Neurovascular Network Explorer 2.0: A Simple Tool for Exploring and Sharing a Database of Optogenetically-evoked Vasomotion in Mouse Cortex In Vivo
Summary
A graphical user interface for exploring and sharing a database of optogenetically-induced vascular responses in mouse somatosensory cortex in vivo measured by 2-photon microscopy is presented. It allows browsing the data, criteria-based selection, averaging, localization of measurements within a 3D volume of vasculature and exporting the data.
Abstract
The importance of sharing experimental data in neuroscience grows with the amount and complexity of data acquired and various techniques used to obtain and process these data. However, the majority of experimental data, especially from individual studies of regular-sized laboratories never reach wider research community. A graphical user interface (GUI) engine called Neurovascular Network Explorer 2.0 (NNE 2.0) has been created as a tool for simple and low-cost sharing and exploring of vascular imaging data. NNE 2.0 interacts with a database containing optogenetically-evoked dilation/constriction time-courses of individual vessels measured in mice somatosensory cortex in vivo by 2-photon microscopy. NNE 2.0 enables selection and display of the time-courses based on different criteria (subject, branching order, cortical depth, vessel diameter, arteriolar tree) as well as simple mathematical manipulation (e.g. averaging, peak-normalization) and data export. It supports visualization of the vascular network in 3D and enables localization of the individual functional vessel diameter measurements within vascular trees.
NNE 2.0, its source code, and the corresponding database are freely downloadable from UCSD Neurovascular Imaging Laboratory website1. The source code can be utilized by the users to explore the associated database or as a template for databasing and sharing their own experimental results provided the appropriate format.
Introduction
The brain is considered one of the most intricate organs and the desire to untangle its complex function is unflagging. It is being studied at different scales from the molecular to the behavioral level using a wide palette of tools2,3,4,5,6,7,8. The amount of non-homogeneous experimental data grows with unprecedented speed. The awareness of the need for experimental data sharing, organization and standardization grows with the amount of acquired data. It has become evident that neuroinformatics will play a critical role in integrating experimental data across scales into models of brain function and dysfunction9,10.
To this end some studies, especially larger-scale studies, were able to earmark resources to make their results available via extensive databases11,12,13,14,15. However, a vast amount of experimental data from individual studies and regular-sized laboratories never reached the wider research community. This is mainly for two reasons: first, more dedicated time is needed to build a database and create tools that would enable the user to interact with the database; and second, more money is needed to support these tasks. Motivated by these challenges, a MATLAB based graphical user interface (GUI) engine called the Neurovascular Network Explorer 2.0 (NNE 2.0)16 was developed as a simple and low-cost tool for databasing, sharing and exploring of vascular imaging data. This manuscript provides a manual for operation of NNE 2.0 and the associated database of experimental data.
NNE 2.0 is already a second-generation software engine. The first generation, called Neurovascular Network Explorer 1.0 (NNE 1.0)17 was built to interact with a database of sensory-evoked vasodilation in rat primary somatosensory cortex (SI) in vivo measured by 2-photon microscopy18. NNE 1.0, its source code as well as the associated database are freely downloadable as a zipped file called ‘NNE 1 Tian’ from UCSD Neurovascular Imaging Laboratory website1. More information about NNE 1.0 and the associated database can be found in17.
The second generation, the NNE 2.0, interacts with a database of optogenetically-evoked dilation of individual vessels in mice SI in vivo measured by 2-photon microscopy20. The user can browse, select and visualize data based on selection categories such as cortical depth, branching order, vessel diameter, animal subject or a particular arteriolar tree. The GUI further performs simple mathematical operations such as averaging and peak-normalization in selected categories. NNE 2.0 enables to view and browse through images capturing 3D volumes of vasculature as well as identify the location of the functional measurement within the vascular trees. This feature can be used to reconstruct vascular morphologies in 3D and populate them with real single-vessel vaso-motion measurements. These reconstructions can in turn be incorporated into computational models of brain function21,22. NNE 2.0, its source code and the associated database are freely downloadable as a zipped file called ‘NNE 2.0 HDbase v1.0’ from UCSD Neurovascular Imaging Laboratory website1.
