Mouse Models Of Helicobacter Infection And Gastric Pathologies
Summary
Mice represent an invaluable in vivo model to study infection and diseases caused by gastrointestinal microorganisms. Here, we describe the methods used to study bacterial colonization and histopathological changes in mouse models of Helicobacter pylori-related disease.
Abstract
Helicobacter pylori is a gastric pathogen that is present in half of the global population and is a significant cause of morbidity and mortality in humans. Several mouse models of gastric Helicobacter infection have been developed to study the molecular and cellular mechanisms whereby H. pylori bacteria colonize the stomach of human hosts and cause disease. Herein, we describe protocols to: 1) prepare bacterial suspensions for the in vivo infection of mice via intragastric gavage; 2) determine bacterial colonization levels in mouse gastric tissues, by polymerase chain reaction (PCR) and viable counting; and 3) assess pathological changes, by histology. To establish Helicobacter infection in mice, specific pathogen-free (SPF) animals are first inoculated with suspensions (containing ≥105 colony-forming units, CFUs) of mouse-colonizing strains of either Helicobacter pylori or other gastric Helicobacter spp. from animals, such as Helicobacter felis. At the appropriate time-points post-infection, stomachs are excised and dissected sagittally into two equal tissue fragments, each comprising the antrum and body regions. One of these fragments is then used for either viable counting or DNA extraction, while the other is subjected to histological processing. Bacterial colonization and histopathological changes in the stomach may be assessed routinely in gastric tissue sections stained with Warthin-Starry, Giemsa or Haematoxylin and Eosin (H&E) stains, as appropriate. Additional immunological analyses may also be undertaken by immunohistochemistry or immunofluorescence on mouse gastric tissue sections. The protocols described below are specifically designed to enable the assessment in mice of gastric pathologies resembling those in human-related H. pylori diseases, including inflammation, gland atrophy and lymphoid follicle formation. The inoculum preparation and intragastric gavage protocols may also be adapted to study the pathogenesis of other enteric human pathogens that colonize mice, such as Salmonella Typhimurium or Citrobacter rodentium.
Introduction
Helicobacter pylori is a spiral-shaped, Gram-negative, human gastric pathogen present in all populations across the world, with infection rates in developing countries estimated to be in the order of 80%1. Although most H. pylori-infected individuals are asymptomatic, some develop more severe diseases, ranging from peptic ulceration to gastric cancer2. H. pylori-associated cancers are broadly characterized either by malignant changes in epithelial cells (GECs) or by the formation of extra-nodal lymphoid tissues in the stomach, resulting in gastric adenocarcinoma or mucosa-associated lymphoid tissue (MALT) lymphoma, respectively. H. pylori is highly adapted to survive in the harsh ecological niche of the stomach due to the presence of various virulence factors and mechanisms facilitating its adherence, growth and metabolism in this niche. In particular, virulent strains of H. pylori possess the 40 kb cag Pathogenicity Island (cagPAI) that encodes approximately 30 genes required for the production of a Type 4 secretion system (T4SS)3,4. cagPAI-positive H. pylori strains are associated with the induction of higher levels of chronic inflammation in the host, which has been implicated as an essential precursor of gastric adenocarcinoma5.
In vivo animal models, particularly mice, have been highly informative by allowing researchers to investigate the relative contributions of host, bacterial and environmental factors on H. pylori infection and disease outcome6. Studies have previously demonstrated that prolonged H. pylori infection of mice on the C57BL/6 genetic background results in the development of chronic gastritis and gland atrophy, both hallmarks of H. pylori infection7. Furthermore, infection with the related feline/canine bacterial species, H. felis, has been shown to induce MALT formation in mice with similar pathology and disease progression as seen in human MALT lymphoma8,9. The most commonly used H. pylori isolate in mouse colonization studies is the “Sydney Strain 1” (SS1) strain10, which is cagPAI+ but has a non-functional T4SS (T4SS−)11. Other widely used strains include H. pylori B128 7.13 (cagPAI+/T4SS+)12 and X47-2AL (cagPAI–/T4SS−)13. For H. felis infections, the strain CS1 (“Cat Spiral 1”, cagPAI–/T4SS−) is generally used14.
Herein, we provide a protocol describing the preparation of Helicobacter inocula for in vivo infection, the procedure for intragastric gavage of mice, as well as methods for the processing of tissues for the study of histopathological changes in the stomach. In particular, this article will focus on the histological methods used to visualize bacterial colonization and assess histopathological changes, including MALT formation, in the gastric mucosa of infected mice. Some of the methods described here may be adapted to the study of other gut pathogens such as S. Typhimurium or C. rodentium.
Protocol
1. Growth and Preparation of Bacterial Inocula
- Thaw glycerol stocks of H. pylori or H. felis15 from -80 °C and subculture on horse blood agar (HBA) plates comprising: Blood Agar Base No.2 (see Table of Materials); a modified “Skirrow’s antibiotic selective supplement” (consisting of vancomycin, 10 μg/mL; polymyxin B, 25 ng/mL; trimethoprim, 5 µg/mL; amphotericin B, 2.5 μg/mL); and 5–10% (v/v) horse blood15,16. The bacteria grow well under microaerobic conditions in 2.5 or 3.5 L anaerobic jars containing the appropriate gas packs (see Table of Materials), at 37 °C.
NOTE: H. pylori strains grown under these conditions must be subcultured after 1–1.5 days of incubation, whereas for H. felis, at least 2 days of incubation is generally required. Suitable culture and storage media have been described in detail previously15. - Prepare bacterial inocula for mouse infection from early-to-mid logarithmic phase cultures. Harvest bacteria gently from agar plates by flooding each plate with 1–2 mL of brain heart infusion (BHI) broth. Aspirate suspensions from plates with Pasteur pipettes.
