Generating Transgenic Plants with Single-copy Insertions Using BIBAC-GW Binary Vector
Summary
Using a pBIBAC-GW binary vector makes generating transgenic plants with intact single-copy insertions, an easy process. Here, a series of protocols is presented that guide the reader through the process of generating transgenic Arabidopsis plants, and testing the plants for intactness and copy number of the inserts.
Abstract
When generating transgenic plants, generally the objective is to have stable expression of a transgene. This requires a single, intact integration of the transgene, as multi-copy integrations are often subjected to gene silencing. The Gateway-compatible binary vector based on bacterial artificial chromosomes (pBIBAC-GW), like other pBIBAC derivatives, allows the insertion of single-copy transgenes with high efficiency. As an improvement to the original pBIBAC, a Gateway cassette has been cloned into pBIBAC-GW, so that the sequences of interest can now be easily incorporated into the vector transfer DNA (T-DNA) by Gateway cloning. Commonly, the transformation with pBIBAC-GW results in an efficiency of 0.2–0.5%, whereby half of the transgenics carry an intact single-copy integration of the T-DNA. The pBIBAC-GW vectors are available with resistance to Glufosinate-ammonium or DsRed fluorescence in seed coats for selection in plants, and with resistance to kanamycin as a selection in bacteria. Here, a series of protocols is presented that guide the reader through the process of generating transgenic plants using pBIBAC-GW: starting from recombining the sequences of interest into the pBIBAC-GW vector of choice, to plant transformation with Agrobacterium, selection of the transgenics, and testing the plants for intactness and copy number of the inserts using DNA blotting. Attention is given to designing a DNA blotting strategy to recognize single- and multi-copy integrations at single and multiple loci.
Introduction
When generating transgenic plants, usually the objective is to have the integrated transgene(s) stably expressed. This can be achieved by intact single copy integrations of a transgene. Multiple integrations can lead to increased expression of a transgene, but also to gene silencing. Silencing of transgenes is more likely if inserted sequences are arranged in tandem or inverted repeats1,2,3,4. Binary vectors are used as shuttles in Agrobacterium-mediated transformation experiments to deliver the sequences of interest into plant genomes. The number of integrations into a plant genome is dependent on the copy number of the binary vector in Agrobacterium tumefaciens5,6. Many commonly used binary vectors are high copy vectors, and therefore yield a high average transgene copy number: 3.3 to 4.9 copies in Arabidopsis5.
The number of T-DNA integrations can be lowered by using binary vectors that have a low-copy number in A. tumefaciens, such as BIBAC7, or by launching a T-DNA from the A. tumefaciens chromosome5. The average number of transgene integrations in such cases is below 25,8,9,10. Due to being single-copy in A. tumefaciens, and also in Escherichia coli, BIBAC-derivatives can maintain and deliver constructs as large as 150 kb11.
GW-compatible BIBAC vectors10,12 allow easy introduction of genes of interest into the vector using Gateway cloning. The use of Gateway technology greatly simplifies the cloning procedure, but also overcomes common problems associated with large low-copy-number vectors13,14, such as a low DNA yield and a limited selection of unique restriction sites available for cloning7,11. The pBIBAC-GW derivatives are available with either resistance to Glufosinate-ammonium (pBIBAC-BAR-GW) or DsRed fluorescence in seed coats (pBIBAC-RFP-GW) for selection in plants (Figure 1)10,12. For both vectors, a kanamycin resistance gene is used as the selection marker in bacteria.
The pBIBAC-GW vectors combine: (1) easy design and genetic manipulation in E. coli, and (2) intact single-copy integrations in planta at high efficiency. The pBIBAC-GW vectors yield on average 1.7 integrations in Arabidopsis with approximately half of the transgenic plants carrying a single integrated T-DNA10.
