Measuring Gene Expression in Bombarded Barley Aleurone Layers with Increased Throughput
Summary
An improved protocol is presented for the measurement of transient gene expression from reporter constructs in barley aleurone cells after particle bombardment. The combination of automated grain grinding with 96-well plate enzyme assays provides high throughput for the procedure.
Abstract
The aleurone layer of barley grains is an important model system for hormone-regulated gene expression in plants. In aleurone cells, genes required for germination or early seedling development are activated by gibberellin (GA), while genes associated with stress responses are activated by abscisic acid (ABA). The mechanisms of GA and ABA signaling can be interrogated by introducing reporter gene constructs into aleurone cells via particle bombardment, with the resulting transient expression measured using enzyme assays. An improved protocol is reported that partially automates and streamlines the grain homogenization step and the enzyme assays, allowing significantly more throughput than existing methods. Homogenization of the grain samples is carried out using an automated tissue homogenizer, and GUS (β-glucuronidase) assays are carried out using a 96-well plate system. Representative results using the protocol suggest that phospholipase D activity may play an important role in the activation of HVA1 gene expression by ABA, through the transcription factor TaABF1.
Introduction
The barley aleurone layer is a well-established model system for the study of hormone-regulated gene expression in plants1. In particular, a number of genes required for germination or early seedling development are activated by gibberellin (GA), while genes associated with stress responses are activated by abscisic acid (ABA). The GA and ABA signaling pathways are intertwined, as the expression of some GA-activated genes is inhibited by ABA, and vice-versa1.
A valuable strategy for understanding the role of particular actors in GA/ABA signaling has been the introduction of effector gene constructs via particle bombardment, followed by transient expression of reporter constructs that allow the resulting effect on downstream gene expression to be determined. The use of reporter genes such as GUS (β-glucuronidase) or Luciferase allows the sensitive and quantitative measurement of gene expression specifically within the cells that have received the effector construct. For example, the introduction of an effector construct encoding the transcription factor TaABF15,6 established that ABA-induced genes such as HVA1 are induced by TaABF1, while GA-induced genes such as Amy32b are repressed. Particle bombardment as an experimental strategy has been used by multiple laboratories to investigate diverse aspects of GA/ABA signaling. Such work has led to the identification of promoter elements important for the activation of both GA-induced2 and ABA-induced genes3, and to the discovery of protein kinases4 and transcription factors5 that regulate the expression of these genes.
The existing protocols2,3,4,5,6 for particle bombardment and subsequent measurement of transient gene expression are quite labor intensive, as each set of bombarded barely grains is homogenized by hand in a mortar and pestle and the enzyme assays are carried out individually. This manuscript reports an improved protocol that partially automates and streamlines the homogenization step and the GUS assays to allow significantly more throughput, permitting a larger number of treatments to be tested in the same experiment, and/or the inclusion of more replicates for each treatment to obtain more statistically robust results. Representative results are shown for the expression of HVA1 and Amy32b reporter constructs, regulated by the transcription factor TaABF1 as well as by GA, ABA, and other regulatory molecules.
Protocol
1. Preparation of Effector and Reporter Gene Constructs
- Build effector constructs that employ a constitutive promoter (e.g. maize ubiquitin, UBI) to drive expression from a desired open reading frame. For example, construct a plasmid containing UBI::TaABF1 to drive strong constitutive expression of TaABF15.
- Build reporter constructs that include the promoter whose activity is to be measured, placed upstream of an open reading frame, such as GUS. For example, construct a plasmid containing HVA1::GUS to measure expression from the HVA1 promoter2,5.
- Build an internal control construct (e.g. UBI::luciferase).
- Using a silica binding column protocol (see Table of Materials), prepare high-quality plasmid for each of the required gene constructs. Using ultraviolet spectroscopy (see Table of Materials), carefully measure the concentration of each plasmid preparation.
2. Preparation of Barley Grains for Bombardment
- Set up a spreadsheet (see Figure 1 for an example) indicating the plasmids that will be bombarded, the appropriate hormone treatments, and other relevant information for each treatment that will be part of the experiment.
- Using a cutting board and a sterile razor blade, cut off the embryos from grains of Himalaya barley (40 for each treatment included in the experiment). Exclude grains that are discolored or substantially smaller than average.