- Alternatively, prepare inocula from Helicobacter bacteria that have been propagated in BHI broth for 16–18 h15. In this case, collect bacteria by low speed centrifugation at 2,200 x g for 10 min at 4 °C.
- Assess the viability and motility of the bacteria by examining wet mount preparations under phase contrast microscopy (100X objective). Prepare wet mounts by resuspending a loopful of bacteria in a 10–20 μL droplet of BHI broth on a glass microscope slide. In the case of H. felis, which is a much larger bacterium than H. pylori, use a haemocytometer to accurately count the numbers of viable bacteria. Confirm culture purity by performing a Gram-stain.
NOTE: Only use H. pylori inocula if the majority of bacteria have a bacillary or spiral shape (the morphology can vary depending on the strain). H. felis inocula should primarily contain helical-shaped bacteria. Do not use inocula if most bacteria have a coccoid morphology, as these forms are not viable and will not establish an infection in mice. - Estimate the number of bacteria in the inoculum by counting under phase contrast microscopy the approximate number of bacteria per field (100X objective) and by using the following guide: 1 bacterium per field = approximately 106 colony-forming units (CFU) of Helicobacter/mL; 10 bacteria per field = approximately 107 CFU/mL; 100 bacteria per field = approximately 108 CFU/mL, etc.
- When using a haemocytometer, calculate CFU/mL using the following formula:
CFU/mL = (average number of bacteria in a 4 x 4 field) x (dilution factor) x (104).
- When using a haemocytometer, calculate CFU/mL using the following formula:
- Adjust the bacterial cell density of the inoculum to approximately 107–108 CFU/mL by dilution in BHI broth, if necessary.
- To ensure maximal bacterial viability, use inocula for intragastric gavage as soon as possible after preparation.
- Always confirm H. pylori cell density and viability by performing viable counting of inocula immediately after the gavage procedure (see below). This is not always possible for H. felis, as it does not usually form isolated colonies on culture media. The numbers of viable Helicobacters in inocula cannot be determined by optical density measurement (A600) alone as this method does not discriminate between viable (i.e., bacillary/spiral/helical) and non-viable (i.e., coccoid) bacteria.
- Use optical density values as a means of estimating the numbers of viable H. pylori bacteria in inocula, but in this case, it is first necessary to generate a growth curve. For this, the A600 values of H. pylori cultures are monitored over time and correlated directly against the numbers of viable bacteria, determined by plate counting.
NOTE: A convenient method for performing such growth curve determinations is to culture bacteria in liquid medium (Section 1.2), using standard flat bottom tissue culture flasks placed in a 10% CO2 incubator. The numbers of CFUs, determined from aliquots of the cultures obtained every 4–6 h over 2–3 days, are then compared to the corresponding A600 values17. Importantly, growth curves must be generated for each H. pylori strain, as these may grow at different rates and, furthermore, not all strains grow well in 10% CO2.
2. Intragastric Gavage of Mice with Helicobacter
NOTE: This method of intragastric gavage can be applied to other bacterial species that colonize the gut e.g. S. Typhimurium, C. rodentium, Listeria monocytogenes.
- Use 6–8-week-old, specific pathogen-free (SPF) and Helicobacter-free male or female mice. Use animals with a C57BL/6 genetic background for infection experiments with H. pylori or H. felis. In the present study, we used wild-type (WT) and genetically modified C57BL/6 mice lacking a key innate immune receptor (termed knock-out or KO animals).
Note: Mice on other genetic backgrounds can also be used for Helicobacter infections, however, colonization levels and disease severity may be impacted by the type of host background18,19. - Aspirate the bacterial inoculum (Step 1.5) into disposable 1 mL syringes and replace the supplied needles with 23 gauge needles onto which are affixed disposable polyethylene catheters (length, 6–8 cm; internal diameter, 0.58 mm). Fasten catheters to the needles by the application of small strips of plastic film (see Table of Materials). Alternatively, replace the needle/catheter assembly by using sterile plastic feeding tubes (20 gauge x 38 mm).
- Physically restrain mice with a firm grip at the scruff of the neck and tail.
NOTE: This procedure can be performed without anaesthesia, or alternatively, with the use of an inhaled anaesthetic, such as methoxyflurane or isoflurane15. - Insert catheter into the center of the open jaw and guide in a caudal direction towards the esophagus. Extend the neck of the mouse to allow ease of access to the stomach through the esophagus (and away from the trachea) until most or all of the catheter is no longer visible and a resistance is felt, corresponding to the base of the stomach. Deliver a specific aliquot, usually 100 μL per inoculation (Figure 1).
NOTE: Mice should be gavaged with ≥ 105 CFU to ensure optimal colonization and disease pathology. - House mice in an SPF animal facility for the duration of the experiment.
NOTE: Severe pathology and adenocarcinoma in WT C57BL/6 mice is only observed at approximately 24 months’ post-infection20. However, this effect may be accelerated in some genetically modified animals, or in mice with other genetic backgrounds. - Upon completion of the gavage procedure, perform a modification of the Miles and Misra technique to determine the numbers of viable H. pylori bacteria administered to mice. For this, the inoculum is serially diluted (from 10−1 to 10−5) in BHI using the method described in detail previously15.
NOTE: In order to ensure isolation of single colonies, HBA plates should be warmed and dried in a biological safety cabinet or 37 °C incubator for 10–15 min prior to use.
3. Harvesting Tissues from Mice Post-experiment
- Euthanize mice by either carbon dioxide inhalation or cervical dislocation, according to the relevant ethics committee for animal experimentation.
- Open the abdominal cavity and excise the stomach using fine, curved scissors.