NNE 2.0 works with a database called 'vdb.mat'. This database is a matrix containing temporal profiles (time-courses) of single vessel diameter changes evoked by an optogenetic stimulus and measured at different locations of arteriolar trees. Each time-course was computed using custom-written software. It calculates the relative change of a vessel diameter from expansion of a fluorescent intensity profile acquired by scanning across the vessel. The fluorescent contrast was presented by intravascular injection of fluorescein isothiocyanate (FITC)-labeled dextran. For more information about the data and analysis procedures, please see20,23. The database has 305 time-courses (i.e. database entries) in total. In addition to the diameter change, each entry to the database holds an array of additional metadata which (1) quantify the time-course (2) describe the measured vessel and (3) identify the measurement location within a 3D volume of cortical vasculature. The metadata include the onset time, peak amplitude, peak amplitude time, cortical depth, branching order, vessel diameter at baseline, path to original reference images and 3D image stacks for each measurement and low-magnification maps of brain surface vasculature. Please see all parameters in the metadata listed and described in detail previously in Table 116.
NNE 2.0 interacts with reference images that are X-Y scans of a plane where the diameter measurement occurred. Each database entry has one corresponding reference image with a reference name displayed in the GUI. Each database entry also has an associated stack of images (3D stack) capturing a 3D volume of the vascular tree within which the measurement occurred. The GUI enables to choose a particular database entry and display the corresponding reference image as well as the 3D stack. It also guides the user to find the matching reference image and frame in the 3D stack (the same features can be found in both images). All stack and reference images in their full resolution (1024 pix x 1024 pix) are included in folders hana_stk and hana_refs, respectively. Low-magnification maps of brain vasculature are included in folder 'maps'. All three folders as well as the database matrix ‘vdb.mat’ are downloaded in the zipped file ‘NNE 2.0 HDbase v1.0’ from the UCSD Neurovascular Imaging Laboratory website1 and saved to the root folder of NNE 2.0 during the installation process.
The GUI has been designed as a set of four panels (Panel 1 (Main Panel) – Panel 4) which open sequentially as the user explores the database and selects specific data based on selection categories. Each panel is divided into two main parts: (1) the right column provides the possibility to interact with the database by selecting parameters and categories of the data and displays important information from the metadata; (2) the left column displays the data in the form of time-courses (diameter change in time) and scatter-plots. There are four types of scatter plots displaying (1) dilation onset time (2) time of the dilation peak (3) maximum diameter change (peak amplitude) and (4) baseline diameter (diameter before stimulation) as function of cortical depth. The user has the possibility to display average time-courses and values for selected data grouped either by cortical depth or branching order. This is to highlight the feature of gradient diameter-change behavior with increasing depth and branching order20. NNE 2.0 allows the user to export the selected subset of data in the format of '.xls', '.csv' or '.mat'.
Protocol
1. Installation of NNE 2.0
- Go to UCSD Neurovascular Imaging Laboratory website1 and left-click at ‘NNE 2.0 HDbase v1.0’ to download the zipped program files to the desired location on your PC.
NOTE: NNE 2.0 requires a Windows-operating system of versions 7-10, at least 2.8 GB of free space to download the zipped file and 6.9 GB to install the program. - Unzip ‘NNE2_HDbase_v1.0.zip’.
NOTE: The unzipped folder NNE2 contains 10 files: ‘hana_refs.tar.gz’, ‘hana_stk.tar.gz’, ‘maps.tgz’, ‘MCRInstaller.exe’, ‘NNE2.exe’, ’NNE2.zip’, ‘NNE2_README.txt’, ’source.zip’, ’users_guide.pdf’ and ‘vdb.mat’. - Install NNE 2.0 by following instructions in ‘NNE2_README.txt’.
2. Running NNE 2.0
- Start the NNE 2.0 with 'NNE2.exe'.
- Panel 1 (Main Panel): Select a subset of data (Figure 1). Images in the left column of the Main Panel show graphs of time-courses and parameters of all entries to the 'vdb.mat' (Figure 1).
- Select the range for Cortical Depth in the right column of the panel. Type in the range of depth in the format [dmin dmax], where dmin is the minimum depth and dmax is the maximum depth.
NOTE: The data was measured at depth from 30-560 µm. - Select Branching Order in the right column of the panel. Left-click on the arrow and choose one of the options from the list (Surface | Diving Trunk | First Order Branches | Higher Order Branches).
- Select the range of Baseline Diameter in the right column of the panel. Type it in the format [diamin diamax], where diamin is the minimum diameter and diamax is the maximum diameter.
- Select subjects (animals according to the date of acquisition) in the right column of the panel. Left-click on the arrow and choose from the available options. Alternatively, choose data from all subjects by left-clicking All in the blue rectangle.
- Press SUBMIT to display and explore the selected data in Panel 2.
