Detection of Enterohemorrhagic Escherichia Coli Colonization in Murine Host by Non-invasive In Vivo Bioluminescence System
Summary
A detailed protocol of a mouse model for enterohemorrhagic E. coli (EHEC) colonization by using bioluminescence-labeled bacteria is presented. The detection of these bioluminescent bacteria by a non-invasive in vivo imaging system in live animals can advance our current understanding of EHEC colonization.
Abstract
Enterohemorrhagic E. coli (EHEC) O157:H7, which is a foodborne pathogen that causesdiarrhea, hemorrhagic colitis (HS), and hemolytic uremic syndrome (HUS), colonize to the intestinal tract of humans. To study the detailed mechanism of EHEC colonization in vivo, it is essential to have animal models to monitor and quantify EHEC colonization. We demonstrate here a mouse-EHEC colonization model by transforming the bioluminescent expressing plasmid to EHEC to monitor and quantify EHEC colonization in living hosts. Animals inoculated with bioluminescence-labeled EHEC show intense bioluminescent signals in mice by detection with a non-invasive in vivo imaging system. After 1 and 2 days post infection, bioluminescent signals could still be detected in infected animals, which suggests that EHEC colonize in hosts for at least 2 days. We also demonstrate that these bioluminescent EHEC locate to mouse intestine, specifically in the cecum and colon, from ex vivo images. This mouse-EHEC colonization model may serve as a tool to advance the current knowledge of the EHEC colonization mechanism.
Introduction
EHEC O157:H7 is a pathogen that causes diarrhea1, HS2, HUS3, and even acute renal failure4 through contaminated water or food. EHEC is a pathogenic enterobacterium and colonizes to the gastrointestinal tract of humans1. When EHEC first adhere to host intestinal epithelium, they inject the colonization factors into host cells through the type III secretion system (T3SS) that functions as a molecular syringe inducing an attaching and effacing (A/E) lesion subsequently to enforce adhesion (colonization)5. These genes involved in A/E lesion formation are encoded by the locus of enterocyte effacement (LEE) pathogenicity island5.
Bioluminescence is a light-producing chemical reaction, in which luciferase catalyzes its substrate luciferin to generate visible light6. This enzymatic process often requires the presence of oxygen or adenosine triphosphate (ATP)6. Bioluminescence imaging (BLI) allows researchers the visualization and quantization of host-pathogen interactions in live animals7. BLI can characterize the bacterial infection cycle in live animals by following the bioluminescent bacteria as they migrate to and invade different tissues7; this reveals a dynamic progression of infection. Moreover, the bacterial load in animals is related to the bioluminescent signal8; thus, it is a convenient indicator to estimate the pathological conditions of experimental animals in a simple and direct way.
The plasmid used here contained the luciferase operon, luxCDABE, which is from the bacterium Photorhabdus luminescens that encodes its own luciferase substrate7,9. By transforming this luciferase-expressing plasmid into bacteria, the colonization and infection processes can be monitored by observing these bioluminescent bacteria in live animals. Overall, BLI and bioluminescence-labeled bacteria allow researchers to monitor the bacterial numbers and location, bacterial viability with antibiotics/therapy treatment, and bacterial gene expression in infection/colonization6,7. Numerous pathogenic bacteria have been reported that express the luxCDABE operon to examine their infection cycle and/or gene expression in infection. These bacteria, including uropathogenic E. coli10, EHEC8,11,12,13, enteropathogenic E. coli (EPEC)8, Citrobacter rodentium14,15, Salmonella typhimurium16, Listeria monocytogenes17, Yersinia enterocolitica18,19, and Vibrio cholerae20, have been documented.
Several experimental models have been developed to facilitate the study of EHEC colonization in vitro and in vivo21,22,23. However, there is a lack of suitable animal models to study the EHEC colonization in vivo, and thus a resulting paucity of details. To facilitate the study of the EHEC colonization mechanism in vivo, it is valuable to build animal models to observe and quantify EHEC colonization in live animals in a non-invasive method.
This manuscript describes a mouse-EHEC colonization model that uses a bioluminescent expressing system to monitor EHEC colonization over time in living hosts. Mice are intragastrically inoculated with bioluminescence-labeled EHEC and the bioluminescent signal is detected in mice with a non-invasive in vivo imaging system13. Mice infected with bioluminescence-labeled EHEC showed significant bioluminescent signals in their intestine after 2 days post infection, which suggested that those bacteria colonized in the host intestine after 2 days post infection. Ex vivo image data showed that this colonization is specifically in the cecum and colon of mice. By using this mouse-EHEC model, the bioluminescent EHEC colonization can be detected in the living host by an in vivo imaging system, to study the detailed mechanisms of enteric bacteria colonization, which may promote further understanding in EHEC-induced physiological and pathological changes.
Protocol
Caution: EHEC O157:H7 is a biosafety level 2 (BSL-2) pathogen according to the Centers for Disease Control and Prevention (CDC) biosafety instruction (https://www.cdc.gov/). Therefore, all experimental procedures involving EHEC must be performed in a BSL-2 facility. Wear lab coats and gloves while performing the experiment. Work in a certified biosafety cabinet (BSC). Disinfect the experimental bench before and after the experimental procedure with 70% ethanol. All instruments or equipment that contact (or potentially contact) EHEC should be disinfected with 70% ethanol or bleach. Contaminated (or potentially contaminated) wastes should be sealed and autoclaved carefully. Wear a mask, eye protection, double gloves or a jumpsuit, if necessary. The 6-week-old C57BL/6 female mice were purchased and maintained at the Laboratory Animal Center of National Cheng Kung University (NCKU). The animal experiments were approved by the Institutional Animal Care and Use Committee of NCKU (Approval number 104-039).
