Generic Protocol for Optimization of Heterologous Protein Production Using Automated Microbioreactor Technology
Summary
This manuscript describes a generic approach for tailor-made design of microbial cultivation media. This is enabled by an iterative workflow combining Kriging-based experimental design and microbioreactor technology for sufficient cultivation throughput, which is supported by lab robotics to increase reliability and speed in liquid handling media preparation.
Abstract
A core business in industrial biotechnology using microbial production cell factories is the iterative process of strain engineering and optimization of bioprocess conditions. One important aspect is the improvement of cultivation medium to provide an optimal environment for microbial formation of the product of interest. It is well accepted that the media composition can dramatically influence overall bioprocess performance. Nutrition medium optimization is known to improve recombinant protein production with microbial systems and thus, this is a rewarding step in bioprocess development. However, very often standard media recipes are taken from literature, since tailor-made design of the cultivation medium is a tedious task that demands microbioreactor technology for sufficient cultivation throughput, fast product analytics, as well as support by lab robotics to enable reliability in liquid handling steps. Furthermore, advanced mathematical methods are required for rationally analyzing measurement data and efficiently designing parallel experiments such as to achieve optimal information content.
The generic nature of the presented protocol allows for easy adaption to different lab equipment, other expression hosts, and target proteins of interest, as well as further bioprocess parameters. Moreover, other optimization objectives like protein production rate, specific yield, or product quality can be chosen to fit the scope of other optimization studies. The applied Kriging Toolbox (KriKit) is a general tool for Design of Experiments (DOE) that contributes to improved holistic bioprocess optimization. It also supports multi-objective optimization which can be important in optimizing both upstream and downstream processes.
Introduction
Modern recombinant gene technology enables the wide use of technical enzymes for various applications in the pharmaceutical industry, animal feeding, organic chemistry, and food processing1,2,3. The production of technical enzymes in bulk quantities is a major topic for industrial biotechnology and for optimized recombinant protein production, and both strain and bioprocess engineering is needed. For the generation of efficiently engineered production strains, different genetic libraries are available, e.g., for balanced gene expression4 or increased secretion efficiency5.
Corynebacterium glutamicum is a major producer of amino acids at the industrial scale6,7 and represents an attractive non-conventional expression host for the secretory production of recombinant proteins8,9. Both the general secretory (Sec) and twin-arginine translocation (Tat) pathway are present in C. glutamicum and were successfully applied for recombinant protein secretion10. Extensive experience in bioprocess engineering regarding amino acid production at the industrial scale, as well as the ability to secrete proteins to g/L amounts11 and great robustness concerning bioprocess inhomogeneities found in large scale cultivations12,13, make C. glutamicum a promising platform organism for the secretory production of heterologous proteins at the industrial scale.
Nutrition medium optimization is known to improve recombinant protein production with microbial systems14,15,16,17 and consequently, the adjustment of medium composition is a rewarding step in bioprocess development with respect to optimal productivity18,19,20,21. Intense research on the application of microtiter plates (MTPs) for microbial cultivation22,23,24 paved the way for the development and design of MTPs for microbial cultivation25,26 and the development of MTP-based microbioreactor (MBR) systems with online monitoring and environmental control27,28. MBRs enable a significant increase in experimental cultivation throughput. Besides, MBR systems stemming from other types of bioreactors, e.g., bubble columns or stirred tank reactors, are available for microbial bioprocess optimization29,30,31,32.
In general, optimization studies benefit from increased experimental throughput, which becomes even more powerful in combination with DOE methodologies, such as to assess interactions between design variables or reduce high-dimensional search spaces. Consequently, the combined use of MBR systems, lab automation, and DOE has proved to be a powerful method in biotechnology8,16,33,34,35.
A protocol for media optimization is presented combining state-of-the-art lab automation, MBR technology with online process monitoring, and Kriging-based data analysis/experimental design. The Kriging methodology is implemented in a MATLAB Toolbox ("KriKit") which can be downloaded and used free of charge36. As application example, maximization of secretory green fluorescent protein (GFP) production with C. glutamicum is shown by optimizing the composition of CgXII minimal medium. GFP titer was chosen as the optimization objective as it can be quantified easily and it is widely applied as model protein for studies on MBR systems37,38,39.
The presented framework is divided into four steps, which are illustrated in Figure 1. The steps are indicated by box frames and correspond to sections of the protocol. The first step (Figure 1A) is to define the project goals and to determine the required methods. The combination of DOE methodologies, MBR technology, and lab automation allows an increased experimental throughput that demands powerful data processing. The second step (Figure 1B) aims to detect sensitive design variables (i.e., medium components) with high influence on the optimization objective. This leads to a reduced number of design variables of interest. The third step (Figure 1C) comprises an iterative optimization for a more detailed investigation of the functional relationship between the remaining design variables and the objective of interest. Using the successively extended data set, the Kriging approach is applied for predicting the experimental outcome at unmeasured locations. The iterative cycle stops as soon as the Kriging model predicts an optimum or plateau with sufficient accuracy. The results are verified in the fourth step (Figure 1D), beginning with a further sensitivity analysis around the identified optimum. If initially, insensitive components are found to be insensitive also in the optimal region, it is reasonable to assume that this holds true during the iterative optimization procedure in the third step. Afterwards, it is advised to verify optimization results by application of orthogonal methods, like an activity assay or SDS-Page.
The generic nature of the presented protocol allows for easy adaption to different lab equipment, other expression hosts, and target proteins of choice, as well as further bioprocess variables like pH value or cultivation temperature. Furthermore, other optimization objectives like protein production rate, specific yield, or product quality can be chosen to fit the scope of other optimization studies.
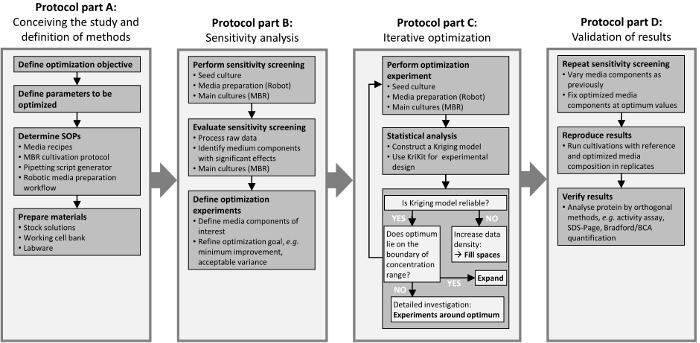
Figure 1: Workflow of optimization study. The four frame boxes correspond to sections of the protocol, "Conceiving the Study and Definition of Methods" (Section 1), "Sensitivity Analysis" (Section 2), "Iterative Optimization" (Section 3), and "Validation" (Section 4). Please click here to view a larger version of this figure.
Protocol
1. Conceiving the Study and Definition of Methods (Figure 1: Part A)
NOTE: Definition of the optimization objective: Is a time course of product formation needed or is only a limited time interval or even a fixed time point relevant? Also, consider potential issues such as stability, effort of analytical quantification, or cultivation time. As an alternative to final protein titer, other objectives could be considered such as biomass or cultivation time. Biomass is reflected by biomass specific product yield, while cultivation time is reflected by space-time-yield. (Minimum) product quality could also be an aim. Multi-objective optimization may be required in certain situations, as discussed elsewhere40. In this study, GFP titer after 17 h of cultivation was chosen as the optimization objective. GFP fluorescence can be followed online using the equipment available in this study, which greatly simplifies determining the concentration of the model protein.
