tubes/tips into bleach. - Add 2 µL of 3% hydrogen peroxide and incubate the tube in the dark with nutation at RT until the desired amount of precipitate is produced.
Note: The incubation time for 1D4 immunohistochemistry in general ranges from 0.5-1 h. - Wash the embryos 4 times with 0.4 mL of PBT solution.
CAUTION: Remove the DAB solution and the first two washes with a pipette and discard them in bleach. - Add 0.2 mL of mounting 70% glycerol/PBS solution to the tube and store at RT or 4 °C.
Note: Keep the tube away from light. Clearer preparations can be obtained by placing the embryos in a 90% glycerol/PBS solution10. However, it is more difficult to dissect embryos cleared in 90% glycerol/PBS solution than those cleared in 70% glycerol/PBS solution10.
6. Staging, Dissecting, Mounting, and Imaging the Embryos (Day 4)
- For staging, transfer the embryos equilibrated with 70% glycerol/PBS onto a glass slide.
- Collect late stage-16 embryos that have abdominal segment 7 (A7), A8, and A9 ISN nerves converging to touch each other or/and have midgut that is subdivided into three bands.
Note: Embryos stained with 1D4 antibody can be staged based on the projection patterns of ISN and ISNb motor axons. After exiting the CNS, A7, A8, and A9 ISN nerves of late stage-16 embryos converge to touch each other and subsequently diverge to extend far out to the dorsal muscles (arrowhead in Figure 1A). In mid-stage-16 embryos, however, A7 ISN nerves extend in parallel with A8/A9 nerves (square bracket in Figure 1B). In addition, the projection axes of A6 ISNb nerves (perpendicular to the ventrolateral muscles) in both hemisegments are roughly aligned with the posterior end of the CNS longitudinal axon fascicles (arrows and a line in Figure 1C). These morphological criteria somewhat vary between different genotypes. Alternatively, embryos can be staged based on gut morphology9,11. Prior to stage 17, the midgut has a heart-like shape, whereas the midgut is subdivided into three bands by stage 1712. - Dissect the embryos.
- Using a 1 mL syringe, move each embryo out of the glycerol drop and place the embryo ventral-face up. Cut off the anterior part at 1/4 of the body length and the most posterior region that is free of motor axons.
- Using a needle probe, move the end-cut embryo out of the glycerol drop, roll to orient it dorsal-side up, and place it horizontally in the field.
Note: When the embryo is moved out of the glycerol drop, a little bit of glycerol associated with the embryo makes it sticky. - Cut the embryo along the dorsal midline using a single very fine tungsten needle13. Make a small cut at the posterior end of the embryo and then continue to cut down the dorsal midline toward the anterior; cutting in the opposite direction is also fine.
- Using a needle probe, move the midline-cut embryo into the glycerol drop, place it dorsal-side up, and detach the gut from the body wall by unfolding each body wall in a ventrolateral (diagonal) direction (2.5X objective).
Note: Make sure that the dorsal midline is fully cut, resulting in the complete separation between the left and right body walls. - Carefully move the midline-cut embryo out of the glycerol drop and then lay two flaps of the body wall down on the glass slide using a fine needle probe (2.5X objective).
- Using a very fine tungsten needle, remove the internal organs from the dissected embryo by pushing them laterally (5X objective).
Note: A more detailed description of another similar dissection procedure can be found on the website5.
- For mounting, add 2-3 µL of 70% glycerol/PBS solution to clean, dissected embryos and, using a fine needle probe, transfer them to a new glass slide onto which 8 µL of 70% glycerol/PBS solution has been spread in the center (2.5X objective).
- Put on a coverslip (18 mm x 18 mm). Seal the edges of the coverslip with regular nail polish (either clear or colored is fine).
- Capture images at high resolution (20X, 40X, and 63X oil-immersion objectives) under a differential interference contrast (DIC) light microscope, according to the manufacturer's instructions.