Stable expression of transgenes is a requirement for most transgenics generated. Stable transgene expression can be achieved by intact, single-copy integrations. Working with transgenic plants carrying intact, single-copy integrations is, however, even more important if for example, the aim is to study the efficiency of chromatin-based processes, such as mutagenesis, recombination, or repair, and the dependence of these processes on the genomic location and the chromatin structure at the insertion site. For our interest, to study the dependence of oligonucleotide directed mutagenesis (ODM) on the local genomic context, a set of reporter lines with intact, single-copy integrations of a mutagenesis reporter gene was generated (Figure 2)10. Using this set of lines, it was shown that the ODM efficiency varies between transgenic loci integrated at different genomic locations, despite the transgene expression levels being rather similar.
Protocol
1. Inserting Sequences of Interest into Binary Vector
- Prepare the Gateway Entry and binary vectors.
- Isolate the Gateway Entry vector containing a DNA fragment or gene of interest using a mini-prep kit according to the suggestions of the supplier.
NOTE: BIBAC-GW vectors require the use of kanamycin (Km) for selection in bacteria, therefore, use an Entry vector with another resistance marker instead of kanamycin. For instance, the pENTR-gm vector, carrying a Gentamicin resistance gene, is a good choice12. - Propagate and isolate the BIBAC-GW vector of interest. Use an E. coli strain that is resistant to the toxicity of the ccdB gene present within the Gateway cassette. Isolate BIBAC-vectors using protocols or kits specifically designed for large plasmids according to the suggestions of the supplier.
- Isolate the Gateway Entry vector containing a DNA fragment or gene of interest using a mini-prep kit according to the suggestions of the supplier.
- Carry out a Gateway reaction.
- Prepare the LR recombination reaction according to the suggestions of the supplier. Mix the following components in a 1.5 mL microcentrifuge tube at room temperature (RT): 100–300 ng Entry clone (supercoiled), 300 ng of BIBAC-GW vector, LR Clonase reaction buffer (final concentration: 1x). Adjust the volume of the mixture to 16 µL with TE (10 mM Tris, 1 mM of EDTA, pH 8.0). Finally, add 4 µL of LR Clonase enzyme mix, and mix by vortexing. Incubate the mixture at 25 °C for 1 h.
- Terminate the LR reaction by adding 2 µL of Proteinase K solution (2 µg/µL) to the mixture prepared in step 1.2.1. Mix and incubate at 37 °C for 10 min.
- Transform E. coli with the Gateway reaction mixture by electroporation.
- Desalt the LR reaction mixture prior to electroporation.
NOTE: This step is critical for successful electroporation. Below a dialysis method is described, but other methods such as precipitation with sodium acetate and ethanol can also be used.- Prepare the setup for filter dialysis of the LR reaction. Pour 20 mL of ultrapure deionized water into a sterile Petri dish. Place a membrane filter disk (pore size = 0.025 µm) on the water surface.
- Pipette the entire LR reaction carefully on top of the membrane and allow the mixture to dialyze at RT for 1 h.
- Add 5 µL of the desalted LR mix to electro-competent DH10B cells in an electroporation cuvette (0.1 cm). Electroporate the cells (1.5 V/cm, resistance 200 Ω, capacitance 25 µF), and immediately add 1 mL pre-warmed Super Optimal Catabolite repression (SOC) medium to the cells, followed by incubation at 37 °C for 45 min, 180 rpm. (SOC medium, 1 L: 20 g of Bacto tryptone, 5 g of Yeast extract, 0.5 g of NaCl, 2.5 mL of 1M KCl, 20 mL of 1 M filter-sterilized Glucose).
NOTE: DH10B cells can be substituted for other E. coli cells that stably maintain large plasmids.
NOTE: The optimal electroporation conditions are dependent on the electroporation device used. - Pellet bacteria at maximum speed for 30 s using a microcentrifuge, remove excess SOC, and resuspend the pellet in approximately 50-100 µL of Luria-Bertani (LB) medium. Spread the bacteria on kanamycin-LB (Km-LB) plates (Km concentration, 40 µg/mL), and incubate the plates at 37 °C overnight. (LB medium, 1 L: 10 g of Bacto tryptone, 5 g of Yeast extract, 10 g of NaCl. For solid medium add agar, 15 g/L).
- Desalt the LR reaction mixture prior to electroporation.