NOTE: Definition of the parameters to be optimized: CgXII medium consists of 16 individual components41 and investigating all of these in a full factorial design would result in 216 ≈ 65,000 experiments. Consequently, the search space needs to be reduced on a rational and experience-driven basis. The selection of media components considered for optimization can be supported by available expert knowledge or literature data.
- Decide which medium component concentrations should not be varied. For the optimization of CgXII medium, the following components were chosen to be fixed:
- Glucose is fixed at 10 g/L. This optimization is trivial because more glucose yields more GFP secreting biomass. The aim was to reveal non-intuitive medium effects.
- 3-(N-morpholino)propanesulfonic acid (MOPS) is fixed at 42 g/L. This provides sufficient buffering capacity during batch cultivation and is not metabolized.
NOTE: The addition of MOPS at this final concentration ensures a starting value of pH 7, even with the different volumes of the other stock solutions added, which are not at pH 7. However, to exclude any deviation in starting pH values, the pH should be checked for medium compositions where all stock solutions are added at their maximum and minimum volumes. - KH2PO4 and K2HPO4 are fixed at 1 g/L each. These provide also buffering capacity, and serve as a phosphate source.
- Urea is fixed at 5 g/L. This serves as a basal nitrogen source and pH-stabilizing agent, sufficient to prevent N-limitation.
- Biotin is fixed at 0.2 mg/L. C. glutamicum ATCC13032 is auxotrophic for biotin.
- Protocatechuic acid (PCA) is fixed at 30 mg/L. It serves as an iron chelating agent.
- Isopropyl-β-D-1-thiogalactopyranoside (IPTG) is fixed at 100 µM. It induces expression of the gfp gene.
- Select high and low concentrations of media components for initial sensitivity screenings. Here, components from three typical groups of media components were chosen. The investigated low and high concentrations for each component are:
- Standard nitrogen source: (NH4)2SO4 (8-32 g/L); the variation of nitrogen source was reported to be promising for optimization of biomass-specific GFP signal8.
- Trace elements: FeSO4 ∙ 7 H2O (4-16 mg/L), MnSO4 ∙ H2O (4-16 mg/L), ZnSO4 ∙ 7 H2O (0.4-1.6 mg/L), CuSO4 ∙ 5 H2O (125-501 µg/L), NiCl2 ∙ 6 H2O (8-32 µg/L) and CoCl2 ∙ 6 H2O (52-208 µg/L). The composition of trace elements is inherited from the first publication of CgXII medium41. In addition, Na2MoO4 ∙ H2O (26-104 µg/L) and H3BO3 (20-80 µg/L) were included as trace elements, as they are used in a published variant of CgXII medium42.
- Macro elements: MgSO4 ∙ 7 H2O (0.2-0.4 g/L), CaCl2 ∙ 2 H2O (5.3-21.2 mg/L). These were found among other divalent cations to enhance secretory GFP production in another study14, where C. glutamicum strain R was used with a different medium background and the CgR0949 Tat signal peptide.
- Preparation of materials and definition procedures
NOTE: It is advised to define and fix the experimental methods at a detailed level. This standardization ensures that different results can be traced back to intrinsic biological variability or different medium compositions.
NOTE: Media stock solutions: Prepare individual stock solutions for all investigated media components. Prepare highly concentrated stocks to allow for the dilution of different volumes for all components. This means that after combining all stock solutions at their highest volume according to the design plan, this cannot exceed the final cultivation volume, cf. paragraph "Generation of medium stock solution pipetting list" step 1.3.3. If the addition of a highly concentrated solution results in a very low addition volume, e.g. < 1/100th of the final cultivation volume, dilute the stock solution accordingly. For instance, in this study a pipetting volume of less than 10 µL was defined to be infeasible. Typically, trace element solutions are concentrated as high as 1,000fold with respect to final standard concentration in the medium. It is advantageous to concentrate media components as multiples (X-fold) with respect to concentrations in the reference recipe. By doing so, infeasible pipetting volumes of fractional decimals are avoided. Detailed recipes for all stock solutions of CgXII medium are given in the supplementary document.- Working cell bank (WCB)
Note: For each growth experiment, one WCB aliquot is used. If more aliquots are needed, use 100 mL brain heart infusion (BHI) medium in 1,000 mL baffled shake flasks or inoculate multiple shake flasks, which are pooled before the addition of glycerol solution.- Prepare single colonies of the recombinant expression strain C. glutamicum pCGPhoDBs-GFP43 on agar plates (37 g/L BHI powder, 20 g/L agar, 25 mg/L kanamycin). Plating material can either come from fresh transformation or from a cryopreserved aliquot. Incubate at 30 °C until the appearance of single colonies; this takes usually one to two days.
- Inoculate a shaker flask culture (50 mL BHI medium with 25 mg/L kanamycin, 500 mL baffled flask, 250 rpm, 25 mm shaking diameter, 30 °C) with colony material and incubate overnight (approximately 16 h).
- Combine one volume of resulting cell suspension with one volume of 500 g/L sterile glycerol solution and distribute in 2 mL aliquots in sterile cryopreservation vials. Store at -80 °C.
- MBR cultivation protocol
Note: For the employed BioLector MBR system in this study, scattered light (biomass) and GFP fluorescence are intensity measurements, which need a certain gain value assigned. The higher the gain, the higher the optical signal is amplified; this is also why scattered light and fluorescence are measured in arbitrary units (a.u.). Determine suitable gain values for biomass and GFP detection in preliminary experiments, along with suitable shaking frequency and filling volume to avoid oxygen limitation at higher biomass concentrations during later process stages. The employed cultivation conditions ("Flowerplates", i.e., flower-shaped 48-well MTPs, shaking frequency of 1,200 rpm, filling volume of 1,000 µL, 10 g/L glucose as main carbon source) ensured oxygen unlimited cultivations. For maximum oxygen transfer rates resulting from other combinations of filling volumes and shaking frequencies in flower-shaped 48-well MTPs, datasheets from the supplier are available. Also, growth defects of individual cultures may occur due to low amounts of secondary substrates (e.g., nitrogen, trace elements). Therefore, check the biomass concentrations at the end of cultivations. In this study, no such growth effects were observed.- Define the cultivation protocol for the MBR system ("BioLector") as follows:
- Filterset 1: Biomass, gain 14. Filterset 2: pH, gain is preset. Filterset 3: pO2, gain is preset. 4: GFP, gain 80.
- Shaking frequency: 1,200 rpm.
- Temperature: 30 °C.
- Cycle time: 15 min.
- Experiment time: manual (Cultivation will not be stopped automatically).
- Generation of medium stock solution pipetting list
Note: Almost any liquid handling robot system is capable to read pipetting actions from external files. Basically, the minimal information needed is the transfer volume and position of reagent source, as well as the destination of each represented by a labware position on the robotic deck and a specific cavity within the labware. However, different syntaxes for different liquid handling systems must be considered. Figure 2 shows an example file structure for the employed pipetting system in this study.- Four types of stock solutions are pipetted into each cultivation well:
- Stock solutions of media components that are varied ("Variation Stocks").
- Water to compensate for different cumulative volumes of above stocks.
- A stock solution ("Rest Stock") containing all components that are fixed. This stock solution can be composed from stocks containing the different components that are, e.g., stored at different temperatures or sterilized by different methods.