- Place all the embryoless grains into a 50 mL tube. Add 40 mL of water and rock for 10 minutes.
- Pour out the water carefully (to avoid losing grains) and add 40 mL of 10% bleach. Rock for 20 minutes.
- Pour out the bleach and add 40 mL of sterile water. Rock for 5 minutes. Repeat this sterile water wash four more times.
- Add 40 mL of Imbibing Solution (20 mM sodium succinate, 20 mM calcium chloride, pH 5.0) and 40 μL of chloramphenicol stock solution (10 mg/mL).
- Transfer most of the Imbibing Solution (with chloramphenicol) from the tube into a Vermiculite Plate (see Table of Materials). Then transfer the grains onto the surface of the wet filter paper in the Vermiculite Plate. Spread out the grains evenly. Pour in enough imbibing solution to keep the grains partially (but not completely) submerged. Place the lid on top of the Vermiculite Plate.
- Incubate the embryoless grains at 24 °C for about 48 h with fluorescent lighting. Add additional Imbibing Solution as necessary.
- Open up a 90 mm plastic petri dish and pour 20 mL of Imbibing Solution into the dish itself and 20 mL into the inverted lid. Add 20 μL of chloramphenicol (10 mg/mL) to each. Sterilize two pairs of fine point forceps by dipping them in alcohol.
- Place a few of the embryoless grains into the lid of the petri dish. Using the forceps, gently remove the (transparent) seed coat from each grain. While removing the seed coat from the smooth side of the grain, do not damage the underlying (aleurone) cells. Transfer the peeled grain to the petri dish containing Imbibing Solution.
- After peeling the seed coat from all of the grains, return them to the Vermiculite Plate. Incubate the peeled embryoless grains at 24 °C for 16 – 20 hours.
- Just before using the grains for bombardment, remove surface moisture by shaking them between filter papers in a petri dish.
3. Preparation of Plasmid DNA for Bombardment
- Label a 1.5 mL tube for each treatment that will be included in the bombardment experiment. To each tube add 2.5 µg of UBI::luciferase internal control plasmid, 2.5 µg of a GUS reporter plasmid, and the desired amount (typically 0.025 µg to 2.5 µg) of an effector plasmid. Add water to bring the total volume to 5 µL.
- Prepare 1.6 µm gold microcarriers (see Table of Materials) following published protocols6.
- For each treatment, add 50 µL of suspended microcarriers into the 1.5 mL tube containing plasmid DNA. Coat the microcarriers with plasmid DNA according to manufacturer's protocols6.
- At the final step, when the plasmid-coated microcarriers are suspended in 48 µL 100% ethanol, pipet 8 μL of suspended microcarriers onto each of five macrocarriers and allow them to air dry.
4. Particle Bombardment of Embryoless Barley Grains
- Set up the particle bombardment apparatus (see Table of Materials)6.
- Place a 1550 psi rupture disk into the retaining cap and mount it on the instrument.
- Place a macrocarrier containing the desired plasmid combination (together with a stopping screen) into the microcarrier launch assembly. Insert the assembly into the top slot of the bombardment chamber. Set the distance between the rupture disc retaining cap and the macrocarrier cover lid at the standard distance (6 mm), and place the stopping screen support at the (standard) middle position, allowing a macrocarrier travel distance of 11 mm (Figure 2A).
- Arrange 8 embryoless grains in a tight radial pattern (with the thinner ends pointing inward) on a filter paper circle that is taped down flat to the lid of a 90 mm petri dish (see Figure 2B).
- Place the dish with the grains into the bombardment chamber, at a distance of 6 cm from the stopping screen.
- Evacuate the bombardment chamber to 95 kPa and then bombard the sample.
- After releasing the vacuum, transfer the bombarded grains to a labeled 60 mm petri dish containing 4 mL of Imbibing Solution containing 1 mg/ml of chloramphenicol.
- Repeat until all of the bombardments have been completed. Incubate (on a shaking platform at 100 rpm) at 24 °C for 24 hours.