NOTE: Sera can also be collected by cardiac puncture to aid in the investigation of systemic responses to Helicobacter infection. Additionally, the collection of spleens and paragastric lymph nodes are useful in studying adaptive immune responses. - Cut the stomach along the greater curvature and remove residual food by gentle washing in sterile phosphate buffered saline (PBS) in a 50 mL tubes.
- Wash the stomach again in sterile PBS and then record the wet weight using tared 6 cm plastic Petri dishes.
- Flatten the stomach and dissect sagittally into two equal tissue fragments, each comprising: the antrum, body and non-glandular forestomach regions (Figure 2). Remove the non-glandular region and weigh one half of each stomach before adding to either 1 mL of sterile BHI (for viable counting) or snap freezing in liquid nitrogen (for DNA/RNA extraction).
Note: Tubes containing tissue in BHI must be stored on ice until they are ready to be processed. Snap frozen stomach tissues can also be used to extract RNA or proteins for qPCR (quantitative PCR) or western blotting analyses, respectively. - Add the other stomach half to a 15 mL tube containing 10% formalin. Immerse tissues in 10% formalin for 10 s and then flatten to the top side of tubes. Allow tissues to fix before re-immersing in 10% formalin solution for a minimum of 24 h.
NOTE: Tissues can remain in 10% formalin for many weeks prior to processing for histology. Prolonged storage of tissue may, however, affect its architecture and/or antigenicity resulting in sub-optimal results in downstream analyses.
4. Confirmation of Bacterial Colonization in the Stomach Post-infection
- Viable Counting of H. pylori in the Stomach
- Supplement sterile HBA plates with additional antibiotics (200 μg/mL bacitracin and 10 μg/mL naladixic acid) prior to performing colony counts from infected mouse stomachs16.
- Homogenize stomach sections either manually, using autoclavable polypropylene micropestles, or using a mechanical dissociation instrument (see Table of Materials).
- Prepare duplicate serial dilutions (10−1 to 10−2) of the resulting gastric homogenates in sterile BHI.
NOTE: Dilutions should be decided based on the typical bacterial loads obtained for a given H. pylori strain used for infection, as well as the duration of infection. Undiluted samples can also be used. - Divide pre-dried HBA plates (see Note above) into three or four segments. Using an adaptation of the Miles and Misra technique, add 10–100 mL of each dilution onto a segment of the agar plate and spread using sterile plastic loops15.
- Allow the plates to dry and then place them in an inverted position (lid side down) in anaerobe gas jars. To maintain humidity in the jars, include a Petri dish containing water.
- Incubate jars at 37 °C until colonies form (typically 4–7 days).
- Enumerate segment(s) containing between 10 and 100 isolated colonies.
NOTE: H. pylori colonies and H. felis growth on plates can be distinguished from those of other members of the murine gastric microbiota using standard urease, catalase and oxidase tests. H. pylori and H. felis are positive for all three tests. - Calculate the bacterial loads as (CFU/g of tissue), using the following formula:
[(Average number of colonies counted) × (dilution factor) x (volume plated)]/(stomach weight).
- Detection of H. felis Infection in Gastric Tissues by the Polymerase Chain Reaction (PCR)
- Extract DNA from mouse stomachs using standard DNA isolation protocols, or a commercially available kit.
- Determine the DNA concentration of samples using a fluorometric quantitation technique (see Table of Materials).
- Set up PCR reactions targeting a 325-base pairs (bp) fragment of the H. felis urease B gene (ureB)21. Each reaction should contain: 100 ng of genomic DNA; 1 mM each of forward (5’-AAA ATC CAC GAA GAC TGG GG-3’) and reverse (5’-CTT TTA TCC AAG TGG TGG CAC ACC-3’) primers; 200 mM dNTPs; 0.5 units of Taq Polymerase and the appropriate amounts of buffer and nuclease-free water.
NOTE: This oligonucleotide pair has been designed to recognize and bind to homologous sequences in both H. pylori and H. felis ureB genes but when subjected to the PCR conditions below, not those present in the ureB genes of enterohepatic Helicobacter spp. - Perform PCR amplification using the following thermal profile: heating at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 61 °C for 30 s and 72 °C for 1 min, before holding at 20 °C.
- Run PCR products on a 2% agarose gel for 30 min at 100 V.
5. Histological Analyses of Helicobacter-infected Mouse Stomach Sections
- Processing of Stomach Tissues
- Remove stomach tissues from formalin and place in clean Petri dishes.
- Cut stomach tissues with a scalpel into several equal-sized longitudinal strips (each 2–3 mm thick) and place in labelled embedding cassettes containing foam padding.
- Fix stomach tissues by placing embedding cassettes in a jar filled with 80% ethanol.
NOTE: Stomachs can be processed immediately or stored for up to 1–2 days prior to proceeding to the next step. - Process stomachs on an automated tissue processor, programmed with the following settings:
Dehydration: 70% ethanol – 1 cycle, 20 min; 90% ethanol – 1 cycle, 20 min; 100% ethanol – 2 cycles, 20 min each + 1 cycle, 40 min + 1 cycle, 1 h.
Clearing: xylene – 2 cycles, 30 min each + 1 cycle, 45 min.
Impregnation: paraffin wax at 60 °C – 1 cycle, 45 min + 1 cycle, 1 h + 1 cycle, 1.25 h. - Remove the processed samples from the machine and store at room temperature for paraffin embedding.
- Paraffin Embedding of Processed Gastric Tissues
- Place stainless steel base molds on the stage of the embedding unit to warm the bases of the molds.
NOTE: Embedding machine should be set at 60 °C for efficient paraffin embedding. - Place waxed cassettes containing the samples into a warm wax bath/hot plate area of the embedding unit until wax fully dissolves.