- Inoculum, which should be added as the last component and put onto the worktable just before addition to avoid settling of the cells.
- Per cultivation well, calculate the volumes to be transferred for all media components according to:
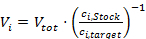
The volume to be added for certain Variation Stocks maybe zero, i.e., when this specific component is omitted. - Revise all calculated volumes Vi for suitable numbers. Pipetting volume increments in volume steps should not be too small. If necessary, adjust the concentrations of Variation Stocks. For instance, the minimal pipetting volume here was defined as 10 µL, and the minimal increment was defined as 5 µL. In general, these volumes should be defined based on experimentally determined precision and accuracy for the used liquid handling station15,44.
- Calculate the volume of Rest Stock containing the fixed components for all wells as follows:
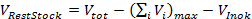
where the maximum cumulative volume of Variation Stock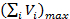 is used. Consequently, calculate the required concentrations of the fixed components in Rest Stock as follows:
is used. Consequently, calculate the required concentrations of the fixed components in Rest Stock as follows:
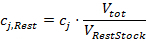
Prepare Rest Stock accordingly. - Per cultivation well, calculate the volume of water to be added as follows:

- Per cultivation well, list all volumes to be added in the following order of addition: VH2O, VRestStock, Vi, VInok. Format the pipetting list according to the specifications of the liquid handling station, as shown for an example in Figure 2.
- Four types of stock solutions are pipetted into each cultivation well:
- Working cell bank (WCB)
- Seed culture, automated media preparation, and start of main culture
- Create a protocol for the liquid handling robot. See Figure 3 and Figure 4 for an example protocol for a "Janus" system, implemented in corresponding "WinPREP" software. The protocol should consider the following aspects:
- Include a sufficient runtime of sterile housing prior to media preparation.
- Include an initial excessive cleaning and washing of all pipetting tips and tubing.
- Choose appropriate labware containers for stock solutions. Here, deep well plates with 12 columns are suitable for storage of Variation Stocks, as all eight pipetting tips can dip in a parallel arrangement into the well columns. This greatly speeds up media preparation. If 15 mL or 50 mL reagent tubes are used as reservoirs, only one pipetting tip can dip into it at once. For other stocks like water and Rest Stock, 100 mL troughs are used, as these stock solutions require higher volumes in total.
- Provide a sufficient total volume for each stock solution to compensate for waste volumes, height pipetting offsets, etc.
- Insert a user prompt before the inoculation step, ensuring that this step is carried out immediately prior to the seed culture procedure.
- Prepare all stock solutions in a sterile manner and store until use, in appropriate containers, e.g., sterile 15 mL and 50 mL test tubes.
- Sterilize the deep well plates for stock solution storage on a worktable, e.g., by wiping with 70% ethanol and subsequent drying in a laminar flow hood.
- Start the seed culture by inoculating 50 mL BHI medium containing 25 mg/L kanamycin with one aliquot from the WCB. Before the MBR cultivation, prepare fresh, vital inoculum from exponentially growing seed cultures in a sufficient amount.
- Place all necessary labware on the robotic worktable and pour stock solutions in the corresponding labware.
- Start the robotic workflow for media preparation, so that the last step (inoculation), is reached in time with the start of seed culture. The total runtime of the robotic workflow needs to be evaluated previously. Here, the total runtime was approximately 1.5 h.
- Sample the seed culture after approximately 2 h, then each hour, to monitor the growth by optical density (OD600). After approximately 5 h, the culture reaches 3-4 OD600 and is used to inoculate the main cultures.
- Place the seed culture on the liquid handler worktable and continue the media preparation protocol. Seal cultivation MTP after inoculation.
- Place the sealed cultivation MTP in the BioLector device and start the pre-defined cultivation protocol.
- Dispose the remaining stock solutions, according to biosafety regulations if necessary, and seed the culture from the robotic worktable. Clean the re-usable labware and start the liquid handler decontamination protocol.
- Create a protocol for the liquid handling robot. See Figure 3 and Figure 4 for an example protocol for a "Janus" system, implemented in corresponding "WinPREP" software. The protocol should consider the following aspects:
- Product quantification and preprocessing of raw data for analysis
- Stop the main culture after a 17 h runtime.
- Transfer the measurement data from BioLector MBR device to the connected computer using the BioLection software package according to the manufacturer's user manual.
- Use the "Data Management → Transform Data" function of the "BioLection" software package that accompanies the MBR system to convert the raw data file into an easy access spreadsheet format. Copy the GFP signal data for all cultivation wells from the timestamp column closest to 17 h. Identify the GFP signals from the reference cultivation wells and the average. Normalize all remaining GFP signal by the average reference signal.
- Quantify the GFP titer using additional methods (if required)
- Transfer the cell suspensions from all cultivation wells into prelabeled reaction tubes and obtain the cell-free supernatant after centrifugation for 10 min at maximum speed using a benchtop centrifuge.
- Transfer 200 µL of each cell-free supernatant into a black 96-well MTP with transparent bottom, and read the GFP specific fluorescence at 488/520 nm using an appropriate microplate reader. Normalize the GFP signal from all wells with average signal from reference cultivations, as in step 1.5.3.
NOTE: The absolute GFP fluorescence signals cannot be compared for different measurement devices. Therefore, the 5 reference strains from all main cultivations serve as an internal standard. The improvement of GFP titer can be expressed in relation to the internal standard. This allows for comparison of results from different experiments and different GFP quantification methods (see next two steps). - Determine the protein content of the cell-free supernatants with standard protocols, e.g., Bradford or BCA assay. In combination with the results from step 2, the determination of a protein specific GFP titer is possible.
NOTE: After fluorescence measurement, use the sample directly from this microplate. Use of multi-channel pipettes greatly facilitates the necessary liquid handling steps. - Perform an SDS-Page visualization of cell-free supernatants following standard protocols to verify that most of the increase protein content is due to increased GFP secretion.
NOTE: SDS-Page is more laborious than the previously mentioned methods, especially with a high sample load. For verification purposes, it is often sufficient to run SDS-Pages of cell-free supernatants from reference medium and final optimized medium cultivations15.
2. Sensitivity Analysis (Figure 1: Part B)
NOTE: The goal of this part is to identify the important factors that have a significant effect on the objective.
- Choose an initial concentration range of media components. The reference medium composition should lie inside the chosen concentration ranges.
- Choose an appropriate DOE. Such designs can be found in literature, e.g., from the NIST/SEMATECH e-Handbook of Statistical Methods (available at www.itl.nist.gov/div898/handbook/pri/section3/pri3347.htm). The chosen design depends on number of media components of interest (here, 11) and the number of performed experiments (here, 32). In the given example, design "2IV11-6" was chosen that results in 32 experiments and allows estimation of the effect of increasing the concentration of one of the eleven components on the objective of interest. To be more specific, the main effects are not confounded with pair-wise factor interactions. They can be confounded with higher order interactions that are, however, most likely not significant.
- Use the remaining wells (here, 16) for performing multiple replicates with the reference medium to assess the reproducibility of the process. Replicates should be equally spread over the plate to discover positional effects. The mean value of the measured output from the reference experiments is used for normalization. That is, each measured output of the sensitivity analysis is divided by the mean value of the reference output.