5. Extraction of Soluble Protein from the Bombarded Grains
- Using forceps, remove the 8 bombarded grains from a 60 mm petri dish and blot them dry on a paper towel. Divide the grains into two labeled 2.0 mL tubes (4 grains per tube) each containing 800 μL Grinding Buffer (100 mM Na phosphate pH 7.2, 5 mM DTT, 10 µg/mL leupeptin, 20% glycerol) and a 5 mm stainless steel bead. Repeat this process for all of bombarded grains.
- Place (up to 48) 2.0 mL tubes into the two racks of the bead homogenizer (see Table of Materials). Mount the racks on the homogenizer.
- Shake the samples at 30 Hz for 3 minutes.
- Remove the homogenizer racks from the homogenizer and place them on ice (5 min) to re-cool the samples.
- Mount the racks back on the homogenizer – in the opposite orientation. Shake the samples again at 30 Hz for 3 minutes.
- Cool the racks on ice, then repeat Steps 5.3 to 5.5. This will add up to a total of 12 minutes of shaking.
- Centrifuge the grain extracts at 16,000 x g at 4 °C for 10 minutes.
- Immediately after the centrifugation, decant the clear supernatants into a second set of microcentrifuge tubes. Give each tube a quick vortex. Store the tubes on ice.
- After transferring the supernatant, recover the steel beads from the pellets so they can be washed and reused.
6. Measurement of Luciferase Activity from Grain Extracts
- For each tube of grain extract to be assayed, prepare a 12 mm x 75 mm glass test tube containing 200 μL of Luciferase Assay Mixture (45 mM Tris sulfate pH 7.7, 15 mM MgCl2, 20 mM DTT, 1.5 mM EDTA, 1 mM luciferin, 1 mM ATP).
- Add 100 μL of the first grain extract to the first assay tube. Vortex quickly to mix well. Immediately place the tube into the tube holder of the luminometer and close the door. When the measurement is finished, remove the tube from the luminometer – leaving the door open for placement of the next tube.
- Repeat this process until all the samples have been measured.
7. Measurement of GUS Activity from Grain Extracts
- Add 200 μL of GUS Assay Buffer (50 mM Na phosphate pH 7.2, 2.5 mM 4-methylumbelliferyl–D-glucuronide, 10 mM DTT, 2 mM EDTA, 10 µg/mL leupeptin, 20% methanol, 0.02% sodium azide) to each well of a 96 well plate.
- Place 50 μL of each grain extract into the corresponding well. Mix with the tip after adding. Seal the top with sealing film.
- Incubate the 96 well plate at 37 °C in the dark for 20 hours.
- Centrifuge the 96 well plate at 4,000 x g at 4 °C for 10 minutes and place it on ice.
- Add 250 μL of 0.2 M Na2CO3 to each well of a black 96 well plate.
- Add 6.25 μL of each centrifuged GUS assay mixture into the corresponding well (containing 0.2 M Na2CO3) of the black 96 well plate. Mix with the tip after adding. Include wells with 6.25 μL of 10 μM, 40 μM, and 100 μM 4-methylumbelliferone as standards.
- Place the black plate into a plate fluorometer and read the fluorescence of methylumbelliferone. Take the readings at an excitation wavelength of 360 nm, and an emission wavelength of 460 nm. Set the gain of the instrument such that a well containing 6.25 μL of 40 μM 4-methylumbelliferone and 250 μL of 0.2 M Na2CO3 gives a reading of 1000 fluorescence units.
8. Data Analysis
- Enter the values for the luciferase and GUS assays into a spreadsheet. Exclude from consideration any sample with a luciferase reading of lower than 20,000 relative luminescence units s-1, as this indicates that the gene constructs were not successfully delivered to the aleurone cells.
- For each successfully bombarded grain extract (representing a group of four embryoless grains), calculate the normalized GUS expression from the raw GUS and luciferase data as follows: normalized GUS = [(GUS expression – GUS blank) x (2,000,000)] / (luciferase expression – luciferase blank).
NOTE: This normalizes the amount of GUS activity to what would have been observed in a bombardment that gave a luciferase reading of 2,000,000. - Report the relative level of expression of a particular reporter construct in the presence of effector constructs or hormone treatments being tested by comparing to the level of expression in the absence of effectors or hormones.