- Select the range for Cortical Depth in the right column of the panel. Type in the range of depth in the format [dmin dmax], where dmin is the minimum depth and dmax is the maximum depth.
- Panel 2: Explore the selected subset of data and continue further data refinement (Figure 2).
- Select the type for group-averaging of the data by left-clicking the appropriate button in the right column on the top. Select: Avg By Cortical Depth or Avg By Branching Order.
NOTE: The actual choice is highlighted in green below (Figure 2). - Select data based on vessel morphology or subject. Left-click Select all data for tree (a single diving arteriole and its branches) or Select all data for Subj (animal subject).
- Left-click Soumettre to display selected data on the left in graphs of (1) individual time-courses (2) group-averaged time-courses and scatter plots of (3) onset times (4) time-to-peaks (5) peak amplitudes and (6) baseline diameters
- Left-click on a trace in the graph of Individual timecourses in the left column to select a time-course.
NOTE: The selected time-course gets highlighted in the graph (magenta) and its onset time, time-to-peak, peak amplitude and baseline diameter will be marked by red circles in the graphs below. Red points in the scatter plots are average values. - Note the identifiers of the subject (Subject ID) and tree (Tree ID) for the selected time-course in the right column at the bottom.
- If desired, change the type of group-averaging by left-clicking the appropriate choice on the top of the right column followed by the Soumettre button and repeat steps from 2.3.4.
- Right-click anywhere in Panel 2 with a cross cursor to view and explore all traces for the selected subject (Subject ID) or tree (Tree ID) in Panel 3.
- Select the type for group-averaging of the data by left-clicking the appropriate button in the right column on the top. Select: Avg By Cortical Depth or Avg By Branching Order.
- Panel 3: Explore the final subset of data and export them (Figure 3).
- Select a time-course in the top graph of the left column by left-clicking on a trace: the selected trace will be highlighted in the graph (magenta) and descriptive parameters of the database entry will be displayed on top of the graph.
NOTE: The average time-course is displayed in thick black (Figure 3). - Note corresponding onset time, time-to-peak, peak amplitude and baseline diameter in the graphs below.
- Left-click the EXPORT SET button in the right column to save traces displayed in the top graph into the folder where NNE 2.0 is running from.
NOTE: This action saves three files: 'vdb_subset.xls', 'vdb_subset.csv' and 'vdb_subset.mat' containing vectors of diameter change and time vectors; 'vdb_subset.mat' contains also descriptive parameters and information from 'vdb.mat'. - To inspect all data for 'subject' instead of 'tree' close Panel 3 by pressing [x], restart NNE 2.0, repeat the selection of categories in Panel 1 (steps 2.2.1-2.2.5) and select all data for subject in Panel 2 (step 2.3.2).
- Right click anywhere in Panel 3 with a cross cursor to go to Panel 4 to explore reference images and 3D stacks for all traces in the top graph of Panel 3.
NOTE: Panel 4 will open if option all data for 'tree' was selected in Panel 2. If all data for 'subject' were selected instead, the user will be prompted to change his selection and directed to Panel 1.
- Select a time-course in the top graph of the left column by left-clicking on a trace: the selected trace will be highlighted in the graph (magenta) and descriptive parameters of the database entry will be displayed on top of the graph.
- Panel 4: Localize the functional measurement within a reference image and within a 3D image stack of vasculature (Figure 4).
- Select a time-course by left-clicking on it in the graph on top of the left column.
NOTE: The selected trace will be highlighted in the graph (magenta) and its descriptive information from metadata will be displayed on top. - Explore the corresponding reference image which is loaded automatically from 'hana_refs' folder at the bottom right of the left column.
- Explore the corresponding 3D image stack loaded automatically from 'hana_stk' folder at the bottom left of the left column. Scroll through stack using the arrows or slider below the figure.
NOTE: When the stack image reaches the level of the reference image – i.e. the diameter measurement level ('Stack index' = 'Ref'), the stack image is highlighted and indicated as 'Frame level'. - Click EXPORT SET in the right column to export the highlighted time-course into a file 'ref_stacks_trace.xls' which is saved to the folder where NNE 2.0 is running from.
NOTE: The file contains the time vector, diameter change vector, subject ID, entry index, location of reference image, location of 3D stack and the stack image number for the frame level. - Close Panel 4 by [x] to go back to Panel 1.
- Select a time-course by left-clicking on it in the graph on top of the left column.