- Calculate the main and if possible the combinatorial effects using appropriate statistics software. The classic design of experiment is based on a polynomial approximation45. For instance, the MATLAB function mvregress can be used for the estimation of the polynomial coefficients. The mvregress function is part of the statistics and machine learning toolbox.
- Identify the relevant effects of various media components on the objective using a ttest. In the example, NH4+ has a significant negative effect, and Ca2+ and Mg2+ show the strongest positive effects (Figure 5). Because GFP signal was normalized, the coefficients represent the average relative change in the GFP signal when increasing concentration of the particular component from its center value,
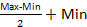 , to its maximum value.
, to its maximum value. - Fixed components without a relevant effect are at their reference value during the optimization.
3. Iterative Optimization (Figure 1: Part C)
NOTE:The MATLAB tool KriKit was used for the interpretation and statistical data analysis36. KriKit allows the user to construct a data-driven Kriging model. This Kriging model predicts the functional relationship between the media components and the objective. It also provides information on the prediction uncertainty. High uncertainty indicates noisy data and/or insufficient data density.
- DOE
NOTE: New experiments are iteratively designed, based on the results of the previous run.- In the first iteration, design new experiments for detailed investigation of identified media components of interest. Experimental results from Section 2 cannot be transferred to the iterative optimization (Section 3), as the concentrations of components without relevant effect are now fixed to the respective reference value. Consequently, experiments from the sensitivity analysis and the iterative optimization are not comparable.
- Otherwise, follow the scheme illustrated in Figure 1, frame "Iterative optimization". If the potential optimum lies inside the defined concentration range, new experiments are designed using the expected improvement40,46. The experimental design based on the expected improvement is integrated in the KriKit toolbox. If the optimum lies on the boundary, expand the concentration range.
- Perform experiments at the designed sample points according to experimental methods defined in Section 2.
- Statistical analysis
- Construct a Kriging model using the combined data from all iterations, including the current.
- Investigate the model output by visualization using the comprehensive tools of KriKit (2/3D interpolation, movie analysis, screening plot, etc.).
- If an optimal area was estimated with sufficient accuracy, stop optimization. If not, continue with step 3.1.
NOTE: Figure 6 illustrates the iterative optimization for the test study. The concentration range was successively expanded until a plateau was found (iteration 1-6). The seventh iteration was used for exploring the boundaries in more detail.
4. Verification of Results (Figure 1: Part D)
NOTE: After finishing the iterative optimization, the initial assumptions need to be checked for validity.
- Redo sensitivity analysis (Section 2) for optimal medium composition. That is, the concentrations of components that are investigated in Figure 1 (part C) are fixed to their optimal values. Concentrations of other components are varied according to an appropriate DOE.
NOTE: Similar results in both screenings indicate that the concentration levels of the investigated components do not alter the effect of the other components. If the screening shows significant differences to the results from part B, components with a changed effect should be added to the pool of investigated components and part C should be repeated. - In the case of indirect measurement (e.g., fluorescence as indicator for product concentration), apply orthogonal measurement approaches (e.g., activity assay, Bradford or BCA protein quantification, SDS page) to confirm the change in the objective of interest by comparing results from the reference medium and the optimized medium15.
Representative Results
The introduced protocol was applied for maximizing the titer of secreted GFP. Specifically, the GFP titer after 17 h of cultivation was chosen as the optimization objective. Online fluorescence detection of GFP allowed simple product quantification. However, the normalization of GFP signal with data from a reference cultivation is indispensable to ensure reproducibility and comparability of results. A pre-selection of media components was performed on a rational basis as described in Section 1. Experiments were performed following the instructions of Section 1: the parameters of the wet lab procedures were defined for the whole study ensuring consistency and reproducibility of results.
As described in Section 2, an initial screening was performed for identifying relevant components showing a significant impact on the optimization objective for the further, more detailed study. The MTP-based MBR system allows 48 experiments to be performed in parallel. Taking into account the maximal possible number of parallel experiments on one MTP (48) and the total number of media components (11) makes the 2IV11-6 fractional design an appropriate choice. This experimental design comprises 32 experiments and allows the estimation of the main effect for each of the investigated media components. The remaining cultivation wells (16) were used for multiple replicates of experiments with the reference medium to assess reproducibility and positional effects. That is, each experiment is conducted once (no replicates), except of the reference experiment (five replicates).
Table 1 summarizes the results of the screening analysis. In the considered concentration range, varying the majority of the media components did not show a noticeable effect on the objective. Component NH4+ shows a strong negative effect, while Ca2+ and Mg2+ show the strongest positive tendency. The effect of Mg2+ is not significant for the current concentration range but might be for a broader concentration range. Consequently, it was decided to omit NH4+ from the medium and to investigate the effect of Ca2+ and Mg2+ in further experiments.
Section 3 describes the iterative optimization procedure that is used for maximizing the GFP fluorescence signal while varying the concentrations of Ca2+ and Mg2+. In iteration 1, the hypothesis that NH4+ can be omitted was tested. The concentration range for Ca2+ and Mg2+ was adopted from the screening analysis. The minimum concentration of NH4+ was set to zero and the maximum concentration was adopted from the screening experiment. In the following experiments, the component concentrations were distributed over a 3 x 3 x 3 grid inside the defined concentration range, resulting in 27 experiments. During all cultivations, five replicates of reference medium were included, which served as internal standard and to ensure that no positional effects over the MTP occurred. For the remaining 16 wells, the concentrations of NH4+, Ca2+, and Mg2+ were randomly distributed inside the given ranges.
Figure 5A visualizes the results of the first iterations. Axis labels refer to the component concentrations used in the original reference medium, indicated by x Ref. The blue surfaces represent the Kriging interpolations that were calculated using the KriKit software. Each surface is associated with a relative concentration level for NH4+ (dark blue: 0 x Ref, checkered: 1 x Ref, light blue: 2 x Ref). This visual representation reveals that it is favorable to omit NH4+. Interpolation surfaces also show the positive effects of both Mg2+ and Ca2+, as all planes rise with increasing concentrations.
Based on the results of iteration 1, it was decided to expand the concentration range of Ca2+ and Mg2+ by doubling the maximum concentrations and shifting the experimental design window to the upper right corner, see Figure 5B. Inside this range, the concentrations were distributed on a 6 x 6 grid. This ensures an even distribution over the full concentration range, leading to optimal Kriging interpolation results. Figure 5B shows the Kriging interpolation plot based on the combined data measured in both iterations (red dots and yellow squares). For both, Ca2+ and Mg2+, the positive effect of increasing their concentrations continues. Consequently, the procedure was repeated by doubling the maximum concentration and thus, the experimental design window was moved to explore the boundaries of the upper right corner.
Figure 6A gives an overview of the remaining optimization procedure. The analysis of the collected data set up to iteration 3 revealed a limitation of the positive effect of Mg2+, i.e., an optimal concentration range of Mg2+ was identified. It was therefore decided to further expand the concentration range only for Ca2+ (iteration 4). This procedure was repeated twice (iteration 5 and 6) until a saturation of the GFP signal was found. This saturation is explained by precipitation of Ca-salts for the applied concentrations of Ca2+, which are not available to the cells.
As experimental results are always perturbed by noise, the resulting Kriging interpolation appears irregular and visual inspection might lead to false conclusions. However, the optimal concentration range of media components for the saturated GFP signal can be reliably identified with the statistical z-test, which is also implemented in KriKit. The z-test uses directly the intrinsic statistical information provided by the Kriging method, i.e., prediction values and prediction uncertainties. Figure 6B shows the identified plateau, as determined and visualized using the KriKit toolbox. The KriKit toolbox is freely available36 and comes with a detailed tutorial that explains how to use its features.