Representative Results
The technique described here can be used to introduce any test gene construct into aleurone cells of barley grains. The level of expression from the test gene may then be conveniently measured (Figure 2). The higher throughput protocol described here greatly increases the efficiency of the seed grinding and enzyme assay steps. This method has been used to assess the ability of the transcription factor TaABF15,6 to activate expression of the ABA-induced gene HVA17. In the representative results presented here, an HVA1::GUS reporter construct was introduced into aleurone cells together with a UBI::TaABF1 effector construct (Figure 3A). While a TaABF1/HVA1 ratio of just 0.01 was sufficient to cause a 3-fold induction of the HVA1 reporter gene in the absence of exogenous ABA, higher levels of the TaABF1 construct resulted in progressively greater HVA1 expression, in a dose-dependent manner (Figure 3B,C).
This experimental protocol may also incorporate the use of hormones or other compounds during the post-bombardment incubation period to assess the role of those molecules in the regulation of gene expression. Previous work by other investigators has suggested the importance of phospholipase D in some aspects of ABA signaling. Specifically, the product of phospholipase D activity, phosphatidic acid, has been implicated as an intermediate in some responses to ABA8,9. The fact that 1-butanol, an inhibitor of phospholipase D (PLD), has been shown to prevent ABA-induced gene expression in aleurone cells suggests that PLD might be an integral part of ABA signaling in this tissue. The bombardment protocol was used to test whether 1-butanol could inhibit either ABA-induced or TaABF1-induced HVA1 expression. As expected, either exogenous ABA or TaABF1 could strongly induce expression of the HVA1::GUS reporter construct (Figure 3C). In either case, the presence of 1-butanol strongly inhibited the HVA1 induction. This suggests that PLD could play an important role in the activation of HVA1 gene expression by ABA, through TaABF1.
In addition to its role in developing and germinating grains, ABA is also critical for regulating stomatal aperture in leaves. In the guard cells of stomata, the production of phosphatidic acid is stimulated by nitric oxide (NO), which is in turn induced by hydrogen peroxide (H2O2). For some responses, NO and H2O2 can substitute for ABA9. To determine whether this is also the case in aleurone cells, the bombardment protocol was used to test whether the NO donor sodium nitroprusside (SNP) or H2O2 could substitute for ABA in inhibiting the expression of the GA-induced Amy32b gene. While ABA could strongly inhibit GA-induced expression for the Amy32b::GUS reporter construct, neither H2O2 nor SNP caused any significant reduction (Figure 3D). These results, therefore, do not support a role for NO and H2O2 in aleurone cell ABA responses.
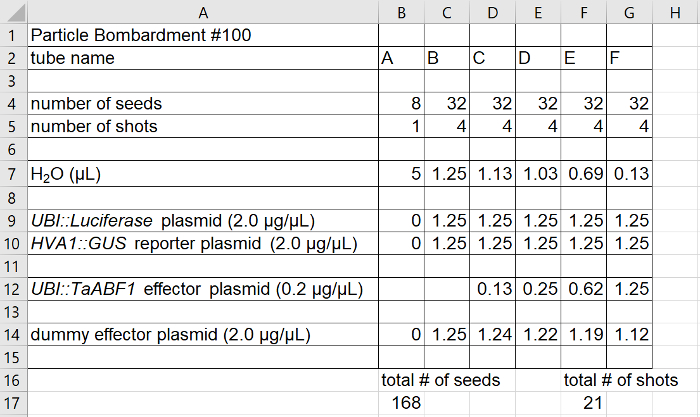
Figure 1: Spreadsheet for a typical bombardment experiment. The amount of each plasmid preparation to be placed in the tube for each bombardment is indicated. In this experiment, four shots (8 grains each) were used for each treatment to give a total of eight possible samples (4 grains each) for each treatment. In order to maintain the same total DNA concentration in each bombardment, a "dummy" effector plasmid was used to substitute for the UBI::TaBF1 effector plasmid in some cases. Treatment A is a "blank" bombardment to establish the background level of GUS and luciferase activity in aleurone cells. The results of the bombardment experiment outlined in this spreadsheet are shown in Figure 3B. Please click here to view a larger version of this figure.