Representative Results
NNE 2.0 and the associate database serve to browse and view the data of the database, sort out the data based on selection criteria, download the selected data, and find the vascular measurements within the corresponding vascular tree.
Panel 1 features selection of data based on categories: 'Cortical Depth', 'Branching Order', 'Baseline Diameter' and 'Subjects' – Figure 1). Please note that in this study, there are no entries for surface arterioles ('Surface') in the 'Branching Order' selection category. If this option is selected, a warning dialog 'No records found – Search too restrictive' appears and prompts the user to select a different option. The same warning will appear if there are no measurements satisfying the selected criteria in Panel 1. In this case the user should close Panel 1 by pressing [x] and restart the program.
All diameter change time-courses fulfilling criteria chosen in Panel 1 can be viewed in Panel 2 (Figure 2). The user can explore all individual time-courses, time-courses group-averaged by cortical depth or branching order and the corresponding values of 'Onset time', 'Peak amplitude', 'Time-to-peak' and 'Baseline diameter' plotted as functions of depth. The user selects one time-course from the individual time-courses graph and explores the shape of the curve as well as the corresponding numerical characteristics in the scatter plots.
All data acquired in the same animal or arteriolar tree can be explored in Panel 3 (Figure 3). In the same manner as in Panel 2, the user selects one time-course from the individual time-courses graph and explores the shape of the curve as well as the corresponding numerical characteristics in the scatter plots. If desired, the user can export all the data from Panel 3 in the format of '.xls', 'csv' and '.mat'. These files are created or overwritten each time the 'Export' action is taken. Before overwriting the files, a warning dialog 'About to overwrite vdb_subset.xls' pops out prompting the user to rename previously exported results. The user should make sure none of the exported files are open during the 'Export' action. If one of the files is open, a warning dialog 'Error exporting Excel file: make sure vdb_subset.xls is not open' will appear. In this case, the user should close the exported file and restart NNE 2.0.
All data acquired within a single arteriolar tree can be explored in the context of 3D vasculature in Panel 4 (Figure 4). Selecting a time-course will automatically display the associated reference image and the 3D image stack which are loaded from folders 'hana_refs' and 'hana_stk', respectively. The measured vessel is highlighted in its reference image with a red semi-transparent rectangle in the middle of the image. The scanning path is marked as a red line crossing the measured vessel. If more vessels are scanned within one measurement (red line crossing multiple vessels – Figure 4), the user needs to take into account the branching order found in 'vdb.mat' or displayed in the GUI on top of the time-courses graph ('B. Order') to understand which scan belongs to the particular measurement. Branching order '0' labels diving trunks, '1' labels branches connected directly to diving trunks, '2' labels branches directly connected to 1st order branches, etc. To identify the appropriate arteriolar tree starting with a diving arteriole at the surface of the brain (seen in the top images of the 3D stack), the user should refer to a low-magnification map saved in the folder 'maps'. This map is unique for each animal subject and can be located using the corresponding subject ID (e.g. '022014.jpg'). This map is an image of the whole brain exposure with surface vasculature. The diving segments of measured arterioles are labeled with the tree identifiers ('Tree ID') (Figure 5). The user can export a single selected time-course along with the information about the corresponding reference image, 3D stack and position of the measurement within the stack into 'ref_stacks_trace.xls'. Similarly, as for 'Export' in Panel 3, 'ref_stacks_trace.xls' should be closed before the 'Export' action is taken. The same type of warning dialogs will appear before overwriting the exported file or when the file is open during the 'Export' action. Please note that if there is a missing reference image for the selected database entry (9 entries in total), a warning 'NO REFERENCE IMAGE FOUND: index = ' will be displayed instead of the reference image in Panel 4. No export is available for those entries and the user is prompted to choose a different time-course. If the user selects an entry for which the 3D stack does not exist or no reference image/stack frame was found (31 and 52 entries, respectively) a note *NO STACK MATCH* will be displayed on top of a blank image in the place of the 3D stack image in Panel 4.