If more than two relevant components are found, 3D-visualization reaches its limit. KriKit provides several other possible visual representation methods such as movies or "screening plot". If the potential optimum lies inside the defined concentration range, new experiments are automatically designed using the expected improvement40,46. The experimental design based on the expected improvement is integrated in the KriKit toolbox. More detailed information can be found in the software documentation.
After the iterative procedure, a verification of the results was conducted, as described in Part D. The validity of initial assumptions was checked by performing an additional sensitivity analysis using the optimal medium composition. That is, all initial media components of interest were varied, but Ca2+ and Mg2+ were set to their optimal concentration levels. In this study, the optimal concentrations 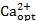 = 32 x Ref and
= 32 x Ref and 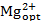 = 6.8 x Ref were chosen. Table 2 shows the results of the validation screening. Similar to the initial sensitivity screening (cf. Table 1), NH4+ still has a significant negative influence and remaining effects are still negligible.
= 6.8 x Ref were chosen. Table 2 shows the results of the validation screening. Similar to the initial sensitivity screening (cf. Table 1), NH4+ still has a significant negative influence and remaining effects are still negligible.
Due to easy access, the GFP fluorescence signal from the cultivation suspension was used to quantify the extracellular GFP titer during all experiments. For verification reasons, GFP fluorescence was validated against other measurements. Because GFP is secreted via the Tat-pathway, the fluorescence signal cannot discriminate between intra- and extracellular GFP. Thus, cultivations were reproduced using the reference medium and the optimized medium. Besides the fluorescence measurement from cell-free cultivation supernatants, protein content was quantified by the Bradford assay and (semi)-qualitative GFP improvement visualized by SDS-Page15. All resulting measurement signals were approximately doubled for cultivations with optimized medium compared to reference medium and validated the approximately 100% improvement of secretion performance of optimized medium. Consequently, GFP specific fluorescence of cultivation suspension can be considered a suitable metric for the optimization objective, i.e., the extracellular GFP titer.
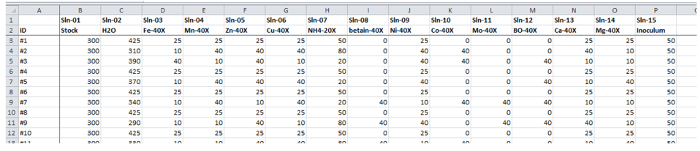
Figure 2: Screenshot from the volume pipetting list for sensitivity analysis. Entries in the first column assign a unique identifier to all volumes of a row; this identifier is the MTP well number of the target cultivation MTP on the liquid handler worktable, cf. Figure 4C. Remaining columns encode volumes for different solutions ("Sln-01" to "Sln-15") to be pipetted. The cumulative volume of one row corresponds to the final cultivation volume of the corresponding well. Please click here to view a larger version of this figure.
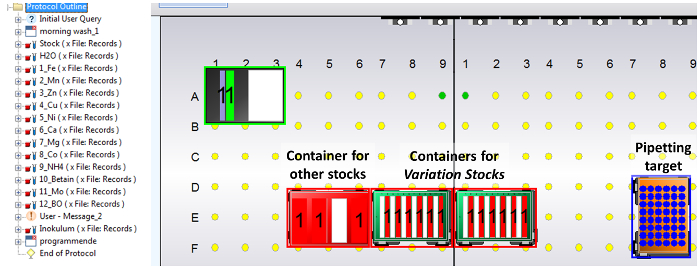
Figure 3: Screenshot from the liquid handling control software "WinPREP". Left: Row-ordered commands, including a transfer command for each stock solution to be pipetted. Before the final command for inoculum addition, a user prompt is inserted to ensure the seed culture is placed at the table just in time. Right: Schematic of the worktable, including the source labware for Variation Stocks (two deep well plates with 12 column-like wells), the reagent trough for Rest Stock, water and inoculum, and the media preparation target cultivation MTP. Please click here to view a larger version of this figure.
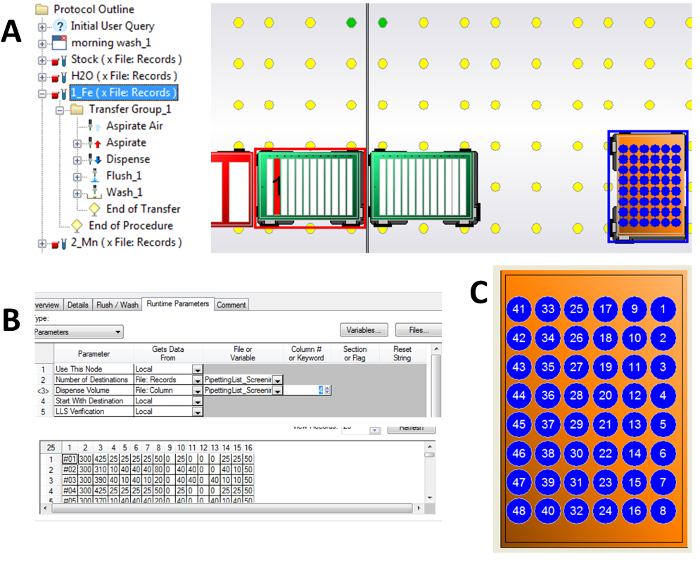
Figure 4: Compilation of detailed screenshots for setup of pipetting of a stock solution. (A) Unwrapped command for pipetting of Fe stock. Source labware and source well within are marked on the worktable by the read frame and red column of the corresponding deep well plate. Destination labware and destination wells within are marked by the blue frame around and blue wells of the target cultivation MTP. (B) Detailed example view on assignment of pipetting volumes for this step (Fe stock solution). Number of destinations is read from the pipetting list, which has 48 rows. The dispense volumes for all destination wells for Fe stock solution is found in column 4 in the pipetting list. Note that the first column in the pipetting list contains identifiers and not volumes to be transferred, see Figure 2. (C) Details on destination well numbering. Volumes written in the row #1 of the corresponding pipetting list will be pipetted into well marked as #1, and so on. Wells #01, #08, #41 and #48 correspond to wells A01, A08, F01 and F08 for the alpha-numeric coding, which is also printed into the cultivation MTP itself. Please click here to view a larger version of this figure.
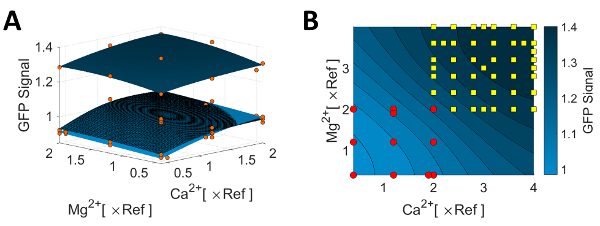
Figure 5: Detailed results from the first iteration. (A) Kriging interpolation based on experimental data of iteration 1. Red dots indicate the data set. For comparison, all three interpolation surfaces are overlayed in one plot (dark blue: 0 x Ref, checkered: 1 x Ref, light blue: 2 x Ref). An alternative representation of the results can be found elsewhere15. (B) Kriging interpolation based on the experiments performed in iteration 1 (red dots) and iteration 2 (yellow squares). Parts of the data presented in this figure have been previously published15. Please click here to view a larger version of this figure.