NOTE: It is recommended that the sample be embedded shortly after wax dissolves. This ensures that hardening of tissues does not occur. - Fill half of the stainless-steel molds with paraffin wax. Using warm forceps, remove stomach tissue strips from the cassettes and gently push the stomach strips through the paraffin to the base of the molds. Carefully orientate tissue strips at a right angle to the base of the molds so that their sliced ends are facing upwards.
NOTE: The following steps should be performed as quickly as possible to avoid hardening and separation of the paraffin layers within embedded blocks. The proper orientation of stomach tissues is critical for downstream analyses. - Place molds on the cold plate of the embedding unit to fix the specimens in place and re-orientate tissues if necessary.
NOTE: If the tissues become dislodged and the paraffin begins to harden, place molds back onto the hot plate to melt the wax and re-embed tissues in molds. - Place half of the labelled embedding cassettes (which were used for the tissue processing) on to the top of the molds and gently fill with warm wax. Do not allow paraffin to overflow.
- Gently place molds on to a cold plate and allow to cool.
- Once the paraffin has fully set, separate the embedded blocks from molds. Clean excess paraffin wax on cassette edges using a hot plate (set above 80 °C) or a scraper. Blocks can be stored at room temperature until sectioning is performed.
- Place stainless steel base molds on the stage of the embedding unit to warm the bases of the molds.
- Sectioning of Tissues
- Chill paraffin-embedded tissue blocks on ice and heat a water bath filled with ultrapure water to 40–45 °C.
- Secure the blade in the holder of the microtome and set the clearance angle between 1–5° to prevent contact between the block face and knife facet, before inserting paraffin blocks.
NOTE: Ensure blocks are clear of excess paraffin acquired from the embedding, as this may hinder the fit of the block. - Orientate the blade for a straight cut across the block. Gently cut 2–3 thin sections to ensure correct positioning of the block.
- Trim blocks by a thickness of approximately 10–30 mm. This step ensures that a maximal surface area of each tissue strip will be cut.
- Cut 10 µm sections and discard any that contain holes caused by trimming.
- Carefully pick up sections using tweezers and float them in the water bath for flattening. Use tweezers to separate each section.
- Collect sections from the water bath and place onto charged glass slides (see Table of Materials).
- Store slides upright in a slide rack and place in an incubator at 37 °C. Dry sections overnight.
- Store sections at room temperature indefinitely for subsequent analyses.
- Haematoxylin and Eosin (H&E) Staining of Stomach Tissues
- Dewax slides using 3 washes of xylene for 5 min each, followed by 3 washes in 100% ethanol, for 3 min each. Ensure fresh solutions are used at each stage.
- Rinse slides in tap water for 30–60 s.
- Remove excess water by gently tapping the bottom of the slides on a paper towel. Stain with filtered Haematoxylin for 3 min. Ensure that sections are sufficiently covered with the solution.
- Rinse slides under running tap water until water runs clear.
- Dip slides in Scott’s Tap water for 8–10 s. Do not expose slides to the solution more than 10 s as this will result in darker and intense stains. Staining of sections can be viewed under a microscope. Efficient staining at this stage will result in a ‘baby blue’ color.
NOTE: Prepare Scott’s Tap water by dissolving 2 g of sodium hydrogen carbonate (NaHCO3) and 20 g of magnesium sulphate (MgSO4) in 1 L of distilled water. - Rinse slides in tap water for 30–60 s.
- Remove excess water as described in step 3, and then stain with filtered 1% aqueous Eosin for 3 min.
- Rinse slides under running tap water until water runs clear.
NOTE: Staining can be assessed using a light microscope. If staining is too dark, slides can be dehydrated in 100% ethanol for longer than the specified time. If darker staining is required, slides can be stained with Eosin for 1–2 min longer, prior to proceeding to subsequent steps. - Dehydrate slides using 3 washes of 100% ethanol for 30 s each, followed by 3 washes of xylene for 2 min each. Ensure fresh solutions are used at each stage.
- Mount slides with mounting medium. Add a drop of mounting medium in the center of a clean coverslip prior to gently placing slide on top with sections facing downwards.
NOTE: Do not dry slides prior to cover slipping. - Place slides on a flat surface and allow to air dry. Slides can also be dried in a fume hood to accelerate drying time.
- Giemsa Staining of Stomach Tissues
- Dewax slides using 2 washes of histolene for 5 min each, followed by 2 washes of 100% ethanol for 3 min each and then a final wash in 70% ethanol for 3 min. Ensure fresh solutions are used for each wash.
- Rinse slides in tap water for 30–60 s.
- Prepare Giemsa solution by mixing 20% Giemsa stain with 80% distilled water. Stain slides with Giemsa solution for 1 h.
- Place slides in 100 mL of distilled water containing 3–4 drops of acetic acid for 2–3 s.
NOTE: Solution must be mixed well prior to use. At this stage, slides should appear pale blue in color. - Wash slides in 96% ethanol for 30 s.
- Wash slides in 3 baths of isopropanol for 2 min each, followed by 3 baths of histolene for 2 min each. Use fresh isopropanol and histolene for each wash.
- Coverslip slides with mounting medium, as described earlier.
Representative Results
This protocol describes an oral gavage technique to achieve intragastric infection with H. pylori or H. felis in murine mouse models (Figure 1). Following euthanasia, stomachs are removed, weighed and divided into 2 equal halves comprising the antrum, body and non-glandular regions of gastric tissues (Figure 2). The non-glandular region is removed prior to performing any analyses.
Successful colonization of animals is typically confirmed by performing viable counting on H. pylori-infected gastric homogenates, and subsequently enumerating individual colonies on HBA plates (Figure 3). Alternatively, PCR is employed to verify infection with H. felis using specific, validated primers directed at a 325 bp region of the H. felis and H. pylori ureB genes (Figure 4).