Note: Better visualization and phenotypic characterization, especially for ISNb motor axons, can be produced by using a 63X oil-immersion objective.
Representative Results
Precise connections between motor axons and target muscles during neural development depend on selective axon-axon repulsion and target recognition at specific choice points4. In Drosophila, selective repulsion between motor axons is in part regulated by the combined action of class 1 and 2 semaphorins (Semas), including Sema-1a, Sema-2a, and Sema-2b8,14,15,16,17,18. Therefore, loss of Sema-1a function frequently results in the failure of ISNb axons to defasciculate at specific choice points, causing them to exhibit an abnormally thick morphology (arrowhead in Figure 2C). In wildtype embryos, at least 7 motor neurons extend and fasciculate their axons to form an ISNb nerve branch (Figure 2A). When the ISNb nerve branch reaches a choice point, which is located between muscles 6 and 7, two axons selectively defasciculate from the main ISNb bundle and subsequently innervate muscles 6 and 7 (Figure 2B). At the next choice point, which is located between muscles 6, 13, and 30, another three motor axons selectively defasciculate to innervate muscles 13, 14, and 30 (Figure 2B). The remaining two axons extend dorsally to reach the last choice point near the edge of muscle 12, thereby innervating muscle 12 in the opposite direction (Figure 2B). These results clearly show that the filleted preparation protocol provides a useful tool to study the genes required for motor axon guidance.
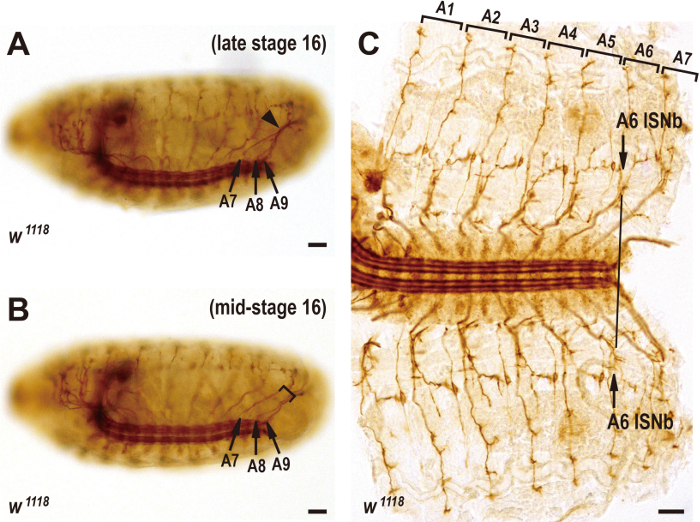
Figure 1: Staging of embryos based on the projection patterns of ISN and ISNb motor axons. (A) In late stage-16 wildtype embryos, A7, A8, and A9 ISN nerves (arrows) converge to touch each other (arrowhead). Anterior is left and ventral is down. The Scale bar = 15 µm. (B) In mid-stage-16 wildtype embryos, the A7 ISN nerve extends in parallel with A8/A9 nerves (square bracket). Arrows indicate A7, A8, and A9 ISN nerves. Anterior is left and ventral is down. The Scale bar = 15 µm. (C) Filleted preparation of late stage-16 wildtype embryos stained with FasII antibody. The projection axes (arrows) of the left and right A6 ISNb nerves are roughly aligned with the posterior end of the CNS longitudinal axon fascicles (black line). Abdominal segments A1-A7 are labelled at the top. Anterior is left. The Scale bar = 15 µm. Please click here to view a larger version of this figure.