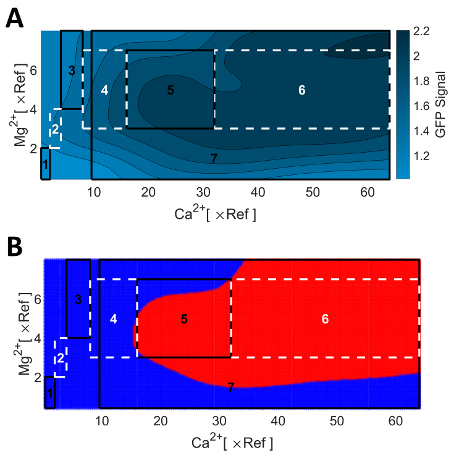
Figure 6: Depiction of iteratively collected optimization results. (A) Final Kriging model prediction. (B) Statistical identification of optimal area (red) based on the statistical z-test, which is provided by KriKit. Boxes indicate successive steps of iterative design and execution of experiments. Parts of the data presented in this figure have been previously published15. Please click here to view a larger version of this figure.
| Component | Normalized Coefficient Mean Value |
| Fe2+ | -0.08 |
| Mn2+ | -0.05 |
| Zn2+ | -0.21 |
| Cu2+ | -0.21 |
| NH4+ | -2.04 |
| Ni2+ | -0.11 |
| Co2+ | -0.10 |
| MoO42- | 0.03 |
| BO33- | 0.06 |
| Ca2+ | 1.00 |
| Mg2+ | 0.45 |
Table 1: Results of the sensitivity analysis. Coefficient values representing the average effect when increasing the respective media component concentration from its center value to its maximum value. For optimal experimental designs, as found in the standard literature and used here, the standard deviation represents directly the experimental variation due to replication of the reference experiment. Coefficient values were normalized by the maxium value (0.0422 for component Ca2+). Normalized and absolute coefficient standard deviation is 0.54 and 0.0226, respectively.
| Component | Normalized Coefficient Mean Value |
| Fe2+ | -1.00 |
| Mn2+ | 1.00 |
| Zn2+ | -3.48 |
| Cu2+ | -0.52 |
| NH4+ | -15.95 |
| Ni2+ | 0.69 |
| Co2+ | -0.51 |
| MoO42- | -0.45 |
| BO33- | -1.11 |
Table 2: Results of the final sensitivity analysis. Coefficient values representing the average effect when increasing the respective media component concentration from its center value to its maximum value. As an optimal experimental design was used, the standard deviation only depends on the experimental variation using the optimized medium composition. Experimental variation increased slightly in comparison to the variation using the reference medium. Coefficient values were normalized by the maxium value (0.0106 for component Mn2+). Normalized and absolute coefficient standard deviation is 3.63 and 0.0385, respectively.
Discussion
The generic nature of the presented protocol allows various adaptions, e.g., for studying other microbial expression hosts9,47,48,49,50,51, or to optimize other properties of the target protein, like glycosylation pattern or disulphide bonds. The protocol may also need to be adapted to the available lab equipment. The integration of an MBR system allows increasing experimental throughput, which enables great savings in time. However, when replacing fully instrumented and controllable bioreactors by MBR systems, scalability of results must be considered8,37,52,53. The use of DOE methodologies and mathematical modeling helps to maximize the information content of measurement data with respect to the studied objective54 by efficient experimental planning and model-based data interpretation15.
Modifications to the Method
Next to multi-purpose and expandable robotic liquid handling systems like the one used in this study, it should be mentioned that there are several smaller liquid handling systems commercially available which are capable to perform this task and can be placed inside of laminar flow work benches. If no automated pipetting system is available, different media compositions according to the DOE plan can also be realized by manual pipetting using single and/or multi-channel pipettes. Since the manual preparation is more error-prone and will require highly focused work for quite a long time, it is recommended to prepare a lower number of different media compositions.
Depending on the capabilities of the employed MBR system, the corresponding cultivation protocol will vary. For instance, if no online measurement of biomass formation is available, it may be sufficient to measure biomass concentration after completion of the growth experiment. In combination with online monitoring of the pH and dissolved oxygen, which is implemented in several MBR systems, the growth saturation can be determined safely. In principle, the growth experiments can be conducted in MTPs alone placed inside shaking incubators, without the use of a MBR system. In this case, proper cultivation conditions have to be ensured: (1) Oxygen-limited cultivations can be avoided by using MTPs with suitable geometries, in combination with proper shaking frequencies and shaking diameters, e.g., square 96 or 24 deep well plates operated at 1,000 rpm at 3 mm throw or at 250 rpm at 25 mm throw, respectively. Importantly, the lower the achievable maximal oxygen transfer rates, the lower the main carbon source should be concentrated. As mentioned above, for this study, the use of 10 g/L glucose was suitable to prevent oxygen limitation for the employed cultivation conditions; (2) Sampling of the MTP cultures for biomass and product quantification should be reduced to a minimum. Each time the MTP is removed from the shaking incubator, oxygen transfer will immediately break-down which may result in unfavorable cultivation conditions; (3) In the opinion of the authors, the use of MTP readers as cultivation devices is not recommended as these devices were not developed for this purpose. For example, shaking mechanics were built for occasional mixing of microplates after the reagent addition and thus, often lack robustness for long runs of continuous shaking lasting for days. Moreover, sufficient power input needed for microbial cultivations cannot be realized in these readers. The integration of optical density readings in short time intervals requires stopping of the shaking motion, resulting in repeated periods of oxygen limitation. Furthermore, evaporation in such systems over long cultivation periods will distort results. For more details on the surprisingly complex topic on using MTPs for microbial cultivations, the reader is referred to the cited literature22,23,24,25,26 and references therein.
Further Considerations
To speed-up iterative optimization steps, it is advised to carefully select the analytical method for product quantification. Fast and simple methods should be preferred at the cost of precision and accuracy, as the iterative experimental design strategy tolerates experimental inaccuracy. However, the final results must be verified against sufficiently precise and accurate product quantification methods that might be more complicated. In general, careful evaluation and decision making about the study procedures require effort in the beginning of the study, but pay out in the long run, after routine methods have been established.
It is highly recommended to define a reference experiment that is compared to all experiments during the optimization. That is, the applied medium component concentrations as well as measured output are normalized via dividing by reference values. This way, each applied and measured value can be interpreted as the x-fold of the reference value. To take into account variations between the plates, five reference experiments are performed on each plate. The mean value of the measured outcome is used for the normalization.
It can generally not be guaranteed that the developed medium is also optimal for other strains. However, the improved medium will most likely also be appropriate for cultivating expression strains with small genetic differences, e.g., when producing enzyme variants with single amino acid substitutions obtained from mutagenesis studies (although even single point mutations have been described to effect cellular metabolism and heterologous expression performance55,56). In this case, the presented protocol can be a first step, followed by protocols for high-throughput expression screenings57. If the protocol is used for medium development with subsequent scale-up to fed-batch cultivations, the optimized medium should be verified for the corresponding bioprocess conditions, as clone screening campaigns at the microscale identified different top performers for different feeding strategies and cultivation media52,58. Furthermore, the introduced KriKit36 can generally contribute to improved holistic bioprocess optimization. Only recently, the tool abilities were extended to also support multi-objective optimization40, which can be important for optimizing both upstream and downstream processes59,60.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The scientific activities of the Bioeconomy Science Center were financially supported by the Ministry of Innovation, Science, and Research within the framework of the NRW-Strategieprojekt BioSC (No. 313/323-400-002 13). The authors thank the Ministry of Innovation, Science, and Research of North Rhine-Westphalia and the Heinrich Heine University Düsseldorf for a scholarship to Lars Freier within the CLIB-Graduate Cluster Industrial Biotechnology. Further funding was received from the Enabling Spaces Program "Helmholtz Innovation Labs" of German Helmholtz Association to support the "Microbial Bioprocess Lab – A Helmholtz Innovation Lab".