- Identify the recombined BIBAC-GW derivatives and isolate plasmid DNA.
- To be able to grow on Km-LB plates, the E. coli cells should contain the recombined BIBAC-GW plasmid in which the ccdB sequence is substituted with the desired insert. Use colony Polymerase Chain Reaction (PCR)15 to check if the bacteria colonies on the plate contain the correct plasmids, having the BIBAC-GW backbone and the insert of interest.
- To identify the pBIBAC-BAR-GW backbone, perform a PCR15 reaction with the primers DM1969 5'-GCGACGAGCCAGGGATAG-3' and DM1970 5'-ATCAGTGCGCAAGACGTGAC-3'. This primer set amplifies a 563 bp fragment of the bar gene.
- To check for the presence of the BIBAC-RFP-GW backbone, perform a PCR15 reaction, using primers M737 5'-CGTGTAAAAAGCTTAGACTG-3' and M892 5'-AACAGATGGTGGCGTCCC-3'. This primer combination amplifies a 791 bp fragment overlapping the cruciferin promotor and rfp sequence.
- Perform PCR15 reactions using gene-specific primers to check for the presence of the insert of interest.
- Inoculate a single positive colony in 2–5 mL of LB medium containing kanamycin (40 µg/mL) for DNA isolation16. Incubate at 37 °C on an orbital shaker at 180 rpm, overnight.
- Isolate the plasmid DNA (see step 1.1.2).
- To be able to grow on Km-LB plates, the E. coli cells should contain the recombined BIBAC-GW plasmid in which the ccdB sequence is substituted with the desired insert. Use colony Polymerase Chain Reaction (PCR)15 to check if the bacteria colonies on the plate contain the correct plasmids, having the BIBAC-GW backbone and the insert of interest.
2. Preparation of A. tumefaciens for Floral Dipping of Arabidopsis
- Transform BIBAC-GW derivatives to A. tumefaciens.
- Prepare electro-competent cells of A. tumefaciens strain C58C1 carrying the pCH32 helper plasmid7. Grow bacteria in the presence of tetracycline (5 µg/mL) and rifampicin (100 µg/mL) to select for pCH32 and ensure growth of only Agrobacterium cells.
- Add 0.25–0.5 µg DNA of a pBIBAC-GW derivative, pre-dissolved in 10–20 µL sterile ultrapure deionized water, to 20 µL competent Agrobacterium cells in electroporation cuvettes (0.1 cm). Keep cells on ice.
- Electroporate the cells (1.5 V/cm, resistance 400 Ω, capacitance 25 µF). Immediately after electroporation, add 1 mL pre-warmed (28 °C) SOC medium to the bacteria and incubate the cells at 28 °C for 60–90 min.
- Spread 100 µL and the rest of the bacteria on separate LB plates containing rifampicin (100 µg/mL), tetracycline (5 µg/mL), and kanamycin (40 µg/mL), and incubate in the dark at 28 °C for 1-2 days.
- Prepare the Agrobacterium suspension.
- Check by PCR a couple of the A. tumefaciens colonies from the plate prepared in step 2.1.4, for the presence of the correct vector (see step 1.4.1).
- Streak a single colony confirmed to contain the binary vector with the appropriate insert on an LB plate containing antibiotics (see step 2.1.4). Grow at 28 °C overnight.
- Repeat the streaking with a single colony obtained in step 2.2.2.
- Inoculate a single colony in 2.5 mL of LC medium supplemented with antibiotics (see step 2.1.4) for preculture. Incubate at 28 °C for at least 8 h or overnight, at 180 rpm. (LC medium, 1 L: 10 g of Bacto tryptone, 5 g of Yeast extract, 0.5 g of NaCl, 2.5 g of MgSO4 · 7H2O, 2 g of Maltose).
- Add the preculture from step 2.2.4. to 250 mL LC supplemented with antibiotics (see step 2.1.4) and grow at 28 °C, at 180 rpm, overnight.