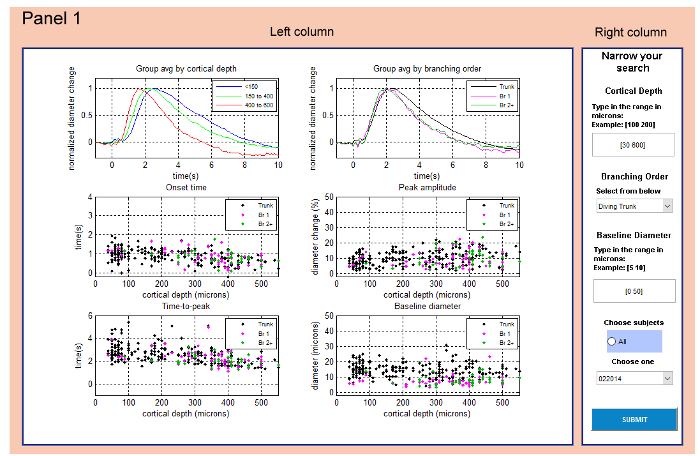
Figure 1: Panel 1 (Main Panel) serves to select a subset of data based on selection categories. All data from 'vdb.mat' are displayed in six graphs in the left column. The right column allows the user to select a subset of this data by choosing Cortical Depth | Branching Order | Baseline Diameter | Subjects. Please click here to view a larger version of this figure.
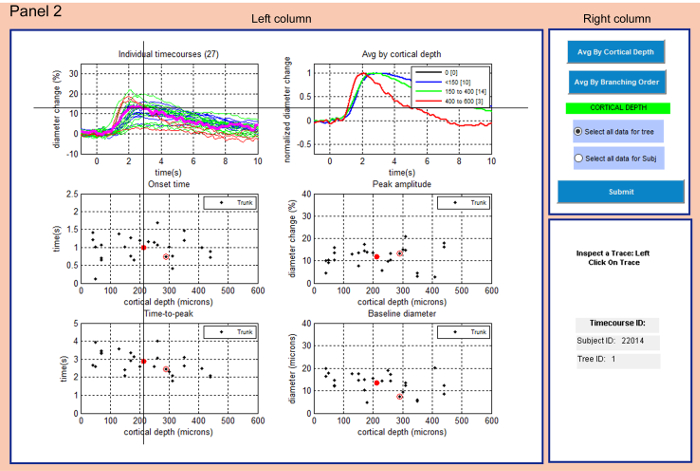
Figure 2: Panel 2 allows examining the selected subset of data and further refining the selection. Right column offers to calculate average of the selected data based on depth categories or branching order (the actual choice is highlighted in green). Left column displays data fulfilling the criteria selection in Panel 1 and the average type selected in the right column. The user can select a time-course in the top left graph (cross cursor) which gets highlighted (magenta). The corresponding times of onset and peak, the peak amplitude and baseline diameter are circled in red and the entry identifiers are displayed in the right column at the bottom. Thick red points in the scatter plots mark the average values for the actual data subset. The selection of 'all data for tree' vs. 'all data for Subj' affects data in the following panels. Please click here to view a larger version of this figure.
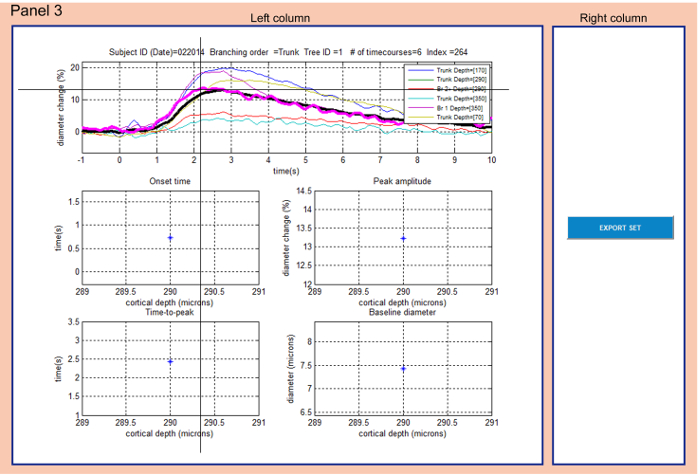
Figure 3: Panel 3 allows exploring the final subset of data and exporting them. The user can select (cross cursor) a time-course in the graph on top of the left column. The selected trace gets highlighted (magenta) and the entry metadata is displayed on top of the graph. Simultaneously the corresponding onset and peak times as well as the peak amplitude and baseline diameters are displayed in the graphs below. The EXPORT SET button in the right column allows exporting all the time-courses from the top graph. The average time-course is plotted in thick black. Please click here to view a larger version of this figure.