Gastric tissues are processed, embedded and sectioned for downstream histological applications. The H&E staining technique is used to assess the histopathology in Helicobacter-infected mice. In the current example, WT C57BL/6 mice display moderate signs of inflammation, including hyperplasia (enlarged mucosa) and gland atrophy at 6 months’ post-infection with H. felis. The presence of cellular infiltrates can also be observed in the sub-mucosa. Interestingly, however, more severe inflammation is observed in KO mice at the same time point, with the additional presence of lymphoid follicles located in close proximity to cellular infiltrates (Figure 5). Finally, H. felis bacteria are observed in Giemsa-stained sections of infected mouse stomachs (Figure 6).
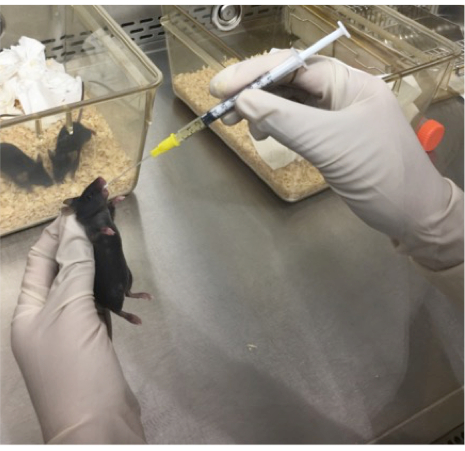
Figure 1: Image demonstrating the oral gavage technique. A disposable 1 mL syringe and flexible catheter are used to deliver ≥105 CFU of bacterial inocula to a mouse via the intragastric route. The mouse was anesthetized using methoxyflurane and held in a firm grip at the neck, allowing for access of the catheter to the stomach via the esophagus.
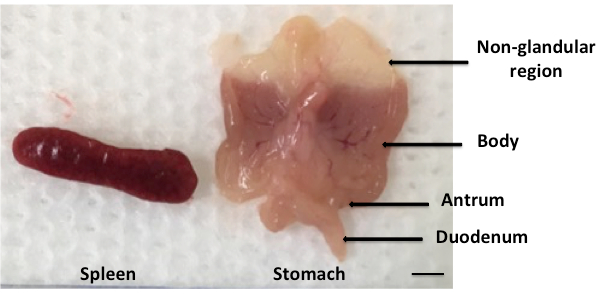
Figure 2: Harvesting of mouse spleens and stomachs post-infection. Mouse stomachs were harvested post-euthanasia and their contents removed by scraping with a scalpel and washing in sterile PBS. The tissues were then weighed and flattened on a cotton sheet to reveal 2 equal halves, each comprising the gastric antrum, body and non-glandular regions; Scale bar = 10 mm.
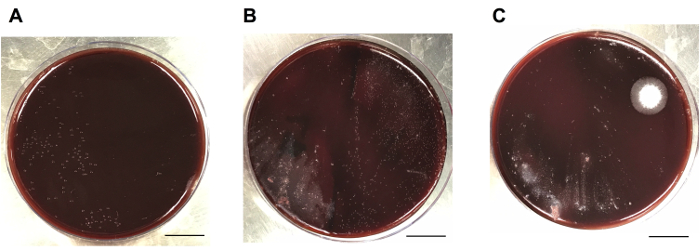
Figure 3: Viable counts on H. pylori-infected mouse stomachs. Mice on the C57BL/6 background were inoculated with 107 CFU of H. pylori SS1 and left for 8 weeks. (A) Dilutions of each gastric homogenate are plated onto a half (or third) of an HBA plate and bacterial loads assessed by enumerating 10-100 individual colonies. The left half of the plate shows a pure culture of H. pylori bacteria. (B) The presence of contaminating bacteria from the mouse gastric microbiota (left) or large numbers of H. pylori colonies (right) can complicate the enumeration of H. pylori CFUs. (C) Common examples of contaminating bacteria in gastric homogenate samples. Scale bar = 1.7 cm. Please click here to view a larger version of this figure.
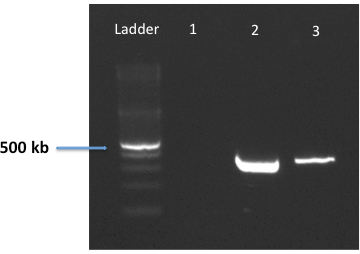
Figure 4: PCR detection of H. felis infection in gastric biopsies using oligonucleotides targeting the ureB gene. A specific oligonucleotide pair was designed to recognize and bind to homologous sequences in both H. felis and H. pylori ureB genes. These primers were validated using genomic DNA from H. pylori SS1 (lane 2) or H. felis (lane 3). Deionized water was included as a negative control (lane 1).
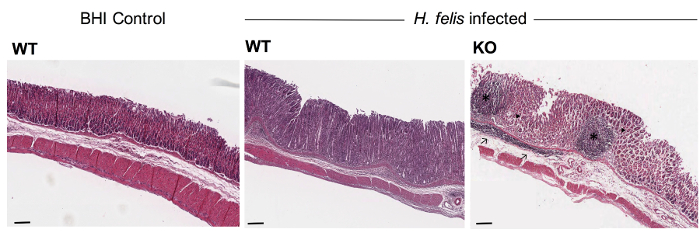
Figure 5: Representative images of H&E-stained stomach sections from WT and KO mice at 6 months’ post-infection with H. felis. Paraffin-embedded tissue sections were stained with H&E. WT mice receiving BHI broth alone (control) had a normal gastric epithelium and no significant inflammation. In contrast, WT animals with chronic H. felis infection displayed moderate levels of inflammation and mucosal thickening which was further exacerbated in H. felis-infected KO animals. Tissue sections from H. felis-infected KO mice exhibited the presence of mucosal lymphoid follicles (*), cellular infiltrates (→), gland atrophy (▶) and hyperplasia. Scale bar = 100 μm. Please click here to view a larger version of this figure.