- Pellet the culture by spinning at 5,500 x g for 12 min. Re-suspend the pellet in 100 mL solution containing 5% sucrose, 0.05% Silwet L-77, 0.5x MS. Pour the suspension into a sterile container for floral dipping of the plants.
3. Arabidopsis Transformation
- Prepare the Arabidopsis plants for transformation.
- Grow Arabidopsis plants in a greenhouse or climate controlled growth chamber until they are flowering (12 pots with 9 plants each per dipping).
- Clip the first bolts to allow more secondary bolts to emerge. Plants are ready for dipping 4–6 days after clipping, when the plants have many immature flower heads and not many fertilized siliques.
- Floral dipping
- Dip inflorescences for 5–10 s in Agrobacterium suspension prepared in step 2.2.6. Use gentle agitation.
- Wrap the above-ground parts of the plants in cling film to keep the humidity high, and cover the plant pots with a box to keep the plants in the dark. Incubate the plants for 2 days in a greenhouse/growth chamber.
- Remove the box and the cling film and grow the plants to maturity in a greenhouse/growth chamber.
NOTE: To increase the efficiency of transformation, the same plants can be re-dipped 7 days after the first dipping. - Harvest the seeds. Pool and analyze the seeds (T1) of plants, transformed with the same construct, as a single set.
- Screen for transgenic plants.
- To screen for transgenic plants transformed with a pBIBAC-RFP-GW derivative, analyze the seeds using fluorescence microscopy. In order to detect DsRed expression in seed coats, image the seeds at an excitation of 560 nm and emission of 600–650 nm. Separate the fluorescent seeds from non-fluorescent counterparts using forceps.
- To screen for transgenic plants transformed with a pBIBAC-BAR-GW derivative, sow the seeds in trays filled with soil (~2,500 seeds/0.1 m2). To ensure an even spreading of seeds over trays, suspend seeds in 0.1% agar in 0.5x Murashige Skoog medium (MS), and spread the seeds using a 1 mL pipette.
NOTE: To stimulate the seeds to germinate in a synchronous manner, incubate the seeds for at least 2 days at 4 °C. This can be done before or after sowing the seeds.- Spray the seedlings with 0.5% Glufosinate-ammonium solution 2 weeks and 3 weeks after sowing in trays. Use 500 mL of Glufosinate-ammonium solution per 1 m2.
- Transfer surviving seedlings to individual pots. A typical image of a tray with seedlings before and after (second) Glufosinate-ammonium treatment are shown in Figure 3.
- Analyze the Glufosinate-ammonium-resistant plants by PCR for the presence of the construct of interest (see step 1.4.1. for primers).
- Isolate the genomic plant DNA for PCR using the method described by Edwards et al.17
4. Characterizing Transgenics for the Number and Integrity of T-DNA Integrations
- Strategy of restriction digestions
NOTE: Determine the number of T-DNA integrations and their integrity by DNA blotting using restriction enzymes. This method allows to identify single, but also repeated integrations at the same or different loci in the genome.- Use a series of restriction digestions to identify the different integration patterns possible:
- Select an enzyme that cuts once in the middle of the T-DNA, to be able to independently probe sequences upstream and downstream of the restriction site (Figure 4A and Figure 7A-C). See Figure 4A, and the figure legend for the expected results and interpretation.
- Select an enzyme or combination of enzymes cutting out the entire sequence of interest at once (Figure 4B and Figure 7A-D). Any deviation from the known length indicates truncation of the integrated cassette.
NOTE: Take care to only use restriction enzymes that are not sensitive to cytosine methylation.
- Use a series of restriction digestions to identify the different integration patterns possible:
- Prepare the genomic DNA samples.
- Isolate the genomic DNA from plants carrying the construct of interest. A CTAB DNA miniprep method can be used for DNA isolation18. For DNA blot analysis of Arabidopsis DNA, 2–2.5 µg of genomic DNA is needed. Dissolve the DNA in 50 µL of TE.
- Check the DNA integrity by gel electrophoresis16. Intact genomic DNA migrates as one discrete band at the top of the gel. DNA degradation can be recognized as the presence of a smear. To avoid DNA damage due to repeated freezing-thawing, genomic DNA samples are kept at 4 °C.