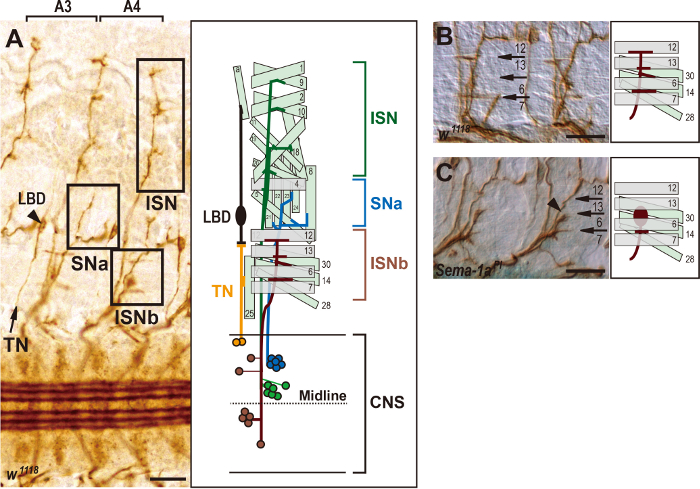
Figure 2: Motor axon projection patterns in wild-type and Sema-1a mutant embryos. (A–C) Filleted preparation of late stage-16 embryos stained with FasII antibody. Anterior is left and ventral is down. Schematic diagrams showing the axonal projection patterns represented in each panel. The Scale bars = 15 µm. (A) A bright field photograph of two sequential abdominal hemisegments, A3 and A4, in wildtype embryos. Three bilaterally symmetric longitudinal axon bundles are present in the CNS. The normal innervation patterns of ISNb, SNa, ISN, and TN motor axons are indicated by the boxes and an arrow in the periphery. The lateral bipolar dendrite neuron (LBD), which innervates the alary muscles, is indicated by an arrowhead. In the schematic diagram, the nerve branches and innervation patterns of the ISNb (in brown), SNa (in blue), ISN (in green), and TN (in yellow) motor neurons and their target muscles are presented. The cell body positions of these motor neurons in the CNS are also shown in their own colors. (B) In wildtype embryos, the ISNb axons project to the ventrolateral muscles, which are numbered, and innervate them at specific choice points (arrows in the DIC image). (C) In Sema-1a loss-of-function mutants, ISNb axons frequently fail to defasciculate from one another between muscles 6 and 13 (arrowhead in the DIC image). Please click here to view a larger version of this figure.
Discussion
The details of motor axon guidance defects are scored faster and with better accuracy by the filleted preparation of DAB-stained embryos than by laser scanning confocal microscopy of fluorescently labeled ones. Therefore, the filleted preparation of fixed and 1D4-stained embryos is best suited for the functional characterization of guidance cue molecules. Four major classes of guidance cues, including netrins, Slits, semaphorins (Semas), and ephrins, and their cognate receptors have evolutionarily conserved roles across worms, flies, and vertebrates20. Among roundabout (Robo) family molecules that act as receptors for Slits, Drosophila Robo was first identified to mediate repulsive guidance function in the CNS6,21. Semas are a large family of guidance molecules that bind to plexin-containing receptor complexes22. The axon guidance function of Semas was first discovered in the grasshopper CNS14. Subsequently, the class 1 transmembrane semaphorin Sema-1a was extensively characterized in Drosophila. Sema-1a, which functions not only as a ligand for plexin A, but also as a receptor for an unknown ligand, plays an important role in axon-axon repulsion at specific choice points during embryonic neural development15,16,17,18 (Figure 2C).
There are a few common troubleshooting steps that may occur with the current protocol. First, in steps 3.9-3.13, the embryos can be devitellinized with low efficiency if too many or too-small embryos are used in the methanol devitellinization procedure. In this case, it is recommended to use a bed volume of 40-80 µL of fixed embryos for devitellinization at step 3.5. Second, in steps 3.9-3.13, weak X-Gal staining can happen in case of longer exposure to methanol or incomplete heptane removal, since methanol used as a fixative destroys β-Gal activity. In this case, it is best that the devitellinization procedure using methanol in steps 3.9-3.13 is performed as quickly as possible. In addition, residual heptane that is solubilized in methanol may interfere with the staining; therefore, it is best to remove as much methanol as possible in steps 3.11-3.13.