1. Bioluciferase-labeled EHEC Bacteria Generation
- Mix 50 ng deoxyribonucleic acid (DNA), 1 μL each of 10 μM forward and reverse primers, 2 μL 2.5 mM deoxynucleotides (dNTPs), 2 μL 10x buffer, 0.2 μL DNA polymerase, and sterile double-distilled water (ddH2O) to a final volume of 20 μL to amplify the anti-kanamycin cassette, nptII gene24. The DNA template is from pBSL18024, which is purchased from the National BioResource Project (NBRP). The PCR conditions are listed in Table 1 and the primer sequence is available in Table 2.
- Ligate the PCR products to a commercial cloning vector following the kit protocol (see Table of Materials).
- Excise the nptII gene fragment from the cloning vector by NsiI and SmaI and clone into pBBR1MCS4, which is digested by the NsiI and ScaI from pAKlux29 to create the kanamycin resistant plasmid, pWF27813.
- Excise the luxCDABE operon, from the luciferase expressing plasmid9, pAKlux2, by SpeI–HF and ScaI and clone to SpeI-HF and SmaI digested pWF278 to generate kanamycin resistant and luciferase expressing plasmid, pWF27913.
NOTE: For the plasmid extractions and the excised fragment purifications, use the commercial plasmid extraction and the gel extraction kit, respectively. For enzyme digestion, mix 100 ng DNA, 1 μL 10x buffer, 1 μL of each selected restriction enzyme, and sterile ddH2O to a total volume of 10 μL, and incubate at 37 °C for 2.5 – 3 h. - Transform the pWF279 plasmid into E. coli O157:H7 EDL933 competent cells by electroporation with 2,500 V for 4 ms.
- Incubate the transformed bacteria cells in 1 mL Luria-Bertani (LB) medium at 37 °C for 1 h.
- Plate the bacteria on an LB agar plate supplemented with 50 µg/mL of kanamycin at 37 °C for 16 – 18 h.
- Check the bioluminescent signal of the plate by an in vivo imaging machine the next day. Pick a single colony from the plate and culture it in 3 mL LB supplemented with 50 µg/mL kanamycin at 37 °C for 16 – 18 h.
- Prepare bacterial stock medium by diluting 100% glycerol in sterile ddH2O to make the 30% glycerol solution.
- Freeze the kanamycin resistant E. coli O157:H7 EDL933 bacteria harboring the luciferase plasmid as a bacterial stock in a screw cap cryovial as a 1:1 ratio of bacteria culture and 30% glycerol solution at -80 °C. The final concentration of glycerol is 15%.
2. Bioluminescent EHEC Bacteria Preparation for Oral Inoculation
NOTE: The timeline flowchart of the experimental procedures for EHEC preparation and mouse oral gavage is presented in Figure 1 to aid in experimental preparation.
- Streak E. coli O157:H7 EDL933 bacteria harboring luciferase plasmid onto an LB agar plate with 50 µg/mL kanamycin from -80 °C stock. Grow the bacteria for 16 – 18 h at 37 °C.
- Pick a single colony from the overnight plate and culture in 3 mL LB medium with 50 µg/mL kanamycin for 16 – 18 h in a 37 °C incubator at 220 rpm.
- Subculture the bacteria (1:100 dilution) into kanamycin (50 µg/mL) containing LB broth for 2.5 – 3 h in a 37 °C incubator at 200 rpm. (For instance, add 2 mL overnight cultured bacteria to 200 mL LB medium with kanamycin (50 µg/mL) supplemented).
- Incubate the bacteria for 2.5 – 3 h and measure the optical density value at 600 nm (OD600) until the value is between 0.9 to 1. The bacterial cell number is at a density of about 108 colony forming units (CFU)/mL.
- Centrifuge the re-cultured bacteria at 8,000 x g for 30 min, 4 °C.
- Discard the supernatant without agitating the pellet of bacteria and wash the pellet with 100 mL 0.9% sterile normal saline by gentle agitation.
- Repeat steps 2.5 and 2.6 once.
- After the wash, centrifuge the bacterial culture at 8,000 x g for 30 min, 4 °C and discard the supernatant gently.
- Condensate the pellet at 100-fold with 0.9% sterile normal saline. For instance, if 200 mL bacteria are centrifuged, add 2 mL normal saline to suspend the pellet.
- Confirm the bacteria CFU (it should be approximately 109 CFU/100 μL after condensation in normal saline) by plating the concentrated bacteria through a 10-fold serial dilution25.
3. Mouse Oral Gavage of EHEC
- Treat 6-week-old female C57BL/6 mice with streptomycin water (5 g/L) for 24 h.
- After 24 h, switch to regular drinking water for another 24 h before gavage. After treating with regular water for 24 h, the mice are ready for oral gavage of EHEC.
- Fill the syringe with 100 μL EHEC bacteria by pulling back on the plunger. Ensure no air bubbles are in the syringe. Remove the bubbles by snapping the syringe with fingers.
- Lift the animal gently and place it on the top of cage with care.
- Grasp the mouse by the tail carefully, and the animal will grip the top of cage and attempt to move away.
- Gently restrain the mouse by grasping the loose skin of the neck and back of the animal with thumb and forefinger to prevent the head of the mouse from moving.
- Hold the mouse in a vertical position to insert the gavage needle and ensure the head of mouse is immobilized and vertical.