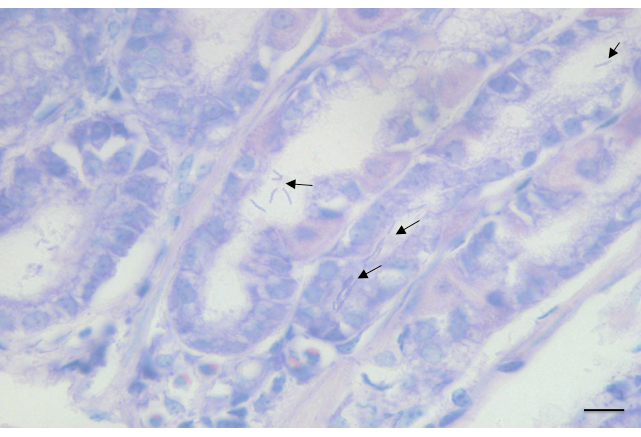
Figure 6: Representative images of Giemsa-stained gastric sections from C57BL/6 WT mice at 3 months’ post-infection with H. felis. Paraffin-embedded tissue sections were stained with Giemsa. Arrows indicate the presence of H. felis in the gastric glands. Scale bar = 200 μm.
Discussion
This protocol describes the use of an in vivo mouse model for Helicobacter infection. The critical steps of the procedure are the: 1) preparation of Helicobacter inocula containing viable and motile bacteria; 2) delivery of the appropriate numbers of bacteria to the mouse via intragastric gavage; 3) detection of bacterial infection by colony counting and/or PCR; and 4) processing of gastric tissues to enable the assessment of histopathology in infected stomachs. Further suggestions for modifications, troubleshooting and technical considerations are discussed below.
The method of growing Helicobacter spp. using Blood Agar Base no. 2 supplemented with horse blood has been well established in our laboratory. However, alternate agar bases such as Brucella agar and Columbia blood agar can also be used22. It is important to ensure that only sterile glassware that is free of detergent is used to prepare the growth medium. Furthermore, to obtain optimal growth, H. pylori bacteria should be routinely subcultured on agar plates that have residual moisture and are not dry. When preparing Helicobacter spp. inocula for infection, it is vital to subculture H. pylori and H. felis strains every 1–1.5 or 2 days, respectively, to ensure bacterial viability. At every subculture, bacteria should be assessed for their viability and motility by phase contrast microscopy. A urease assay can also be routinely employed to discriminate between gastric Helicobacter spp. and other bacteria23, however, it is important to realize that this assay detects both viable and non-viable Helicobacter bacteria. Following inoculation of animals, viable counts on Helicobacter suspensions must be performed to quantitate numbers of viable bacteria used for infection. As H. felis does not reproducibly form isolated colonies on agar medium15, estimation of bacterial numbers is performed using phase contrast microscopy. Quantification of bacterial numbers by optical density measurement (A600) alone is inaccurate as this method does not discriminate between viable and non-viable bacteria. This method should not be used in Helicobacter research without rigorous optimization, as described above (Section 1.5).
When performing Helicobacter infection studies, it is crucial to consider the optimal mouse and Helicobacter strain, as well as the length of infection, to suit the purpose of the experiment. It is also essential to regularly confirm that the animals used for experimentation are indeed Helicobacter-free using genus-specific PCR primers24. The presence of other enteric Helicobacter species, such as Helicobacter bilis, Helicobacter hepaticus or Helicobacter muridarum, may alter the disease susceptibility of mice and introduce confounding factors into in vivo studies25,26. It is also advisable to include a mock treatment control group of animals (i.e., fed broth only) in initial screening experiments to investigate the effects of the normal microbiota on Helicobacter colonization and pathogenesis.
Post-euthanasia, H. pylori colonization in murine stomachs can be measured by viable counting. HBA plates used for colony counts should be supplemented with bacitracin and naladixic acid in addition to the modified Skirrow’s selective supplement, to restrict the growth of bacterial species from the normal gastric microbiota and hence prevent contamination27. H. felis does not always form colonies, but instead tends to form swarming growth on agar plates15,28. Therefore, PCR and qPCR are normally employed to determine the presence and levels of H. felis, respectively29,30. In section 4.2, we introduced a simple and quick PCR method to confirm colonization by H. felis in the murine stomach using a pair of primers, which have been validated to target a 325 bp region of H. felis and H. pylori ureB genes. Using the PCR conditions described above, it is possible to discriminate between infection by these gastric Helicobacter spp. and urease-producing enterohepatic Helicobacters. Other genes that have been validated for PCR detection of gastric Helicobacter infection include the 16s rRNA and flagellin B (flaB) genes15,29,30.
Finally, we have described the use of two powerful staining techniques: H&E staining, to assess histopathological changes in the stomach post-infection; and Giemsa staining, to detect H. felis infection. To obtain optimal staining, it is essential to ensure that tissues have been preserved, processed and embedded in the correct orientation. Additionally, only freshly prepared solutions and filtered stains must be used during this process. Tissue sections can be stored indefinitely and utilized for more specific analysis of gastric pathology via immunofluorescence or immunohistochemistry. Some other common measures of gastric inflammation and disease include: immune cell recruitment (anti-CD45 staining); mucosal thickening/destruction (Periodic Acid Schiff/Alcian blue staining); epithelial cell proliferation (proliferating cell nuclear antigen, PCNA/Bromodeoxyuridine, BrDU staining); or cellular apoptosis (TUNEL staining). The lymphoid follicles observed in the H&E-stained tissues of H. felis-infected mice can be confirmed by immunohistochemistry, using antibodies directed against B (B220+) and T cell (CD3+) antigens31.