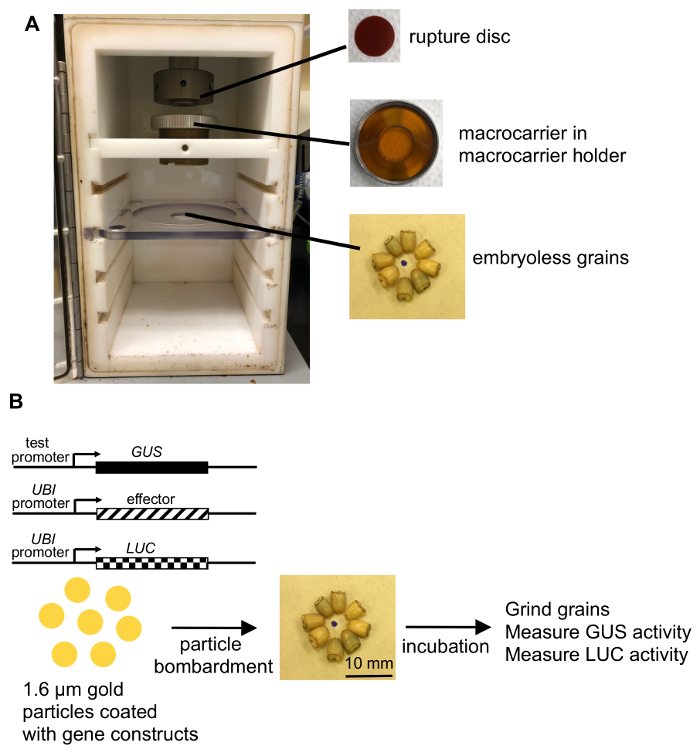
Figure 2: Diagram of the protocol. (A) Schematic of the bombardment device. (B) Gold particles coated with a reporter construct carrying the promoter to be tested, an effector (if needed), and an internal control (UBI::luciferase) are introduced into the barley aleurone cells by particle bombardment. After incubation (typically for 24 h), the grains are ground and assayed for both GUS activity and luciferase activity. Please click here to view a larger version of this figure.
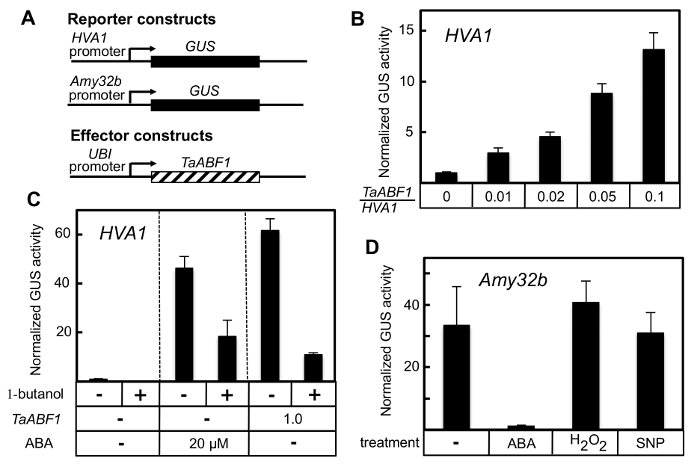
Figure 3: Regulation of HVA1 and Amy32b promoters by TaABF1, ABA, and potential mediators of ABA signaling. (A) Reporter and effector constructs used in the experiments. (B) The effector construct UBI::TaABF1 was co-bombarded into aleurone cells with the HVA1::GUS reporter and the internal control construct (UBI::luciferase). The amount of UBI::TaABF1 effector (relative to the HVA1::GUS reporter) varied from 0 to 0.1, as indicated below the bars. GUS and luciferase activities were measured after 24 h of incubation. Data shown are the means (± SE) of 6 – 8 biological replicates. (C) The HVA1::GUS reporter was co-bombarded into aleurone cells with the internal control construct (UBI::luciferase) in the presence or absence of the effector construct UBI::TaABF1. The amount of TaABF1 effector (relative to the reporter) was 1.0. GUS and luciferase activities were measured after 24 h of incubation with or without 20 µM ABA and 0.4% 1-butanol. (D) The Amy32b::GUS reporter was co-bombarded into aleurone cells with the internal control construct (UBI::luciferase). GUS and luciferase activities were measured after 24 h of incubation in the presence of 1 mM GA with or without 20 µM ABA, 1 mM H2O2, or 100 µM sodium nitroprusside (SNP). Values for GUS activity are relative to the expression of the Amy32b::GUS reporter in the absence of GA. Please click here to view a larger version of this figure.