- Insert the gavage needle into the mouse mouth following the roof of the mouth and move down into esophagus and toward the stomach.
- When the inserted needle is half or two-thirds length in the mouse, inject the 100 μL EHEC bacteria, which contain approximately 109 CFU cells.
4. Visualization
- After the oral gavage, detect the bioluminescent signals on 1 and 2 days.
- Before examining the bioluminescent signals of animals, anesthetize them by putting them into a chamber of 2.5% isoflurane with 1.5 L/min oxygen.
- Wait for 2 – 5 min until all mice become unconscious and stop moving. Animals are ready for in vivo detection of bioluminescence.
- Detect and image bioluminescent signals of animals by an in vivo imaging system. Please see Section 5 "Data Acquisition" for software operation. During the imaging process, all animals are under a continued supply of 2.5% isoflurane with 1.5 L/min oxygen.
- Click file>live, and manually focus on the mouse.
- Select the exposure time and laser intensity.
- Click acquire>capture.
- For ex vivo imaging, euthanize the mice by cervical dislocation and remove the entire intestine of infected mice. Place the intestines in a 9-cm Petri dish and image by the in vivo imaging system. The setting is the same as the in vivo imaging except the Field of View is set as A/B. Please see Section 5 "Data Acquisition" for details.
NOTE: Method for cervical dislocation: Restrain the mice on the top of cage by grasping the tail with one hand so that the animals grip the cage. Place a marker pen or the thumb and first finger of the other hand against the back of the neck at the base of the skull. Quickly push forward with the hand or object while restraining the head and pull backward with the hand holding the tail.
Check closely to confirm respiratory arrest, and no heartbeat.
5. Data Acquisition
NOTE: The software used for data acquisition is listed in the Table of Materials.
- For image acquisition, open the Acquisition Control Panel of the software (Figure 2).
- Select "Luminescent," "Photograph," and "Overlay."
- Set Exposure Time as "Auto." Set Binning as "Medium."
- Set ƒ/stop as 1 for luminescent and 8 for photograph. ƒ/stop controls the amount of light received by the CCD detector.
- Set the Field of View based on the field of images that are of interest to acquire. Option "D" can fit five 6-week-old mice and image them all at a time. "C" can image three 6-week-old mice in one field.
- Once the mice/samples are ready for imaging, click "Acquire" for image acquisition.
- Open the image data that was acquired.
- Open the Tool Palette panel (Figure 3).
- Select ROI Tools. We recommend the Circle (the left-most one) to range the bioluminescent area on images (Figure 4).
- Click "Measure ROIs" (pencil icon) to measure the surface bioluminescent intensity (Figure 3). The ROI Measurements panel and the quantification values appear (Figure 5).
- Use the Configure Measurement on the left corner of the ROI Measurements panel to select the values/information needed (Figure 6), otherwise click "Export" to export this data table and save as .csv file (Figure 5).
- Use the values of column "Total Flux (p/s)" as the bioluminescent intensity quantification in the .csv file.
Representative Results
We administered bioluminescence-labeled EHEC (~ 109 bacterial cells) to 6-week old female C57BL/6 mice by oral gavage. After oral inoculation of EHEC to mice within 1 h, the animals were examined for bioluminescent signal by the in vivo imaging system as shown in Figure 7. The results showed a strong bioluminescent signal in gavage mice with bioluminescence-labeled EHEC. We examined the signals on 2 days post infection. As shown in Figure 8A, the mice inoculated with bioluminescence-labeled wild-type EHEC EDL933 showed intense bioluminescent signals even after 2 days post infection, which suggested EHEC colonized in hosts by 2 days. We also intragastrically infected bioluminescence-labeled EDL933ΔrfaD (ΔrfaD) to mice (Figure 8A). This mutant, defected in lipopolysaccharide (LPS), has been shown to reduce colonization in the host in our previous study. As shown in Figure 8A, there is no bioluminescent signal detected in ΔrfaD-infected mice, which suggests that there are no or less bacteria cells colonized in the mice. Quantification of the fluorescent signal is shown in Figure 8B. Next, the location of these bioluminescence-labeled bacteria was determined. The infected mice were sacrificed humanely and their whole intestine removed. The intestines of mice 2 days post infection were positioned on 9 cm Petri dishes and imaged ex vivo (Figure 9A). The intestinal tissues of bioluminescence-labeled EDL933 infected mice revealed a significant increase in bioluminescent signals in the cecum and colon, which suggest that these bioluminescent EHEC colonized in the cecum and colon of infected mice for 2 days at least. In contrast, mice infected with bioluminescence-labeled ΔrfaD (Figure 9A), revealed decreased bioluminescent signal in their intestinal tissue, which is consistent with the in vivo image (Figure 8A). Quantification of the fluorescent signal is shown in Figure 9B.
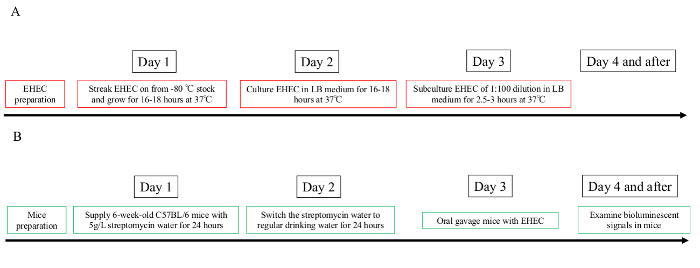
Figure 1: Timeline of the experimental preparation flow chart.