The current filleted preparation protocol can be applied to loss-of-function and gain-of-function genetic screens to identify novel genes that are involved in motor axon pathfinding and target recognition. However, a disadvantage of this protocol is that it is quite time-consuming to dissect fixed embryos to generate filleted preparations. To overcome this problem, phenotypic characterization of the axonal projections can be monitored in a whole-mount preparation of 1D4-stained embryos, although this method provides low-resolution visualization of the axonal projections in the periphery. Nevertheless, the whole-mount preparation protocol has proven to be an efficient approach for screening axon guidance genes4. An alternative protocol using the live dissection of fluorescently labeled embryos was recently developed for systematic screening11. The live dissection protocol allows for relatively rapid preparation for phenotypic analysis11. However, this protocol is not adequate for embryos older than the beginning of stage 1711.
The filleted preparation protocol described here can be used to examine protein expression patterns with different antibodies. In addition, this protocol can also be applied to Drosophila embryos at various stages of development.
Divulgations
The authors have nothing to disclose.
Acknowledgements
I thank Alex L. Kolodkin, as I learned this filleted preparation protocol in his laboratory. I also thank Young Gi Hong for technical assistance. This study was supported by NRF-2013R1A1A4A01011329 (S.J.).
Materials
| Bovine Serum Albumin | Sigma-Aldrich | A7906 | |
| Triton X-100 | Sigma-Aldrich | X100 | t-Octylphenoxypolyethoxyethanol |
| 16% Paraformaldehyde Solution | Ted Pella | 18505 | |
| Sodium Chloride | Sigma-Aldrich | S5886 | |
| Potassium Chloride | Sigma-Aldrich | P5405 | |
| Sodium Phosphate Dibasic | Sigma-Aldrich | 30435 | |
| Sodium Phosphate Monobasic | Sigma-Aldrich | 71500 | |
| X-Gal Substrate | US Biological | X1000 | X-Gal (5-Bromo-4-chloro-3-indolyl-b-D-galactoside galactopyranoside) |
| Dimethyl Sulfxide | Sigma-Aldrich | D4540 | |
| Magnesium Chloride | Sigma-Aldrich | M8266 | |
| Potassium hexacyanoferrate(II) trihydrate | Sigma-Aldrich | P9387 | |
| Potassium hexacyanoferrate(III) | Sigma-Aldrich | 244023 | |
| Hydrogen Peroxide | Sigma-Aldrich | 216763 | |
| 3,3'-diaminobenzidine Tetrahydrochloride | Sigma-Aldrich | D5905 | |
| Agar | US Biological | A0930 | |
| Sucrose | Fisher Scientific | S5-3 | |
| Tegosept (Methy 4-Hydroxybenzoate) | Sigma-Aldrich | H5501 | |
| Culture Dish (60 mm) | Corning | 430166 | |
| Tricon Beaker | Simport | B700-100 | This is used to make a plastic beaker cage for embryo collection. |
| Yeast | Societe Industrielle Lesaffre | Saf Instant Yeast Red | |
| Cotton Swab (Wooden Single Tip Cotton PK100) | VWR | 14220-263 | |
| Eppendorf Tube (1.5 ml) | Sarstedt | #72.690 | |
| Bleach | The Clorox Company | Clorox | |
| Heptane | Sigma-Aldrich | 246654 | |
| Methanol | J.T. Baker | UN1230 | |
| Normal Goat Serum | Life Technologies | 16210-064 | |
| Anti-FasciculinII Antibody | Developmental Studies Hybridoma Bank | 1D4 anti-Fasciclin II | |
| Goat Anti-mouse-HRP Antibody | Jackson Immunoresearch | 115-006-068 | AffiniPure F(ab')2 Fragment Goat Anti-Mouse IgG+IgM (H+L) (min X Hu, Bov, Hrs Sr Prot |
| Glycerol | Sigma-Aldrich | G9012 | |
| Slide Glass | Duran Group | 235501403 | |
| Coverslip | Duran Group | 235503104 | 18 x 18 mm |
| 1 ml Syringe | Becton Dickinson Medical(s) | 301321 | |
| Tungsten Needle | Ted Pella | #27-11 | Tungsten Wire, ø0.13mm/6.1m (ø.005"/20 ft.) |
| Nutator (Mini twister) | Korean Science | KO.VS-96TWS | Alternatively, BD Clay Adams Brand Nutator (BD 421125) |
References
- Bate, M. The embryonic development of larval muscles in Drosophila. Development. 110 (3), 791-804 (1990).