Discussion
The introduction of effector gene constructs via particle bombardment, followed by transient expression of reporter constructs is a valuable strategy for dissecting the role of particular actors in GA/ABA signaling and in the resulting hormone-regulated gene expression.
However, existing protocols for carrying out such experiments in barley aleurone cells2,3,4,5,6 are very labor intensive. Each group of bombarded barely grains must be hand homogenized in a mortar and pestle, and the resulting extracts are assayed individually for GUS and luciferase activity. The current protocols therefore limit the number of separate treatments that can be included in the same experiment, and restrict the number of biological replicates that can reasonably be included for each treatment. These limitations make it more difficult to carry out comprehensive and simultaneous comparisons between multiple treatments. Such comparisons can be especially difficult if the difference in gene expression between the treatments is modest and a relatively large number of biological replicates is needed to establish statistical significance.
A streamlined protocol is presented here that partially automates the grain extract preparation and the GUS assays to allow substantially higher throughput. This new version of the protocol makes it feasible to include a much larger total number of samples in an experiment, allowing for both the inclusion of more different treatments per experiment, and more replicates for each treatment. For the preparation of grain extracts after bombardment and incubation, we have substituted a largely automated grinding protocol that utilizes a bead homogenizer that can process 48 samples simultaneously (in about 20 minutes). Thus, a single investigator can prepare soluble protein extracts from nearly 200 samples in less time that it would take a team of four investigators to hand grind only 100 samples. The modified GUS assay, carried out in 96 well plates, also requires much less labor that the original protocol that employed individual tube assays.
In preparing the plasmid mixtures for bombardment (Step 3.1), it is possible to use an effector to reporter ratio of 1.0, especially for preliminary tests to determine if a particular effector has any effect at all. However, because of the strength of the maize ubiquitin (UBI) promoter, a strong effect can often be observed with much lower ratios, as seen in Figure 2B and in previously published reports4,5,6. Other critical steps in this protocol include peeling of the seed coat from the barley grains and the use of carefully defined settings in the bombardment apparatus. If the seed coats are not fully removed, the gold microcarriers will be prevented from entering the subtending aleurone cells. Successful delivery of the gold microcarriers into the aleurone cells is also quite dependent on the specific type of rupture disc used, the position of the microcarrier cover lid, stopping screen, and tissue to be bombarded. The conditions reported here typically result in successful delivery of the gene constructs (see Step 8.1) to a high percentage (≥70%) of bombarded barley grain samples. It should be expected that other biological materials would require different conditions for optimal gene delivery.
Although it is not described here, it would be possible to further streamline the procedure by carrying out the flash luciferase assays in 96 well plates. However, as the light production from luciferase must be measured in a luminometer immediately after mixing the protein extract with the substrate (luciferin), this would require the use of a plate flash luminometer with injectors, an instrument that is much more expensive than a single tube luminometer.
The representative results reported here confirm that the transcription factor TaABF1 can activate HVA1 expression in the absence of exogenous ABA5,6. Activation of HVA1 by either TaABF1 or exogenous ABA was prevented by the PLD inhibitor 1-butanol, suggesting that production of phosphatidic acid by PLD may be required for HVA1 activation. These results for HVA1 regulation by TaABF1 and ABA are similar to those obtained by Zou et al.10 for the activation of HVA22 by the transcription factor LtWRKY21 and by ABA. The finding that H2O2 or SNP do not substitute for exogenous ABA in suppressing aleurone expression of Amy32b, confirms that some aspects of ABA signaling in cereal grains are distinct from what occurs in stomatal guard cells. The finding that H2O2 does not downregulate Amy32b expression is consistent with the results observed by Ishibashi et al.11 for amylase enzyme activity.
The increased throughput protocol is now being used to carry out a comprehensive study of the effects of TaABF1 phosphorylation on its ability to regulate downstream gene expression. As other members of the ABF family of transcription factors can be phosphorylated at a number of different sites12,13,14,15,16, multiple putative phosphorylation sites on TaABF1 need to be tested. The higher throughput allowed by this protocol will permit a large number of mutant versions of TaABF1 (with potentially phosphorylatable serine and threonine residues modified) to be interrogated, each with a large number of biological replicates.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors thank Greyson Butler and Margaret Barrett for help in carrying out the experiments, Judy Stone for advice on grain homogenization, and Lynn Hannum for advice on fluorometry. This work was supported by the National Science Foundation (IOB 0443676), by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM0103423, and by grants from the Colby College Division of Natural Sciences.