Overview of the timing needed to prepare bioluminescent EHEC bacteria and pretreat mice with streptomycin. (A) EHEC preparation. (B) Mice preparation. Please click here to view a larger version of this figure.
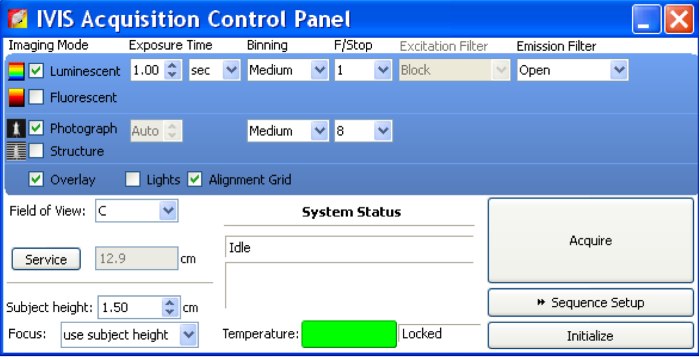
Figure 2: In vivo imaging system acquisition control panel.
Before imaging samples, open IVIS Acquisition Control Panel. Select "Luminescent," "Photograph," and "Overlay." Set Exposure Time as "Auto." Set Binning as "Medium." Set ƒ/stop as 1 for luminescent and 8 for photograph. ƒ/stop controls the amount of light received by the CCD detector. Once samples are ready for imaging, click "Acquire" to acquire images. Please click here to view a larger version of this figure.
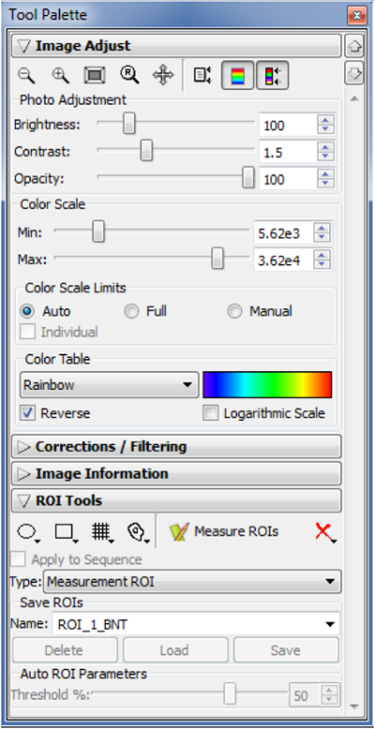
Figure 3: Tool Palette panel.
After image acquiring, use the Tool Palette panel for quantifying bioluminescent intensity. Open the Tool Palette panel and image the data. Choose one of the ROI Tools to range the bioluminescent signals on images. Please click here to view a larger version of this figure.
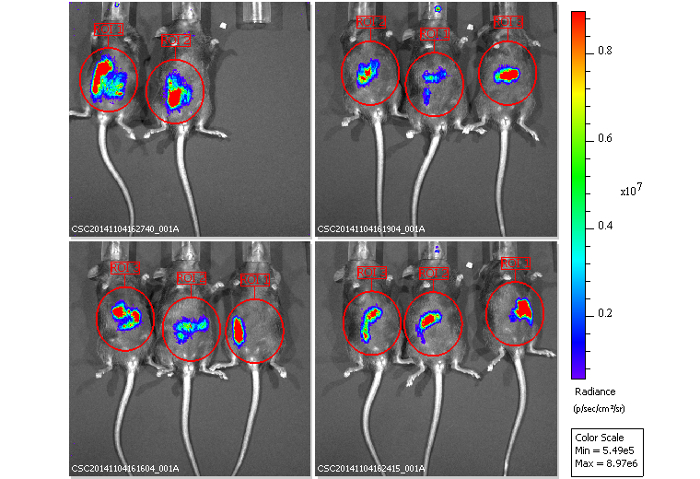
Figure 4: Bioluminescent signal from sample for quantification.
Bioluminescent signal area on images encircled by ROI Tools. All bioluminescent signals shown here are in the red circle. Please click here to view a larger version of this figure.
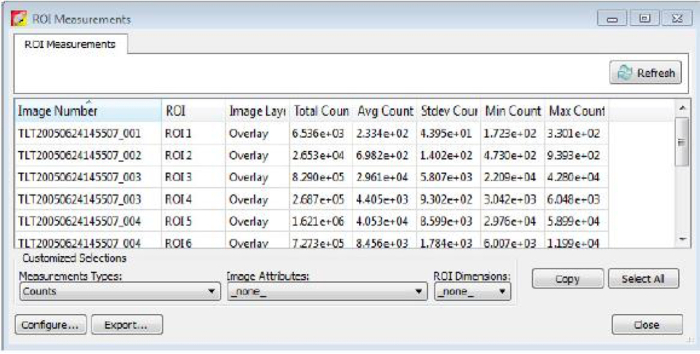
Figure 5: ROI measurements.
After circling bioluminescent signals and clicking "Measure ROIs" on the Tool Palette panel, values are presented as shown. The values of the column Total Flux (p/s) are used for the bioluminescent intensity quantification. Please click here to view a larger version of this figure.