- Landgraf, M., Thor, S. Development of Drosophila motoneurons: specification and morphology. Semin. Cell Dev. Biol. 17 (1), 3-11 (2006).
- Grenningloh, G., Rehm, E. J., Goodman, C. S. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 67 (1), 45-57 (1991).
- Vactor, D. V., Sink, H., Fambrough, D., Tsoo, R., Goodman, C. S. Genes that control neuromuscular specificity in Drosophila. Cell. 73 (6), 1137-1153 (1993).
- . Available from: https://www.its.caltech.edu/~zinnlab/motoraxons.html (2017)
- Seeger, M., Tear, G., Ferres-Marco, D., Goodman, C. S. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron. 10 (3), 409-426 (1993).
- Thor, S., Thomas, J. B. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 18 (3), 397-409 (1997).
- Roh, S., Yang, D. S., Jeong, S. Differential ligand regulation of PlexB signaling in motor neuron axon guidance in Drosophila. Int. J. Dev. Neurosci. 55, 34-40 (2016).
- Campos-Ortega, J. A., Hartenstein, V. . The embryonic development of Drosophila melanogaster. , (1985).
- Patel, N. H., Goldstein, L. S. B., Fyrberg, E. Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. Methods in cell biology, vol 44. Drosophila melanogaster: practical uses in cell biology. 44, 445-487 (1994).
- Lee, H. K., Wright, A. P., Zinn, K. Live dissection of Drosophila embryos: streamlined methods for screening mutant collections by antibody staining. J. Vis. Exp. (34), (2009).
- Hartenstein, V. Stages of Embryonic Development. Atlas of Drosophila. development. , 52 (1993).
- Brady, J. A simple technique for making very fine, durable dissecting needles by sharpening tungsten wire electrolytically. Bull. World Health Organ. 32 (1), 143-144 (1965).
- Kolodkin, A. L. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 9 (5), 831-845 (1992).
- Jeong, S., Juhaszova, K., Kolodkin, A. L. The Control of semaphorin-1a-mediated reverse signaling by opposing pebble and RhoGAPp190 functions in Drosophila. Neuron. 76 (4), 721-734 (2012).
- Winberg, M. L. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 95 (7), 903-916 (1998).
- Yang, D. S., Roh, S., Jeong, S. The axon guidance function of Rap1 small GTPase is independent of PlexA RasGAP activity in Drosophila. Dev. Biol. 418 (2), 258-267 (2016).
- Yu, H. H., Araj, H. H., Ralls, S. A., Kolodkin, A. L. The transmembrane Semaphorin Sema I is required in Drosophila for embryonic motor and CNS axon guidance. Neuron. 20 (2), 207-220 (1998).
- Hartenstein, V. Stages of Embryonic Development. Atlas of Drosophila. development. , 52 (1993).
- Dickson, B. J. Molecular mechanisms of axon guidance. Science. 298 (5600), 1959-1964 (2002).
- Kidd, T. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 92 (2), 205-215 (1998).
- Pasterkamp, R. J. Getting neural circuits into shape with semaphorins. Nat. Rev. Neurosci. 13 (9), 605-618 (2012).