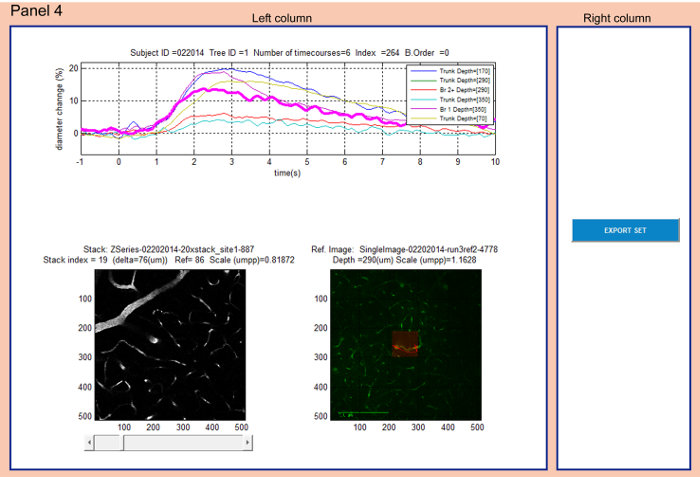
Figure 4: Panel 4 allows localizing the diameter measurements within a reference image and within a 3D stack of vasculature. The time-courses plot on top of the left column is identical with the time-courses plot in Panel 3. The user can select individual time-courses from the top graph in the left column. The corresponding reference image is displayed at the bottom right along with the metadata information: 'Ref. Image' (reference image name), 'Depth' (cortical depth of the measurement) and 'Scale' (scale of the image in microns per pixel). The corresponding 3D stack is displayed at the bottom left along with the descriptive metadata information: 'Stack index' (actual image number in the stack), 'Delta' (vertical distance from the top image), 'Ref' (image number in the stack which corresponds to the reference image and captures the measurement location) and 'Scale' (scale of the image in microns per pixel). Please note that the 'Depth' on top of the reference image was entered manually during the experiment and is approximate. It does not match exactly the value of 'Delta' of the frame level in stack images due to the fact that the brain surface is tilted with regards to the imaging plane. Please click here to view a larger version of this figure.
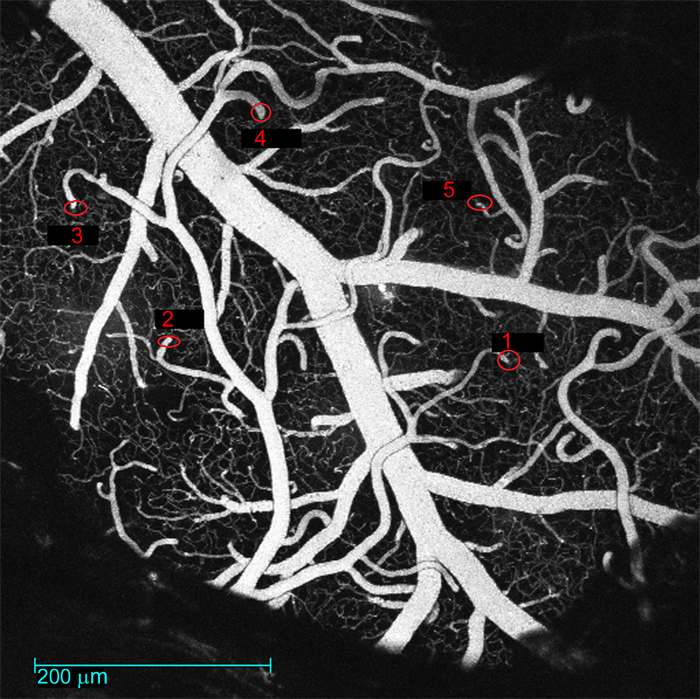
Figure 5: Low-magnification map of the surface vasculature. The image captures the exposed brain region with surface vessels. The diving segments of measured arteriolar trees are labeled with the tree identifiers ('Tree ID'). These maps are saved in the folder 'maps' in the folder where NNE 2.0 is running from. Please click here to view a larger version of this figure.
Discussion
NNE 2.0 was written in order to share the vascular imaging data of a specific study20 but with the intention of developing a simple tool for sharing and exploring data of similar kind by other users. Researchers interested in inspecting the associated database of vascular data may use the GUI to browse the data, select subsets of data, compare them to their own experimental results or process them further using their own computational procedures. Users familiar with MATLAB can utilize directly the database 'vdb.mat' while users employing a different type of programming language can export the data in one of the alternative formats ('.xls' or '.csv').
Researchers who are interested in sharing their own experimental data using NNE 2.0 need to structure the results into a matrix similarly to 'vdb.mat'. Principal entries to this database should be time-courses of any kind in the form of vectors and the corresponding time vectors. Parameters of the database can be modified or added to via modular structure of the GUI. All reference images, image stacks and brain exposure maps (if applicable) should be collected and deposited along with the database and the executable GUI on the internet (e.g. laboratory webpage or a third-party repository).