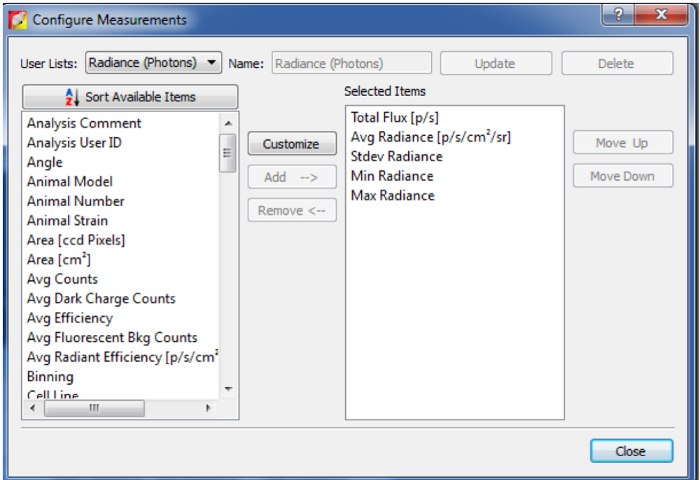
Figure 6: Add different quantification information.
By clicking on Configure Measurement on the left corner of the ROI Measurements panel, you can select other desired quantification values/information. Please click here to view a larger version of this figure.
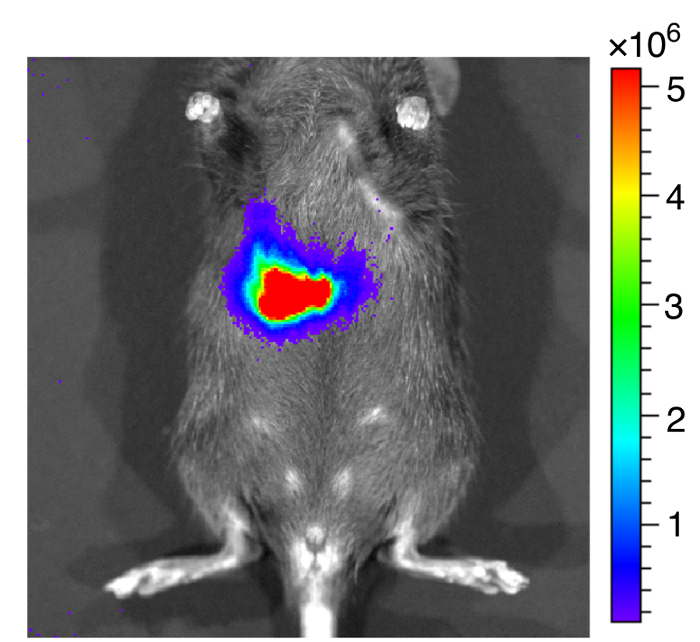
Figure 7: Representative image of mice after inoculated with bioluminescent EHEC.
Representative image of mice inoculated with bioluminescent EHEC by oral gavage within 1 h. The color scale represents the radiance (p/s/cm2/sr). Please click here to view a larger version of this figure.
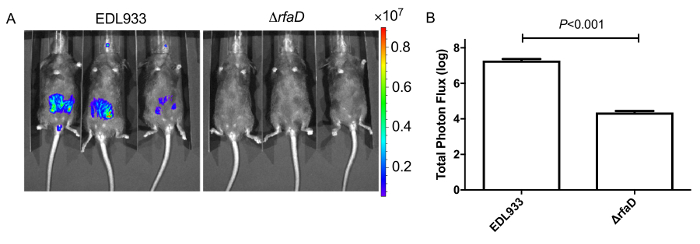
Figure 8: Images of mice inoculated with bioluminescence-labeled EHEC after 2 days.
(A) Represent image of mice inoculated with bioluminescent wild-type EHEC EDL933 and EDL933:ΔrfaD by oral gavage after 2 days post infection. (B) Quantification of bioluminescence intensity of mice infected with EHEC. Error bars indicate the standard deviations. Representative images are shown.All experiments were conducted independently three times with 2 – 3 animals each time, and error bars indicate the standard deviations. P-values denote the results of statistical analysis by t-test. The color scale represents the radiance (p/s/cm2/sr). Please click here to view a larger version of this figure.
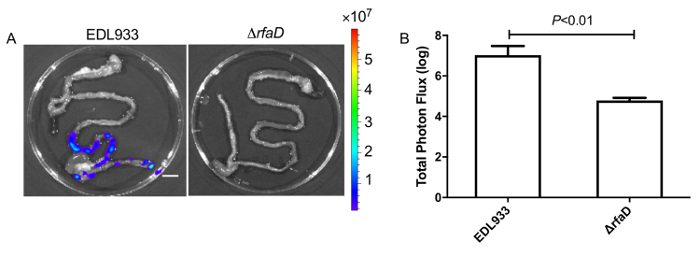
Figure 9: Images of intestinal tissues of infected mice with bioluminescence-labeled EHEC.
(A) 2 days after inoculation with bioluminescence-labeled EHEC, the mice were euthanized and whole intestinal tissues were removed and imaged ex vivo. Representative images are shown.(B) Quantification of bioluminescence intensity of intestinal tissues from mice infected with EHEC. All experiments were conducted independently three times with 2 - 3 animals each time, and error bars indicate the standard deviations. P-values denote the results of statistical analysis by t-test. The color scale represents the radiance (p/s/cm2/sr). Scale bar represents 1 cm. Please click here to view a larger version of this figure.