- Digest the genomic DNA (2–2.5 µg in the case of Arabidopsis genomic DNA) in a total volume of 50 µL, overnight, using buffer conditions suggested by the enzyme supplier.
- In a test tube, mix 2–2.5 µg of Arabidopsis genomic DNA, 5 µL 10x restriction buffer, and 5 U restriction enzyme in a total of 50 µL ultrapure deionized water.
- Add loading dye (1x final concentration) to the restriction samples before loading on a gel. For good visual tracking, use one of the following: comigrating with small fragments (Bromophenol blue, 350–400 bp), or comigrating with larger fragments (Cylene cyanol, 3–4 kbp).
- Run the DNA gel.
- Prepare a long (20 cm) 0.5x TBE agarose gel19. The percent of agarose in the gel depends on the fragment sizes expected. 0.8–1% optimally separates fragments >1 kb in size. Use 1–1.5% agarose for fragments <1 kb. Do not add Ethidium Bromide to the gel. (5x TBE, 1 L: 54 g of Trizma base, 27.5 g of Boric acid, 3.75 g of EDTA).
NOTE: To prevent DNA contamination, use gel trays that are not used to fractionate plasmid and PCR amplified DNA. - Load the samples on gel19.
- Add DNA markers to the gel that are in the size range of the expected fragments. Load approximately 1 µg of marker (50-250 ng of different sized fragment) on the gel to allow visualization by UV light.
- Size-fractionate the DNA at a low voltage (40–50 V/500 mA) overnight.
- Prepare a long (20 cm) 0.5x TBE agarose gel19. The percent of agarose in the gel depends on the fragment sizes expected. 0.8–1% optimally separates fragments >1 kb in size. Use 1–1.5% agarose for fragments <1 kb. Do not add Ethidium Bromide to the gel. (5x TBE, 1 L: 54 g of Trizma base, 27.5 g of Boric acid, 3.75 g of EDTA).
- Prepare for transfer of the DNA from the agarose gel to the nylon membrane.
- Transfer the gel to a separate tray, and stain it for 20–25 min in 0.5x TBE containing Ethidium Bromide (5 µg/mL) by rotating at 40 rpm on an orbital shaker.
- Visualize the gel on a UV transilluminator. Verify the size separation of the genomic DNA, including visibility of discrete satellite bands (Figure 5A). A smear towards lower molecular weight sizes indicates DNA degradation.
- On the UV transilluminator, lay a transparency over the gel, and mark the position of the slots and the marker bands with a marker pen (Figure 5B). This will facilitate determining the size of the hybridized fragments later in case the marker sequences do not hybridize specifically with the probe DNA. By indicating the marker fragments at this step, it is possible to track the size of the hybridizing fragments.
- Place the gel back into the tray, rinse with ultrapure deionized water, and submerge it in 0.25 M HCl for 15 min to fragmentize the DNA within the gel. Wash with ultrapure deionized water. Use enough HCl and water to cover the gel in the tray, and rotate the tray with the submerged gel at 40 rpm on an orbital shaker.
- Incubate the gel in Denaturation buffer for 30 min. Wash with ultrapure deionized water. Use enough buffer and water to cover the gel in the tray, and rotate the tray with the submerged gel at 40 rpm on an orbital shaker. (Denaturation buffer: 0.5 M NaOH, 1.5 M NaCl).
- Incubate the gel in Neutralization buffer for 30 min. Wash with ultrapure deionized water. Use enough buffer and water to cover the gel in the tray, rotate the tray with submerged gel at 40 rpm on an orbital shaker. (Neutralization buffer: 0.5 M Tris, 1.5 M NaCl, 220 mM HCl, pH 7.6).
- Transfer the DNA to a nylon membrane.
- Prepare the setup for capillary transfer of the genomic DNA. Place a plastic plate (approximately the size of the gel or bigger) over a tray filled with 20x saline-sodium citrate (SSC). Fold a piece of thick filter paper over the tray, so that both of its ends are hanging in the SSC. Cut a gel-sized piece of positively charged nylon membrane Hybond N+, and 2 pieces of thick filter paper. (20x SSC: 3 M NaCl, 0.3 M Sodium Citrate).