Researchers interested in using the vascular measurements along with the 3D vascular morphology in modelling studies of brain function can first explore the data using the GUI. After selecting a desired subset of data, they can use the metadata information exported to 'ref_stacks_trace.xls' along with variables in 'vdb.mat', 3D stack images saved in 'hana_stk' and brain exposure maps in 'maps' to reconstruct the vascular morphology in 3D.
The most critical step of the protocol is exporting the data. For correct export of selected data (from Panel 3 and 4) it is important to close all files into which the data should be exported to before the ‘Export’ action is taken. Only then the files will be correctly overwritten with the actual choice of data. Any modifications to the program or troubleshooting need to be performed to the source code.
NNE 2.0 has been developed to share temporal profiles computed from line-scans which were acquired by 2-photon microscopy. Data explored by NNE 2.0 are therefore not raw intensity scans but rather pre-processed time-courses of relative diameter changes. In this way the users are able to process their raw experimental data using their own standard procedures and software and use NNE 2.0 as a template to present and share their results with other researchers. Not only vascular diameter changes but basically any time-dependent signal measured using different measurement techniques can be shared this way. Such signals include fluorescence of calcium24, sodium25, voltage sensitive dyes26, genetically encoded indicators for metabolites27, partial pressure of oxygen (pO2)28, blood oxygenation (BOLD18, spectral imaging29), blood flow (speckle imaging29) or electrophysiology signals30. The prerequisite for using NNE 2.0 is a database in the format of a matrix '*.mat'. This could be achieved either by processing the data directly in MATLAB or using tools to build the matrix from other formats (e.g. 'xlsread(filename)' serves to read excel spread sheets into '.mat' formats). The use of NNE 2.0 without MATLAB is limited to exploring, downloading and further processing of data from the current database 'vdb.mat'.
NNE 2.0 has the potential to help spreading experimental data across the wide neuroscience research community to be explored and compared to other experimental data of similar kind facilitating the development of standards for data acquisition and processing31. NNE 2.0 can also help spreading the data across the neuroinformatics community where it can be used in models of non-invasive imaging signals such as functional magnetic resonance imaging (fMRI)21. While NNE 2.0 cannot compete with more complex databases5,32,33,34, it can provide a seamless and ready-to-use platform for databasing and sharing experimental data without the need of additional extensive investments.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We gratefully acknowledge support from the NIH (NS057198, EB00790, MH111359, and S10RR029050) and the Ministry of Education, Youth and Sports of the Czech Republic (CEITEC 2020, LQ1601). KK was supported by postdoctoral fellowships from the International Headache Society in 2014 and The Scientific and Technological Research Council of Turkey in 2015. MT was supported by postdoctoral fellowship from the German Research Foundation (DFG TH 2031/1).
References
- . Use Our Data Available from: https://neurosciences.ucsd.edu/research/labs/nil/Pages/UseOurData.aspx (2017)
- Craddock, R. C., et al. Imaging human connectomes at the macroscale. Nat Methods. 10 (6), 524-539 (2013).
- Devor, A., et al. Frontiers in optical imaging of cerebral blood flow and metabolism. J Cereb Blood Flow Metab. 32 (7), 1259-1276 (2012).
- Ji, N., Freeman, J., Smith, S. L. Technologies for imaging neural activity in large volumes. Nat Neurosci. 19 (9), 1154-1164 (2016).
- Maze, I., et al. Analytical tools and current challenges in the modern era of neuroepigenomics. Nat Neurosci. 17 (11), 1476-1490 (2014).
- Medland, S. E., Jahanshad, N., Neale, B. M., Thompson, P. M. Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci. 17 (6), 791-800 (2014).
- Osten, P., Margrie, T. W. Mapping brain circuitry with a light microscope. Nat Methods. 10 (6), 515-523 (2013).
- Poldrack, R. A., Farah, M. J. Progress and challenges in probing the human brain. Nature. 526 (7573), 371-379 (2015).
- Kotter, R. Neuroscience databases: tools for exploring brain structure-function relationships. Philos Trans R Soc Lond B Biol Sci. 356 (1412), 1111-1120 (2001).
- Uhlirova, H., et al. The roadmap for estimation of cell-type-specific neuronal activity from non-invasive measurements. Philos Trans R Soc Lond B Biol Sci. 371 (1705), (2016).