| Steps | Temperature | Time | Number of cycles |
| Initial denaturation | 95 °C | 10 min | 1 |
| Denaturation | 95 °C | 30 sec | 35 |
| Annealing | 58.4 °C | 30 sec | |
| Extension | 72 °C | 1.5 min | |
| Final extension | 72 °C | 10 min | 1 |
| Hold | 4 °C | ∞ | 1 |
Table 1: Polymerase chain reaction (PCR) conditions
| Primers name | Sequence |
| nptII F | 5’CCTATGCATAATAATTCCGCTAGCTTCACG3’ |
| nptII R | 5’GCTCCACCGATAATATTCCTGAGTCATACT3’ |
Table 2: Primer sequences used to amplify nptII
Discussion
It has been reported that EHEC transformed with luciferase plasmid has been utilized to examine its localization in hosts or gene expression in vivo8,11,12. The murine model demonstrated here has also been reported to detect the EHEC colonized timing and localization in murine host8. Nevertheless, we provide the detail protocol of how to administer EHEC inoculation to mice intragastrically and how to carefully prepare the bioluminescent bacteria for oral gavage. Notably, for the mouse oral gavage of EHEC (step 3.7), the position of the mouse head is critical when the gavage needle is inserted. If the position is not vertical, it will be difficult to pass the needle, and it could possibly injure the mouse. In step 3.8, when the gavage needle is inside the mouth of the mouse, the tongue will lay outside of the mouth slightly. If resistance is encountered when passing the gavage needle to the esophagus, stop moving the needle forward and withdraw it immediately. Alter the needle position to make sure that the needle is entering the esophagus. The needle could be entering the trachea when resistance occurs, which would lead to injecting bacteria in the lungs instead of the stomach.
Application of green fluorescent protein (GFP) as a biosensor is common in biological experiments. However, using GFP as a reporter to observe the pathogen infection/colonization in live animals by in vivo imaging is not recommended, because the absorbance by hemoglobin, proteins and water are high between 200-650 nm26, which overlaps with GFP (excitation 480 nm, emission 510 nm)27. Therefore, using GFP signal as a reporter for in vivo imaging can be interrupted by hemoglobin, proteins, and water in animals26. The near-infrared (NIR) fluorescence is ideally suited for in vivo imaging because its absorbance window is around 650 – 900 nm28, which is in the region of lowest absorption coefficient of hemoglobin (<650 nm) and water (>900 nm)26,28 . Moreover, when tissue absorbs light, there is a chance to induce autofluorescence. When the wavelengths of excitation and emission range in the GFP window, it induces much more autofluorescence than NIR29. Use of NIR can improve the signal to background ratio compared to that of GFP by eliminating the autofluorescence background29. Bioluminescence does not require energy excitation to generate visible light. It depends on the reaction to catalyze substrate luciferin by its enzyme luciferase and generate light. Since bioluminescence does not require light directly on a sample, the background signal from a sample is very low. Therefore, use of bioluminescence as a reporter is more general and easier for in vivo imaging. In contrast, fluorescence requires light excitation to induce signal light. When tissues absorb light, there is a chance that the fluorescent light will be emitted and induce autofluorescence so that their signal-to-noise is higher compared to that of bioluminescence.
Considering EHEC is naturally less colonized in mice by oral infection2,22, a natural mucosal pathogen of mice, called Citrobacter rodentium, has been utilized to study the colonization mechanism to murine host as a surrogate bacterium22,30. Both of EHEC and C. rodentium colonize the intestinal mucosa and induce the formation of A/E lesions in host22,30. They also contain the LEE pathogenicity island, which encodes a T3SS and several effector proteins that induced A/E lesion22,30. Therefore, the use of luciferase expressing plasmid as a reporter in C. rodentium to detect the colonization pathology and study the colonization mechanism via an in vivo imaging system has also been reported14,15. Nevertheless, while C. rodentium infection of mice is a useful model to investigate the function of T3SS and the mechanism of A/E lesion, C. rodentium does not contain Shiga toxin (Stx)30, which is a dominant virulence factor that causes kidney failure in EHEC, particularly serotype O157:H73. Although a Stx-expressing C. rodentium strain has been constructed recently31, which is more realistic to EHEC infection, it does not include other potential EHEC virulence factors that are crucial for colonization and/or infection. Furthermore, C. rodentium shares 67% of its genes with EHEC32, which suggests that EHEC may use a virulence distinct from C. rodentium during colonization and/or infection.
The luciferase expressing plasmid used in here, pWF27913, was modified from pAKlux29 whose backbone is pBBR1MCS433. Although pBBR1MCS4 have been tested and replicated in various bacteria33, it is crucial to ensure the origin of replication (ORI) of this plasmid is suitable for the bacterial host before using this plasmid-based luciferase system for the experiment and thus confirming that this luciferase expressing plasmid can replicate in the bacterial host. We use antibiotic stress to maintain the stability of plasmid in the bacteria. When bacteria enter animals in the absence of antibiotics, the bioluminescent signal of wild-type EHEC has been detected for at least 2 days. However, we didn't following infection for longer than 2 days because we had already seen a significant difference in luminescent intensity between EHEC WT and EHEC rfaD (that encodes a gene required for EHEC synthesizing intact LPS) mutant at 2 days. To maintain the plasmid stably in bacteria under the absence of antibiotics, a plasmid pCM1710,34 can be used for this purpose. pCM17 encodes a two-plasmid partitioning system and a post-segregational killing mechanism to ensure the maintenance of the plasmid in bacteria in the absence of antibiotics10,34. The plasmid pCM17 containing the luxCDABE operon driven by the OmpC promoter can be detected by bioluminescent signal for at least 7 days8. An alternative method to obtain a continuous bioluminescent expression bacteria in the absence of antibiotics is to insert luxABCDE gene into the bacterial chromosome35. Francis et al. used transposon inserted the luxABCDE operon and antibiotics cassette randomly inserted into the chromosome of Streptococcus pneumoniae to obtain the bioluminescence stable strain35.