- Prepare the blotting setup (Figure 6) by placing the gel, slots down on the top of the filter paper on the plastic plate. Place a Hybond N+ membrane on top, followed by 2 layers of filter paper. Pre-wet each layer in 20x SSC before adding them to the assembly. Make sure to remove any air bubbles in between layers as these impede the DNA transfer.
- Cover the assembly with a thick layer of tissue paper. Place a plastic plate with a weight, such as a small bottle, on top. Make sure the pressure is equally divided over the gel. This will ensure proper transfer of the DNA.
NOTE: The weight should be about 200–300 g; heavy weights hamper the DNA transfer. - Cover the area surrounding the assembly, including exposed filter paper, with cling film (Figure 6) to avoid evaporation of the 20x SSC buffer and target the capillary forces towards the nylon membrane. Blot overnight.
- Mark the position of the slots, name, and/or date on top of the membrane with pencil, and remove the membrane from the assembly. Note that the bottom side that has been in contact with the gel, carries the DNA.
- Immediately fix the DNA to the membrane by UV irradiation (2,400 µJ/m2) using a UV Crosslinker.
NOTE: Crosslinking conditions depend on the type of membrane used.
NOTE: At this point the membrane can be stored at -20 °C and used for hybridization with a probe later. Rinse the crosslinked membrane in 2x SSC and seal it in pre-folded heat-sealable polyethylene tubing before placing it at -20 °C.
- Prepare the probe for hybridization.
- Amplify the sequence to be used as the probe for DNA blotting by PCR20. 50-100 ng of a 250 bp-2 kbp PCR fragment is used as a probe. Two separate probes, one hybridizing to the Right border proximal region and the other to the Left border proximal region of the T-DNA, can be used to assess the presence of the entire T-DNA (Figure 4A and Figure 7).
- Dilute 50-100 ng of PCR product in 24 µL of ultrapure deionized water in a test tube.
- Denature the diluted PCR product by boiling it for 5 min in a beaker of water or heat block, then cool directly on ice.
- Thaw premade GCT-mix on ice. (GCT-mix: dGTP, dCTP, dTTP (all 0.5 mM), random hexamers 43.2 ng/µL, Acetylated BSA 1.33 mg/mL, 33 mM of β-mercaptoethanol, 0.67 M Hepes, 0.17 mM Tris pH 6.8, 17 mM MgCl).
- Add 21 µL of the GCT-mix and 2 U of Klenow fragment to the PCR product.
- Add 2 µL of [32P]ATP to the mix and incubate at 37 °C for 1 h.
CAUTION: All steps involving [32P]ATP need to be carried out in an environment designated for radioactive work while using the appropriate protection.
NOTE: Make sure the [32P]ATP is fresh (not more than 1 half-time has passed). - Prepare a Sephadex G-50 (coarse or medium) column21 for purifying the labeled probe from unincorporated (radioactive) nucleotides. Take a 2 mL syringe, and cover the outlet with a small circle of thick filter paper. Add 2 mL of Sephadex G-50 dissolved in TE into the syringe, and remove all liquid from the column by spinning.
- Place the column into a 15 mL plastic tube, load the labeled probe on the column, and spin at room temperature (set the centrifuge at 750 x g, allow the rpm to increase until 750 x g is reached, then stop the centrifuge and allow the rotation to decline to 0 x g) to elute the probe. Add 200 µL of TE to the column and spin to elute the remaining probe; repeat once. In these conditions, labeled DNA fragments are excluded from the Sephadex matrix and elute, while free nucleotides remain in the column.
- Use 300 µL of the labeled probe per hybridization tube. Keep the remaining labeled probe at -20 °C for later usage. However, keep the half time of [32P]ATP in mind.
- Hybridize the DNA blot.