In summary, animal models of bacterial disease provide valuable tools in the field of infection biology. The protocols of intragastric gavage and processing of stomach tissues provided here may be adapted to mouse infection models involving other enteric pathogens.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Ms. A. De Paoli and Ms. Georgie Wray-McCann for technical assistance. The authors acknowledge use of the facilities and technical assistance of Monash Histology Platform, Department of Anatomy and Developmental Biology, Monash University. The laboratory is supported by funding from the National Health and Medical Research Council (NHMRC) to RLF (APP1079930, APP1107930). RLF is supported by a Senior Research Fellowship from the NHMRC (APP1079904). KD and MC are both supported by Monash Graduate Scholarships. KD is also supported by the Centre for Innate Immunity and Infectious Diseases, Hudson Institute of Medical Research, while MC has an International Postgraduate Scholarship from the Faculty of Medicine, Nursing and Health Sciences, Monash University. Research at the Hudson Institute of Medical Research is supported by the Victorian Government’s Operational Infrastructure Support Program.
Materials
| Bacteriological reagents | |||
| Oxoid Blood Agar Base No.2 | Thermo Fischer Scientific | CM0271B | Dissolve in deinonized water prior to sterilization |
| Premium Defibrinated Horse blood | Australian Ethical Biologicals | PDHB100 | |
| Bacto Brain Heart Infusion Broth | BD Bioscience | 237500 | Dissolve in deinonized water prior to sterilization |
| CampyGen gas packs | Thermo Fischer Scientific | CN0035A/CN0025A | |
| Histological reagents | |||
| Formalin, neutral buffered, 10% | Sigma Aldrich | HT501128 | |
| Absolute alcohol, 100% Denatured | ChemSupply | AL048-20L-P | |
| Isopropanol (2-propanol) | Merck | 100995 | |
| Xylene (sulphur free) | ChemSupply | XT003-20L | |
| Mayer's Haematoxylin | Amber Scientific | MH-1L | Filter before use |
| Eosin, Aqueous Stain | Amber Scientific | EOCA-1L | Filter before use |
| Wright-Giemsa Stain, modified | Sigma Aldrich | WG80-2.5L | Dilute before use (20% Giemsa, 80% deionized water) |
| Histolene | Grale Scientific | 11031/5 | |
| DPX mounting medium | VWR | 1.00579.0500 | |
| Molecular biology reagents | |||
| Qubit dsDNA BR Assay Kit | Thermo Fischer Scientific | Q32850 | |
| Oligonucleotides | Sigma Aldrich | The annealing temperature of ureB primers used in this study is 61°C | |
| GoTaq Flexi DNA Polymerase | Promega | M8291 | Kit includes 10X PCR buffer and Magnesium Chloride |
| dNTPs | Bioline | BIO-39028 | Dilute to 10mM in sterile nuclease free water before use |
| Molecular Grade Agarose | Bioline | BIO-41025 | |
| Sodium Hydrogen Carbonate | Univar (Ajax Fine Chemicals) | A475-500G | |
| Magnesium Sulphate Heptahydrate | Chem-Supply | MA048-500G | |
| Antibiotics | |||
| Vancomycin | Sigma Aldrich | V2002-1G | Dissolve in deionized water |
| Polymyxin B | Sigma Aldrich | P4932-5MU | Dissolve in deionized water |
| Trimethoprim (≥98% HPLC) | Sigma Aldrich | T7883 | Dissolve in 100% (absolute) Ethanol |
| Amphotericin | Amresco (Astral Scientific) | E437-100MG | Dissolve in deionized water |
| Bacitracin from Bacillus licheniformis | Sigma Aldrich | B0125 | Dissolve in deionized water |
| Naladixic acid | Sigma Aldrich | N8878 | Dissolve in deionized water |
| Other reagents | |||
| Methoxyflurane (Pentrhox) | Medical Developments International | Not applicable | |
| Paraffin Wax | Paraplast Plus, Leica Biosystems | 39601006 | |
| Equipment and plasticware | |||
| Oxoid Anaerobic Jars | Thermo Fischer Scientific | HP0011/HP0031 | |
| COPAN Pasteur Pipettes | Interpath Services | 200CS01 | |
| Eppendorf 5810R centrifuge | Collect bacterial pellets by centrifugation at 2,200 rpm for 10 mins at 4°C | ||
| 23g precision glide needle | BD Bioscience | 301805 | |
| Parafilm M | Bemis, VWR | PM996 | |
| Portex fine bore polythene tubing | Smiths Medical | 800/100/200 | |
| Plastic feeding catheters | Instech Laboratories | FTP20-30 | |
| 1 ml tuberculin luer slip disposable syringes | BD Bioscience | 302100 | |
| Eppendorf micropestle for 1.2 – 2 mL tubes | Sigma Aldrich | Z317314 | Autoclavable polypropylene pestles used for stomach homogenization |
| GentleMACs Dissociator | Miltenyi Biotec | 130-093-235 | Use a pre-set gentleMACS Programs for mouse stomach tissue |
| M Tubes (orange cap) | Miltenyi Biotec | 30-093-236 | |
| Qubit Fluorometer | Thermo Fischer Scientific | Q33216 | |
| Sterile plastic loop | LabServ | LBSLP7202 | |
| Cold Plate, Leica EG1160 Embedding System | Leica Biosystems | Not applicable | |
| Tissue-Tek Base Mould System, Base Mold 38 x 25 x 6 | Sakura, Alphen aan den Rijn | 4124 | |
| Tissue-Tek III Uni-Casette System | Sakura, Alphen aan den Rijn | 4170 | |
| Microtome, Leica RM2235 | Leica Biosystems | ||
| Charged SuperFrost Plus glass slides | Menzel Glaser, Thermo Fischer Scientific | 4951PLUS4 |
References
- Goh, K. L., Chan, W. K., Shiota, S., Yamaoka, Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 16, 1-9 (2011).