Materials
| BioLector | m2p-labs | G-BL-100 | |
| Flowerplate | m2p-labs | MTP-48-BOH | For cultivation in the BioLector device |
| Sealing foil | m2p-labs | F-GP-10 | Sterile sealing for Flowerplate |
| MATLAB | Mathworks | 2016b | |
| KriKit | Forschungszentrum Jülich | n/a | Freely available, MATLAB installation required |
| Janus pipetting robot | Perkin Elmer | n/a | Includes "WinPrep" software installation |
| 12-column deep well microplate | E&K Scientific | EK-2034 | Container for medium stock solutions |
| 96 well microplates, transparent, F-bottom | Greiner | 655101 | For Bradford protein assay |
| µclear 96 well microplates, black body, transparent F-bottom | Greiner | 655087 | For flourescence measurement in cell-free supernatants |
| Pipette Research plus multi-channel pipettes | Eppendorf | n/a | Facilitates manual liquid handling with microplates |
| TruPAG Precast Gels | Sigma | PCG2002 | For SDS-Page analysis of cell-free supernantants |
| Bradford Reagent | Sigma | B6916 | |
| C. glutamicum pCGPhoDBs-GFP | n/a | n/a | Carries pEKEx2 plasmid with fusion of GFP gene and PhoD signal peptide from B.subtilis as expression insert. Plasmid provides kanamycin resistance. Described and published by Meissner et al. Appl Microbiol Biotechnol 76 (3), 633–42 (2007) |
References
- Choi, J. -. M., Han, S. -. S., Kim, H. -. S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol Adv. 33 (7), 1443-1454 (2015).
- Li, S., Yang, X., Yang, S., Zhu, M., Wang, X. Technology Prospecting on Enzymes: Application, Marketing and Engineering. Comput Struct Biotechnol J. 2 (3), (2012).
- Adrio, J. L., Demain, A. L. Microbial enzymes: tools for biotechnological processes. Biomolecules. 4 (1), 117-139 (2014).
- Yim, S. S., An, S. J., Kang, M., Lee, J., Jeong, K. J. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng. 110 (11), 2959-2969 (2013).
- Hemmerich, J., et al. Use of a Sec signal peptide library from Bacillus subtilis.for the optimization of cutinase secretion in Corynebacterium glutamicum. Microb Cell Fact. 15 (1), 208 (2016).
- Wendisch, V. F. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol. 30, 51-58 (2014).
- Lee, J. -. Y., Na, Y. -. A., Kim, E., Lee, H. -. S., Kim, P. The Actinobacterium Corynebacterium glutamicum, an Industrial Workhorse. J Microbiol Biotechnol. 26 (5), 807-822 (2016).
- Rohe, P., Venkanna, D., Kleine, B., Freudl, R., Oldiges, M. An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb Cell Fact. 11 (1), 144 (2012).
- Liu, X., et al. Expression of recombinant protein using Corynebacterium glutamicum.: progress, challenges and applications. Crit Rev Biotechnol. 36 (4), 652-664 (2016).
- Freudl, R., Burkovski, A. Corynebacterium glutamicum as a Platform Organism for the Secretory Production of Heterologous Proteins. Corynebacterium glutamicum: From systems biology to biotechnological applications. , 161-177 (2015).
- Yim, S. S., et al. Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol Bioeng. 113 (1), 163-172 (2015).
- Limberg, M. H., et al. Metabolic profile of 1,5-diaminopentane producing Corynebacterium glutamicum under scale-down conditions: Blueprint for robustness to bioreactor inhomogeneities. Biotechnol Bioeng. 114 (3), 560-575 (2016).
- Käβ, F. Assessment of robustness against dissolved oxygen/substrate oscillations for C. glutamicum DM1933 in two-compartment bioreactor. Bioprocess Biosyst Eng. 37 (6), 1151-1162 (2014).
- Teramoto, H., Watanabe, K., Suzuki, N., Inui, M., Yukawa, H. High yield secretion of heterologous proteins in Corynebacterium glutamicum.using its own Tat-type signal sequence. Appl Microbiol Biotechnol. 91 (3), 677-687 (2011).
- Freier, L., Hemmerich, J., Schöler, K., Wiechert, W., Oldiges, M., von Lieres, E. Framework for Kriging-based iterative experimental analysis and design: Optimization of secretory protein production in Corynebacterium glutamicum. Eng Life Sci. 16 (6), 538-549 (2016).
- Huber, R., Roth, S., Rahmen, N., Büchs, J. Utilizing high-throughput experimentation to enhance specific productivity of an E. coli. T7 expression system by phosphate limitation. BMC Biotechnol. 11 (1), 22 (2011).
- Kottmeier, K., Müller, C., Huber, R., Büchs, J. Increased product formation induced by a directed secondary substrate limitation in a batch Hansenula polymorpha culture. Appl Microbiol Biotechnol. 86 (1), 93-101 (2010).
- Kennedy, M. J., Krouse, D. Strategies for improving fermentation medium performance: A review. J Ind Microbiol Biotechnol. 23 (6), 456-475 (1999).
- Kleman, G. L., Strohl, W. R. Developments in high cell density and high productivity microbial fermentation. Curr Opin Biotechnol. 5 (2), 180-186 (1994).
- Jones, R., Gadd, G. Ionic nutrition of yeast: Physiological mechanisms involved and implications for biotechnology. Enzy Microb Technol. 12 (6), 402-418 (1990).
- Zhang, J., Greasham, R. Chemically defined media for commercial fermentations. Appl Microbiol Biotechnol. 51 (4), 407-421 (1999).
- Kensy, F., et al. Oxygen transfer phenomena in 48-well microtiter plates: Determination by optical monitoring of sulfite oxidation and verification by real-time measurement during microbial growth. Biotechnol Bioeng. 89 (6), 698-708 (2005).
- Hermann, R., Lehmann, M., Büchs, J. Characterization of gas-liquid mass transfer phenomena in microtiter plates. Biotechnol Bioeng. 81 (2), 178-186 (2002).
- Duetz, W. A. Microtiter plates as mini-bioreactors: Miniaturization of fermentation methods. Trends Microbiol. 15 (10), 469-475 (2007).
- Funke, M., Diederichs, S., Kensy, F., Müller, C., Büchs, J. The baffled microtiter plate: Increased oxygen transfer and improved online monitoring in small scale fermentations. Biotechnol Bioeng. 103 (6), 1118-1128 (2009).
- Lattermann, C., Funke, M., Hansen, S., Diederichs, S., Büchs, J. Cross-section perimeter is a suitable parameter to describe the effects of different baffle geometries in shaken microtiter plates. J Biol Eng. 8, 18 (2014).