In our previous study13, we utilized this model system to examine EHEC colonization in a host and compare the difference of colonization ability between the EHEC wild type (WT) and mutant13. When mice were administered the bioluminescent EHEC rfaD mutant, the bioluminescent signals diminished dramatically compared to that of WT EHEC after 2 days post infection. It provides evidence that this murine model can analyze the mutation effect of EHEC colonization in the host. Furthermore, therapeutic treatments for reducing the colonization of EHEC is a considered, potential solution to EHEC infection since the use of antibiotics is contraindicated5,36. Therefore, it is worth testing whether this model system can be used to examine the efficacy of anti-colonization drugs/treatments against enterobacteria colonization in the host. We believe that by using this model system, it is possible to examine the timing and location of not merely EHEC, but also other enterobacteria colonization in vivo. By using this animal model, the process of EHEC colonization in mice can be monitored and the colonization burden in the host can be quantified to determine spatial and temporal colonization of EHEC in live animals. The visualization and quantification of enterobacteria colonization by using this model makes it a great tool to investigate and analyze the fine mechanisms of enterobacterial colonization, and thus compensate for the deficiency of colonization research and improve current knowledge.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We acknowledge Chi-Chung Chen from the Department of Medical Research, Chi Mei Medical Center (Tainan, Taiwan) for the help in mouse infection, and the support from the laboratory animal center of National Cheng Kung University. This work is supported by the Minister of Science and Technology (MOST) grants (MOST 104-2321-B-006-019, 105-2321-B-006 -011, and106-2321-B-006 -005) to CC.
Materials
| Shaker incubator | YIH DER | LM-570R | bacteria incubation |
| Orbital shaking incubator | FIRSTEK | S300 | bacteria incubation |
| pBSL180 | source of nptII gene | ||
| pAKlux2 | source of luxCDABE operon | ||
| T&A Cloning Kit | Yeastern Biotech | FYC001-20P | use for TA cloning |
| Nsi I | NEB | R0127S | use for plasmid cloning |
| Sca I | NEB | R0122S | use for plasmid cloning |
| Spe I-HF | NEB | R0133S | use for plasmid cloning |
| Sma I | NEB | R0141S | use for plasmid cloning |
| T4 ligase | NEB | M0202S | use for plasmid cloning |
| Ex Taq | TaKaRa | RR001A | use for PCR amplification |
| 10X Ex Taq Buffer | TaKaRa | RR001A | use for PCR amplification |
| dNTP Mixture | TaKaRa | RR001A | use for PCR amplification |
| PCR machine | applied Biosystem | 2720 thermal cycler | for PCR amplification |
| Glycerol | SIGMA | G5516-1L | use for bacteria stocking solution |
| NaCl | Sigma | 31434-5KG-R | chemical for making LB medium, 10 g/L |
| Tryptone | CONDA pronadisa | Cat 1612.00 | chemical for making LB medium, 10 g/L |
| Yeast Extract powder | Affymetrix | 23547-1 KG | chemical for making LB medium, 5 g/L |
| Agar | CONDA pronadisa | Cat 1802.00 | chemical for making LB agar |
| kanamycin | Sigma | K4000-5G | antibiotics, use for seleciton |
| streptomycin | Sigma | S6501-100G | antibiotics, eliminate the microbiota in mice |
| EDL933 competent cell | Homemade | method is on supplemental document | |
| Electroporator | MicroPulser | for electroporation | |
| Electroporation Cuvettes | Gene Pulser/MicroPulser | 1652086 | for electroporation |
| High-speed centrifuge | Beckman Coulter | Avanti, J-26S XP | use for centrifuging bacteria |
| Fixed-Angle Rotor | Beckman Coulter | JA25.5 | use for centrifuging bacteria |
| Fixed-Angle Rotor | Beckman Coulter | JLA10.5 | use for centrifuging bacteria |
| centrifuge bottles | Beckman Coulter | REF357003 | use for centrifuging bacteria |
| centrifuge bottles | Thermo Fisher scientific | 3141-0500 | use for centrifuging bacteria |
| eppendorf biophotometer plus | eppendorf | AG 22331 hamburg | for measuring the OD600 value of bacteria |
| C57BL/6 mice | Laboratory Animal Center of NCKU | ||
| lab coat, gloves | for personnel protection | ||
| isoflurane | Panion & BF Biotech Inc. | G-8669 | for mice anesthesia, pharmaceutical grade |
| 1ml syringe | use for oral gavage of mice | ||
| Reusable 22 G ball-tipped feeding needle | φ0.9 mm X L 50 mm | use for oral gavage of mice | |
| surgical scissors | use for mice experiment | ||
| Xenogen IVIS 200 imaging system | Perkin Elmer | IVIS spectrum | use for bioluminescent image capture |
| Living Image Software | Perkin Elmer | version 4.1 | use for quantifying the image data |
References
- Pennington, H. Escherichia coli O157. Lancet. 376 (9750), 1428-1435 (2010).
- Mayer, C. L., Leibowitz, C. S., Kurosawa, S., Stearns-Kurosawa, D. J. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins (Basel). 4 (11), 1261-1287 (2012).
- Tarr, P. I., Gordon, C. A., Chandler, W. L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 365 (9464), 1073-1086 (2005).
- Obrig, T. G. Escherichia coli Shiga Toxin Mechanisms of Action in Renal Disease. Toxins (Basel). 2 (12), 2769-2794 (2010).
- Nguyen, Y., Sperandio, V. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol. 2, 90 (2012).