- Heat 2x SSC and Hybridization buffer (15 mL per hybridization tube, maximum of 2 blots per tube) to 65 °C. (Hybridization buffer: 10% dextran sulphate, 1% SDS, 1 M NaCl, 50 mM Tris pH 7.5, dissolve at 65 °C, keep aliquots at -20 °C).
- Pre-heat the hybridization oven to 65 °C.
- Place nylon mesh in a tray with a little bit of heated (65 °C) 2x SSC to cover the tray. Place the DNA blot on top of the mesh, with the DNA side up. Roll the blot together with the mesh and insert the roll into a hybridization tube. Pour off the excess 2x SSC.
- In a microcentrifuge tube, boil 150 µL of Salmon Sperm DNA (concentration 10 mg/mL) per hybridization tube for 5 min (see also step 4.6.3). Cool immediately on ice and add to the pre-heated Hybridization buffer.
- Add the Hybridization buffer-Salmon Sperm solution to the tube with the blot. Pre-hybridize at 65 °C for at least 1 h in a rotating wheel, at 12 rpm.
- When the pre-incubation in step 4.7.5. is almost finished, boil 300 µL of the labeled probe for 5 min (see also step 4.6.3) and add immediately to the blot after the incubation.
- Hybridize overnight in the rotating wheel at 63 °C, 12 rpm. Do not pipette the probe directly on the blot, but into the Hybridization buffer-Salmon Sperm solution.
- Wash the blot.
- Preheat the washing solutions (1x SSPE, 0.1% SDS and 0.1x SSPE, 0.1% SDS) to 65 °C. (20x SSPE: 3 M NaCl, 230 mM NaH2PO4, 20 mM EDTA, pH 7.0).
- Dispose the hybridization solution and add about 100–150 mL of 1x SSPE, 0.1% SDS solution to the hybridization tube, close the tube, and rotate by hand. Pour the hybridization and washing solution in the appropriate liquid radioactive waste.
- Add about 100–150 mL of 1x SSPE, 0.1% SDS solution to the tube, close, and incubate the tube for 15 min at 63 °C in the rotating wheel, 12 rpm. Discard the washing solution appropriately.
- Add about 100–150 mL of 0.1x SSPE, 0.1% SDS solution to the hybridization tube, close, and rotate the tube for 5 min at 63 °C, 12 rpm. Discard the washing solution appropriately.
- Take the blot out of the tube and place it in a tray containing sufficient preheated 0.1x SSPE, 0.1% SDS, and shake for 3 min in a shaking water bath at 65 °C. Meanwhile, rinse the mesh in a tray filled with water.
- Take the blot out, place it in between pre-folded plastic (polyethylene tubing), carefully wipe excess liquid off, and let the blot dry briefly. Note that liquid will ruin the phosphorimager screen.
- Seal the blot in plastic on the three sides. Remove all excess liquid surrounding the blot and close the plastic tube by sealing the fourth side. Cut off the surplus of plastic. Make sure the sealed blot is not leaking and that the plastic is dry on the outside.
- Expose the phosphorimager screen.
- Place the sealed blot in a phosphorimager cassette, with the phosphorimager screen facing the DNA side of the blot. Close the cassette and leave for ± 2–4 days, depending on the strength of the radioactive labeling and sensitivity of the phosphorimager.
- Scan the phosphorimager screen using a phosphorimager. Take care to expose the screen as little as possible to light before scanning. Save the image. Erase the screen from signal by exposing it to bright light.
- Analyze the blot.
- The analysis depends on the restriction strategy used in step 4.1. When analyzing the blot prepared according to the strategy shown in step 4.1.1.1 (Figure 4A), count the number of the fragments detected. In this strategy, the number of hybridized fragments refers to the number of T-DNA integrations.
- Compare the number of the fragments detected with a probe for the left (Figure 7B) and the right (Figure 7C) part of the T-DNA.
NOTE: If a different number of hybridizing fragments is detected, then either i) multiple T-DNA copies (either in inverted or direct orientation) or ii) incompletely integrated T-DNAs are present. - Estimate the sizes of hybridized fragments on the blot based on the size of the marker bands, and compare the sizes of hybridized fragments with the expected fragment sizes calculated based on the restriction strategy of tandem insertions (Figure 4A) to identify possible tandem arrangement of the T-DNAs. Use the Figure 4A as a guide to calculate the size of expected fragments.