Materials
| GeneElute HP plasmid Maxiprep kit | Sigma | NA0310-1KT | |
| UV-vis spectrophotometer | Nanodrop | ND-1000 | |
| Himalaya barley grains | / | / | A variety of hulless barley (store in the dark at 4° C) |
| sodium succinate | Sigma | S2378 | Reagent for Imbibing Solution |
| calcium chloride (dihydrate) | Fisher | C79-500 | Reagent for Imbibing Solution |
| Imbibing Solution | home made | / | 20 mM sodium succinate, 20 mM calcium chloride, pH 5.0. Sterilize by autoclaving before use. |
| chloramphenicol | Sigma | C0378 | Prepare a 10 mg/mL stock solution in 70% ethanol. |
| vermiculite | Fisher | NC0430369 | Used for vermiculite plates. |
| filter paper circles (90 mm) | Whatman | 1001 090 | Used for vermiculite and for pre-bombardment grain preparation |
| Vermiculite Plates | home made | / | Add 50 mL of vermiculite to a glass petri dish. Place a 90 mm paper circle on top of the vermiculite. Autoclave. |
| forceps (fine pointed) | Fisher | 13-812-42 | Used for removing seed coat from barley grains. |
| forceps (ultra fine point) | Fisher | 12-000-122 | Used for removing seed coat from barley grains. |
| gold microcarriers (1.6 μm) | BioRad | 1652264 | |
| macrocarriers | BioRad | 1652335 | |
| calcium chloride (dihydrate) | Fisher | C79-500 | Prepare a 2.5 M stock solution and store 1 mL aliquots at -20° C. |
| spermidine | Sigma | S0266 | Prepare a 100 mM stock solution and store 500 μL aliquots at -20° C (use within 2 months). |
| rupture discs (1550 psi) | BioRad | 1652331 | |
| stopping screens | BioRad | 1652336 | |
| macrocarrier holders | BioRad | 1652322 | |
| Biolistic particle delivery system | BioRad | PDS-1000/He | |
| sodium phosphate monobasic monohydrate | Sigma | S9638 | Reagent for 1M sodium phosphate pH 7.2 |
| sodium phosphate dibasic | Sigma | S9763 | Reagent for 1M sodium phosphate pH 7.2 |
| 1M sodium phosphate pH 7.2 | home made | / | Combine 6.9 g of sodium phosphate monobasic monohydrate with 7.1 g of sodium phosphate dibasic. Add water to 100 mL. Add NaOH to get pH 7.2. |
| dithiothrietol (DTT) | Sigma | 43819 | Dissolve in water to 1 M. Store at -20° in 1 mL aliquots. |
| leupeptin | Sigma | L2884 | Dissolve in water to 10 mg/mL. Store at -20° C. |
| glycerol | Sigma | G5516 | Prepare a 50% solution in water. |
| Grinding Buffer | home made | / | Combine 10 mL of 1 M sodium phosphate pH 7.2, 500 μL of 1 M DTT, 100 μL of 10 mg/mL leupeptin, and 40 mL of 50% glycerol. Add water to 100 mL. |
| stainlesss steel beads (5 mm) | Qiagen | 69989 | |
| 2.0 mL tubes | Eppendorf | 22363352 | This specific model of tube is recommended for use with the homogenizer. |
| bead homogenizer (TissueLyser) | Qiagen | 85210 | |
| 12mm x 75 mm glass test tubes | Fisher | ||
| luciferin | Goldbio | LUCK-100 | Prepare a 25 mM stock solution and store 1 mL aliquots at -20° C. |
| ATP | Sigma | A7699 | Prepare a 100 mM stock solution and store 250 μL aliquots at -20° C. |
| Tris base | Sigma | T1503 | Reagent for 1M Tris sulfate pH 7.7. |
| sulfuric acid | Sigma | 258105 | Reagent for 1M Tris sulfate pH 7.7. |
| 1M Tris sulfate pH 7.7 | home made | / | Dissolve 12.1 g Tris base in 100 mL of water. Adjust pH to 7.7 with sulfuric acid. |
| magnesium chloride | Sigma | M9397 | Dissolve in water to 2 M. |
| Luciferase Assay Buffer (LAB) | home made | / | Combine 3 mL of 1 M Tris sulfate pH 7.7, 500 μL of 2 M magnesium chloride, 1 mL of 1 M DTT, and 200 μL of 0.5 M EDTA. Add water to 50 mL. |