- Wiles, S., Robertson, B. D., Frankel, G., Kerton, A. Bioluminescent monitoring of in vivo colonization and clearance dynamics by light-emitting bacteria. Methods Mol Biol. 574, 137-153 (2009).
- Hutchens, M., Luker, G. D. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 9 (10), 2315-2322 (2007).
- Rhee, K. J., et al. Determination of spatial and temporal colonization of enteropathogenic E. coli and enterohemorrhagic E. coli in mice using bioluminescent in vivo imaging. Gut Microbes. 2 (1), 34-41 (2011).
- Karsi, A., Lawrence, M. L. Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid. 57 (3), 286-295 (2007).
- Lane, M. C., Alteri, C. J. S., Smith, S. N., Mobley, L. H. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A. 104 (42), 16669-16674 (2007).
- Roxas, J. L., et al. Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Lab Invest. 90 (8), 1152-1168 (2010).
- Siragusa, G. R., Nawotka, K., Spilman, S. D., Contag, P. R., Contag, C. H. . Real-Time Monitoring of Escherichia coli O157:H7 Adherence to Beef Carcass Surface Tissues with a Bioluminescent Reporter. , (1999).
- Kuo, C. J., et al. Mutation of the Enterohemorrhagic Escherichia coli Core LPS Biosynthesis Enzyme RfaD Confers Hypersusceptibility to Host Intestinal Innate Immunity In vivo. Front Cell Infect Microbiol. 6, 82 (2016).
- Wiles, S., et al. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 6 (10), 963-972 (2004).
- Wiles, S., Pickard, K. M., Peng, K., MacDonald, T. T., Frankel, G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect Immun. 74 (9), 5391-5396 (2006).
- Contag, C. H., Contag, P. R., Mullins, J. I., Spillman, S. D., Stevenson, D. K., Benaron, D. A. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 18 (4), 593-603 (1995).
- Hardy, J., Francis, K. P., DeBoer, M., Chu, P., Gibbs, K., Contag, C. H. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science. 303 (5659), 851-853 (2004).
- Kaniga, K., Sory, M. P., Delor, I., Saegerman, C., Limet, J. N., Cornelis, G. R. Monitoring of Yersinia enterocolitica in Murine and Bovine Feces on the Basis of the Chromosomally Integrated luxAB Marker Gene. Appl Environ Microbiol. 58 (3), 1024-1026 (1992).
- Trcek, J., Fuchs, T. M., Trulzsch, K. Analysis of Yersinia enterocolitica invasin expression in vitro and in vivo using a novel luxCDABE reporter system. Microbiology. 156 (Pt 9), 2734-2745 (2010).
- Morin, C. E., Kaper, J. B. Use of stabilized luciferase-expressing plasmids to examine in vivo-induced promoters in the Vibrio cholerae vaccine strain CVD 103-HgR. FEMS Immunol Med Microbiol. 57 (1), 69-79 (2009).
- Law, R. J., Gur-Arie, L., Rosenshine, I., Finlay, B. B. In vitro and in vivo model systems for studying enteropathogenic Escherichia coli infections. Cold Spring Harb Perspect Med. 3 (3), a009977 (2013).
- Ritchie, J. M. Animal Models of Enterohemorrhagic Escherichia coli Infection. Microbiol Spectr. 2 (4), EHEC-0022-2013 (2014).
- Chou, T. C., et al. Enterohaemorrhagic Escherichia coli O157:H7 Shiga-like toxin 1 is required for full pathogenicity and activation of the p38 mitogen-activated protein kinase pathway in Caenorhabditis elegans. Cell Microbiol. 15 (1), 82-97 (2013).
- Alexeyev, M. F., Shokolenko, I. N. Mini-Tnl 0 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene. 160 (1), 59-62 (1995).
- Wiegand, I., Hilpert, K., Hancock, R. E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 3 (2), 163-175 (2008).
- Pansare, V., Hejazi, S., Faenza, W., Prud’homme, R. K. Review of Long-Wavelength Optical and NIR Imaging Materials: Contrast Agents, Fluorophores and Multifunctional Nano Carriers. Chem Mater. 24 (5), 812-827 (2012).
- Heim, R., Cubitt, A. B., Tsien, R. Y. Improved green fluorescence. Nature. 373 (6516), 663-664 (1995).
- Weissleder, R. A clearer vision for in vivo imaging. Nat Biotechnol. 19 (4), 316-317 (2001).
- Frangioni, J. In vivo near-infrared fluorescence imaging. Current Opinion in Chemical Biology. 7 (5), 626-634 (2003).
- Collins, J. W., et al. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol. 12 (9), 612-623 (2014).
- Mallick, E. M., et al. A novel murine infection model for Shiga toxin-producing Escherichia coli. J Clin Invest. 122 (11), 4012-4024 (2012).
- Petty, N. K., et al. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol. 192 (2), 525-538 (2010).
- Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson , G. T., Farris, M. A., Roop, R. M., Peterson, K. M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 166 (1), 175-176 (1995).
- Galen, J. E., Nair, J., Wang , J. Y., Wasserman, S. S., Tanner, M. K., Sztein , M. B., Levine, M. M. Optimization of Plasmid Maintenance in the Attenuated Live Vector Vaccine Strain Salmonella typhiCVD 908-htrA. Infect Immun. 67 (12), 6424-6433 (1999).
- Francis, K. P., et al. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect Immun. 69 (5), 3350-3358 (2001).
- Goldwater, P. N., Bettelheim, K. A. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS). BMC Med. 10, (2012).