- Montecucco, C., Rappuoli, R. Living dangerously: how Helicobacter pylori survives in the human stomach. Nature Reviews Molecular Cell Biology. 2 (6), 457-466 (2001).
- Akopyants, N. S., et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Molecular Microbiology. 28 (1), 37-53 (1998).
- Censini, S., et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 93 (25), 14648-14653 (1996).
- Peek, R. M., Fiske, C., Wilson, K. T. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiological Reviews. 90 (3), 831-858 (2010).
- O’Rourke, J. L., Lee, A. Animal models of Helicobacter pylori infection and disease. Microbes and Infection. 5 (8), 741-748 (2003).
- Sakagami, T., et al. Atrophic gastric changes in both Helicobacter felis and Helicobacter pylori infected mice are host dependent and separate from antral gastritis. Gut. 39 (5), 639-648 (1996).
- Correa, P. Helicobacter pylori and gastric carcinogenesis. The American Journal of Surgical Pathology. 19, S37-S43 (1995).
- Enno, A., et al. MALToma-like lesions in the murine gastric mucosa after long-term infection with Helicobacter felis. A mouse model of Helicobacter pylori-induced gastric lymphoma. The American Journal of Pathology. 147 (1), 217-222 (1995).
- Lee, A., et al. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 112 (4), 1386-1397 (1997).
- Crabtree, J. E., Ferrero, R. L., Kusters, J. G. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter. 7 (2), 139-140 (2002).
- Israel, D. A., et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. Journal of Clinical Investigation. 107 (5), 611-620 (2001).
- Fox, J. G., et al. Helicobacter pylori-induced gastritis in the domestic cat. Infection and Immunity. 63 (7), 2674-2681 (1995).
- Lee, A., Hazell, S. L., O’Rourke, J., Kouprach, S. Isolation of a spiral-shaped bacterium from the cat stomach. Infection and Immunity. 56 (11), 2843-2850 (1988).
- Ferrero, R. L., Wilson, J. E., Sutton, P. Mouse models of Helicobacter-induced gastric cancer: use of cocarcinogens. Methods in Molecular Biology. 921, 157-173 (2012).
- Ferrero, R. L., Thiberge, J. M., Huerre, M., Labigne, A. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infection and Immunity. 66 (4), 1349-1355 (1998).
- Blanchard, T. G., Nedrud, J. G. Laboratory maintenance of Helicobacter species. Current Protocols in Microbiology. , (2012).
- Kim, J. S., Chang, J. H., Chung, S. I., Yum, J. S. Importance of the host genetic background on immune responses to Helicobacter pylori infection and therapeutic vaccine efficacy. FEMS Immunology and Medical Microbiology. 31 (1), 41-46 (2001).
- Nedrud, J. G., et al. Lack of genetic influence on the innate inflammatory response to helicobacter infection of the gastric mucosa. Frontiers in Immunology. 3, 181 (2012).
- Cai, X., et al. Helicobacter felis eradication restores normal architecture and inhibits gastric cancer progression in C57BL/6 mice. Gastroenterology. 128 (7), 1937-1952 (2005).
- Ferrero, R. L., Labigne, A. Cloning, expression and sequencing of Helicobacter felis urease genes. Molecular Microbiology. 9 (2), 323-333 (1993).
- Stevenson, T. H., Castillo, A., Lucia, L. M., Acuff, G. R. Growth of Helicobacter pylori in various liquid and plating media. Letters in Applied Microbiology. 30 (3), 192-196 (2000).
- Uotani, T., Graham, D. Y. Diagnosis of Helicobacter pylori using the rapid urease test. Annals of Translational Medicine. 3 (1), 9 (2015).
- Riley, L. K., Franklin, C. L., Hook, R. R., Besch-Williford, C. Identification of murine helicobacters by PCR and restriction enzyme analyses. Journal of Clinical Microbiology. 34 (4), 942-946 (1996).
- Chaouche-Drider, N., et al. A commensal Helicobacter sp. of the rodent intestinal flora activates TLR2 and NOD1 responses in epithelial cells. PLoS One. 4 (4), e5396 (2009).
- Fox, J. G. Helicobacter bilis: bacterial provocateur orchestrates host immune responses to commensal flora in a model of inflammatory bowel disease. Gut. 56 (7), 898-900 (2007).
- McGee, D. J., et al. Cholesterol enhances Helicobacter pylori resistance to antibiotics and LL-37. Antimicrobial Agents and Chemotherapy. 55 (6), 2897-2904 (2011).
- Viala, J., et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nature Immunology. 5 (11), 1166-1174 (2004).
- Kong, L., et al. A sensitive and specific PCR method to detect Helicobacter felis in a conventional mouse model. Clinical and Vaccine Immunology. 3 (1), 73-78 (1996).
- Ng, G., Every, A., McGuckin, M., Sutton, P. Increased Helicobacter felis colonization in male 129/Sv mice fails to suppress gastritis. Gut Microbes. 2 (6), 358-360 (2011).
- Ferrero, R. L., Ave, P., Radcliff, F. J., Labigne, A., Huerre, M. R. Outbred mice with long-term Helicobacter felis infection develop both gastric lymphoid tissue and glandular hyperplastic lesions. The Journal of Pathology. 191 (3), 333-340 (2000).