- Aine, C. J., et al. Multimodal Neuroimaging in Schizophrenia: Description and Dissemination. Neuroinformatics. , (2017).
- Amunts, K., et al. BigBrain: an ultrahigh-resolution 3D human brain model. Science. 340 (6139), 1472-1475 (2013).
- Laird, A. R., Lancaster, J. L., Fox, P. T. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics. 3 (1), 65-78 (2005).
- Lein, E. S., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445 (7124), 168-176 (2007).
- Shin, D. D., Ozyurt, I. B., Liu, T. T. The Cerebral Blood Flow Biomedical Informatics Research Network (CBFBIRN) database and analysis pipeline for arterial spin labeling MRI data. Front Neuroinform. 7, 21 (2013).
- Uhlirova, H., et al. Neurovascular Network Explorer 2.0: A Database of 2-Photon Single-Vessel Diameter Measurements from Mouse SI Cortex in Response To Optogenetic Stimulation. Front Neuroinform. 11, 4 (2017).
- Sridhar, V. B., Tian, P., Dale, A. M., Devor, A., Saisan, P. A. Neurovascular Network Explorer 1.0: a database of 2-photon single-vessel diameter measurements with MATLAB((R)) graphical user interface. Front Neuroinform. 8, 56 (2014).
- Tian, P., et al. Cortical depth-specific microvascular dilation underlies laminar differences in blood oxygenation level-dependent functional MRI signal. Proc Natl Acad Sci U S A. 107 (34), 15246-15251 (2010).
- . Use Our Data Available from: https://neurosciences.ucsd.edu/research/labs/nil/Pages/UseOurData.aspx (2017)
- Uhlirova, H., et al. Cell type specificity of neurovascular coupling in cerebral cortex. Elife. 5, (2016).
- Gagnon, L., et al. Quantifying the microvascular origin of BOLD-fMRI from first principles with two-photon microscopy and an oxygen-sensitive nanoprobe. J Neurosci. 35 (8), 3663-3675 (2015).
- Sakadzic, S., et al. Two-photon high-resolution measurement of partial pressure of oxygen in cerebral vasculature and tissue. Nat Methods. 7 (9), 755-759 (2010).
- Nizar, K., et al. In vivo stimulus-induced vasodilation occurs without IP3 receptor activation and may precede astrocytic calcium increase. J Neurosci. 33 (19), 8411-8422 (2013).
- Reznichenko, L., et al. In vivo alterations in calcium buffering capacity in transgenic mouse model of synucleinopathy. J Neurosci. 32 (29), 9992-9998 (2012).
- Langer, J., Rose, C. R. Synaptically induced sodium signals in hippocampal astrocytes in situ. J Physiol. 587 (Pt 24), 5859-5877 (2009).
- Gong, Y., et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 350 (6266), 1361-1366 (2015).
- Tantama, M., Hung, Y. P., Yellen, G. Optogenetic reporters: Fluorescent protein-based genetically encoded indicators of signaling and metabolism in the brain. Prog Brain Res. 196, 235-263 (2012).
- Devor, A., et al. “Overshoot” of O(2) is required to maintain baseline tissue oxygenation at locations distal to blood vessels. J Neurosci. 31 (38), 13676-13681 (2011).
- Devor, A., et al. Stimulus-induced changes in blood flow and 2-deoxyglucose uptake dissociate in ipsilateral somatosensory cortex. J Neurosci. 28 (53), 14347-14357 (2008).
- Rauch, A., Rainer, G., Logothetis, N. K. The effect of a serotonin-induced dissociation between spiking and perisynaptic activity on BOLD functional MRI. Proc Natl Acad Sci U S A. 105 (18), 6759-6764 (2008).
- Lemmon, V. P., et al. Minimum information about a spinal cord injury experiment: a proposed reporting standard for spinal cord injury experiments. J Neurotrauma. 31 (15), 1354-1361 (2014).
- Ascoli, G. A., Donohue, D. E., Halavi, M. NeuroMorpho.Org: a central resource for neuronal morphologies. J Neurosci. 27 (35), 9247-9251 (2007).
- Mennes, M., Biswal, B. B., Castellanos, F. X., Milham, M. P. Making data sharing work: the FCP/INDI experience. Neuroimage. 82, 683-691 (2013).
- Marmarou, A., et al. IMPACT database of traumatic brain injury: design and description. J Neurotrauma. 24 (2), 239-250 (2007).