- Samorski, M., Müller-Newen, G., Büchs, J. Quasi-continuous combined scattered light and fluorescence measurements: A novel measurement technique for shaken microtiter plates. Biotechnol Bioeng. 92 (1), 61-68 (2005).
- Kensy, F., Zang, E., Faulhammer, C., Tan, R. -. K., Büchs, J. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb Cell Fact. 8 (1), 31 (2009).
- Puskeiler, R., Kaufmann, K., Weuster-Botz, D. Development, parallelization, and automation of a gas-inducing milliliter-scale bioreactor for high-throughput bioprocess design (HTBD). Biotechnol Bioeng. 89 (5), 512-523 (2005).
- Bareither, R., Pollard, D. A review of advanced small-scale parallel bioreactor technology for accelerated process development: Current state and future need. Biotechnol Prog. 27 (1), 2-14 (2011).
- Hortsch, R., Stratmann, A., Weuster-Botz, D. New milliliter-scale stirred tank bioreactors for the cultivation of mycelium forming microorganisms. Biotechnol Bioeng. 106 (3), 443-451 (2010).
- Isett, K., George, H., Herber, W., Amanullah, A. Twenty-four-well plate miniature bioreactor high-throughput system: Assessment for microbial cultivations. Biotechnol Bioeng. 98 (5), 1017-1028 (2007).
- Motta Dos Santos, L. F., Coutte, F., Ravallec, R., Dhulster, P., Tournier-Couturier, L., Jacques, P. An improvement of surfactin production by B. subtilis BBG131 using design of experiments in microbioreactors and continuous process in bubbleless membrane bioreactor. Bioresour Technol. 218, 944-952 (2016).
- Islam, R. S., Tisi, D., Levy, M. S., Lye, G. J. Framework for the Rapid Optimization of Soluble Protein Expression in Escherichia coli Combining Microscale Experiments and Statistical Experimental Design. Biotechnol Prog. 23 (4), 785-793 (2007).
- Huber, R., et al. Robo-Lector: A novel platform for automated high-throughput cultivations in microtiter plates with high information content. Microb Cell Fact. 8 (1), 42 (2009).
- . Kriging toolKit (KriKit) Available from: https://github.com/modsim/KriKit (2017)
- Kensy, F., Engelbrecht, C., Büchs, J. Scale-up from microtiter plate to laboratory fermenter: evaluation by online monitoring techniques of growth and protein expression in Escherichia coli.and Hansenula polymorpha fermentations. Microb Cell Fact. 8 (1), 68 (2009).
- Lu, C., Bentley, W. E., Rao, G. A high-throughput approach to promoter study using green fluorescent protein. Biotechnol Prog. 20 (6), 1634-1640 (2004).
- Zanzotto, A., Boccazzi, P., Gorret, N., van Dyk, T. K., Sinskey, A. J., Jensen, K. F. In situ measurement of bioluminescence and fluorescence in an integrated microbioreactor. Biotechnol Bioeng. 93 (1), 40-47 (2006).
- Freier, L., von Lieres, E. Multi-Objective Global Optimization (MOGO): Algorithm and Case Study in Gradient Elution Chromatography. Biotechnol J. , (2016).
- Keilhauer, C., Eggeling, L., Sahm, H. Isoleucine synthesis in Corynebacterium glutamicum.: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol. 175 (17), 5595-5603 (1993).
- Weuster-Botz, D., Kelle, R., Frantzen, M., Wandrey, C. Substrate Controlled Fed-Batch Production of L-Lysine with Corynebacterium glutamicum. Biotechnol Prog. 13 (4), 387-393 (1997).
- Meissner, D., Vollstedt, A., van Dijl, J. M., Freudl, R. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl Microbiol Biotechnol. 76 (3), 633-642 (2007).
- Morschett, H., Wiechert, W., Oldiges, M. Automation of a Nile red staining assay enables high throughput quantification of microalgal lipid production. Microb Cell Fact. 15, 34 (2016).
- Fisher, R. A. . The design of experiments. , (1935).
- Morschett, H., Freier, L., Rohde, J., Wiechert, W., von Lieres, E., Oldiges, M. A framework for accelerated phototrophic bioprocess development: Integration of parallelized microscale cultivation, laboratory automation and Kriging-assisted experimental design. Biotechnol Biofuels. 10, 26 (2017).
- Le Loir, Y., et al. Protein secretion in Lactococcus lactis: An efficient way to increase the overall heterologous protein production. Microb Cell Fact. 4 (1), 2 (2005).
- Anné, J., Vrancken, K., van Mellaert, L., van Impe, J., Bernaerts, K. Protein secretion biotechnology in Gram-positive bacteria with special emphasis on Streptomyces lividans. Biochim Biophys Acta. 1843 (8), 1750-1761 (2014).
- Gupta, S. K., Shukla, P. Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit Rev Biotechnol. 36 (6), 1089-1098 (2016).
- Fu, L. L., Xu, Z. R., Li, W. F., Shuai, J. B., Lu, P., Hu, C. X. Protein secretion pathways in Bacillus subtilis: implication for optimization of heterologous protein secretion. Biotechnol Adv. 25 (1), 1-12 (2007).
- Yoon, S., Kim, S., Kim, J. Secretory Production of Recombinant Proteins in Escherichia coli. Rec Pat Biotechnol. 4 (1), 23-29 (2010).
- Hemmerich, J., et al. Comprehensive clone screening and evaluation of fed-batch strategies in a microbioreactor and lab scale stirred tank bioreactor system: application on Pichia pastoris producing Rhizopus oryzae lipase. Microb Cell Fact. 13 (1), 36 (2014).
- Glazyrina, J., Krause, M., Junne, S., Glauche, F., Strom, D., Neubauer, P. Glucose-limited high cell density cultivations from small to pilot plant scale using an enzyme-controlled glucose delivery system. New Biotechnol. 29 (2), 235-242 (2012).
- Gernaey, K. V., et al. Monitoring and control of microbioreactors: An expert opinion on development needs. Biotechnol J. 7 (10), 1308-1314 (2012).
- Rahmen, N., Fulton, A., Ihling, N., Magni, M., Jaeger, K. -. E., Büchs, J. Exchange of single amino acids at different positions of a recombinant protein affects metabolic burden in Escherichia coli. Microb Cell Fact. 14, 10 (2015).
- Rahmen, N., et al. A particular silent codon exchange in a recombinant gene greatly influences host cell metabolic activity. Microb Cell Fact. 14, 156 (2015).
- Saez, N. J., Nozach, H., Blemont, M., Vincentelli, R. High throughput quantitative expression screening and purification applied to recombinant disulfide-rich venom proteins produced in E. coli. J Vis Exp. (89), e51464 (2014).
- Scheidle, M., et al. High-throughput screening of Hansenula polymorpha clones in the batch compared with the controlled-release fed-batch mode on a small scale. FEMS Yeast Res. 10 (1), 83-92 (2010).
- Baumann, P., Bluthardt, N., Renner, S., Burghardt, H., Osberghaus, A., Hubbuch, J. Integrated development of up- and downstream processes supported by the Cherry-Tag™ for real-time tracking of stability and solubility of proteins. J Biotechnol. 200, 27-37 (2015).
- Baumann, P., Hahn, T., Hubbuch, J. High-throughput micro-scale cultivations and chromatography modeling: Powerful tools for integrated process development. Biotechnol Bioeng. 112 (10), 2123-2133 (2015).