| Luciferase Assay Mixture | home made | / | Combine 15 mL of LAB, 800 μL of 25 mM luciferin, 200 μL of 100 mM ATP, and 4 mL of water. This makes enough assay mixture (20 mL) for 100 luciferase assays. |
| luminometer (Sirius) | Berthold | / | |
| 4-methylumbelliferyl-β-D-glucuronide (MUG) | Goldbio | MUG1 | Dissolve in DMSO to 100 mM. |
| sodium azide | Sigma | S8032 | Prepare a 2% stock solution in water and store 1 mL aliquots at -20° C. |
| 96 well plates (standard) | Fisher | 12565501 | |
| GUS assay buffer | home made | / | Combine 2.5 mL of MUG, 5 mL of 1 M sodium phosphate pH 7.2, 400 μL of 0.5 M EDTA, 1 mL of 1 M DTT, 100 μL of 10 mg/ml leupeptin, 20 mL of methanol, and 1 mL of 2% sodium azide. Add water to 100 mL. |
| TempPlate sealing film | USA Scientific | 2921-1000 | |
| 96 well plates (black) | Costar | 3916 | |
| sodium carbonate | Sigma | S7795 | Prepare a 200 mM solution in water. |
| 4-methylumbelliferone | Sigma | M1381 | Prepare a 100 μM solution in water. Freeze 1 mL aliquots at -20° C. |
| microplate fluouresence reader | Bio-Tek | FLX-800 |
References
- Chen, K., An, Y. Q. C. Transcriptional responses to gibberellin and abscisic acid in barley aleurone. J. Integ. Plant Biol. 48, 591-612 (2006).
- Lanahan, M. B., Ho, T. H. D., Rogers, S. W., Rogers, J. C. A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell. 4, 203-211 (1992).
- Shen, Q., Zhang, P., Ho, T. H. D. Modular nature of abscisic acid (ABA) response complexes; composite promoter units that are necessary and sufficient for ABA induction of gene expression. Plant Cell. 8, 1107-1119 (1996).
- Gómez-Cadenas, A., Zentella, R., Walker-Simmons, M. K., Ho, T. H. D. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell. 13, 667-679 (2001).
- Johnson, R. R., Shin, M., Shen, J. Q. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol. Biol. 68, 93-103 (2008).
- Harris, L. J., Martinez, S. A., Keyser, B. R., Dyer, W. E., Johnson, R. R. Functional analysis of TaABF1 during abscisic acid and gibberellin signaling in aleurone cells of cereal grains. Seed Science Res. 23, 89-98 (2013).
- Shen, Q., Casaretto, J., Zhang, P., Ho, T. H. D. Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare). Plant Mol. Biol. 54, 111-124 (2004).
- Ritchie, S., Gilroy, S. Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 95, 2697-2702 (1998).
- Takemiya, A., Shimazaki, K. Phosphatidic acid inhibits blue light-induced stomatal opening via inhibition of protein phosphatase 1. Plant Physiol. 153, 1555-1562 (2010).
- Zou, X., Seeman, J. R., Neuman, D., Shen, Q. J. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J. Biol. Chem. 279, 55770-55779 (2004).
- Ishibashi, Y., et al. Reactive oxygen species are involved in gibberellin/abscisic acid signaling in barley aleurone cells. Plant Physiol. 158, 1705-1714 (2012).
- Piskurewicz, U., et al. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 20, 2729-2745 (2008).
- Hong, J. Y., et al. Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry. 72, 27-36 (2011).
- Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., Chua, N. H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317-328 (2002).
- Zhou, X., et al. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-Type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSESENSITIVE5. Plant Physiol. 168, 659-676 (2015).
- Zong, W., et al. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 171, 2810-2825 (2016).