NOTE: If the size of hybridized fragments does not agree with calculated ones, then most likely one of the integrations present is not complete.
- Compare the number of the fragments detected with a probe for the left (Figure 7B) and the right (Figure 7C) part of the T-DNA.
- When analyzing the blot prepared according to strategy shown in step 4.1.1.2 (Figure 4B), estimate the size of the hybridizing fragment on the blot based on the size of marker bands, and compare it with the expected size. An intact insertion yields a single fragment with a defined length. Any deviation of the expected length indicates incomplete integration (Figure 7D).
- The analysis depends on the restriction strategy used in step 4.1. When analyzing the blot prepared according to the strategy shown in step 4.1.1.1 (Figure 4A), count the number of the fragments detected. In this strategy, the number of hybridized fragments refers to the number of T-DNA integrations.
- Strip the blot for re-hybridization (optional).
NOTE: The same blot can be hybridized consecutively with different probes. Before continuing with a new probe, strip a previous hybridized probe from the blot.- To remove the probe from the blot, place the blot in a tray with its DNA-side facing down. Pour a surplus of 0.5% SDS into the tray. Boil the membrane for 2–5 min. The duration of the treatment depends on the size and GC-content of the probe used. Longer and GC-richer probes need a longer treatment.
- After stripping, hybridize the blot with another probe, or seal and store at -20 °C.
Representative Results
Using the BIBAC-GW system, reporter constructs for studying ODM in plants were generated10. Constructs were designed in the Gateway Entry vector pENTR-gm12 and inserted into pBIBAC-BAR-GW (Figure 1) using the Gateway LR recombination reaction.
Arabidopsis were transformed with pDM19, a BIBAC-BAR-GW plasmid with an mTurquoise-eYFP reporter carrying a translational stop codon in the eYFP reading frame at position 120 (mTurquoise2-eYFP*40) (Figure 2)10. In total, 126 Arabidopsis plants were transformed (9 plants per pot, 14 pots). Seeds of these plants were pooled, sown on trays with soil, and allowed to grow for two weeks prior to treatment with Glufosinate-ammonium solution. Only seedlings expressing the bar gene (present in BIBAC-BAR-GW) survive Glufosinate-ammonium treatment (Figure 3). In total, 11 transgenics transformed with pDM19 were identified, corresponding to a transformation efficiency of 0.02% of the seeds analyzed.
For the 11 transgenics isolated, DNA blotting was used to determine the number of T-DNA integrations. For that purpose, genomic DNA was cut with either BglII or ScaI (strategy as devised on Figure 4A). Both of these restriction enzymes cut only once into the T-DNA sequence (Figure 7A). Hybridization with probes recognizing the bar and eYFP coding regions allowed detection of the number of respective DNA fragments.
The number of individual DNA fragments on the blots allowed for estimating the number of T-DNA insertions in the reporter lines (Table 1). Single hybridizing fragments with both the Bar and eYFP probe indicated the presence of a single T-DNA integration. From the 11 transgenics analyzed, six carried single integrations. The average number of integrations was 1.2.
For 6 lines carrying a single T-DNA integration, the integrity of the inserted reporter construct was tested using DNA blotting (strategy as devised on Figure 4B). Genomic DNA was cut with BglII and PciI to release a 5.5 kb fragment containing both the Bar and mTurquoise-eYFP fusion gene (Figure 7A). A probe against eYFP was used to detect the expected fragment. All plants tested carried an intact fragment. Note that the fragment examined excludes the Left and the Right T-DNA border, and therefore does not examine the integrity of the entire T-DNA, but only the part containing the transgenes of interest.
The expression of the fluorescent reporter gene was determined in independent single-copy transgenic lines differing only by the genomic location of the T-DNA. Relative transcript levels of the CaMV-35S promoter-driven mTurquoise-eYFP reporter were measured by RT-qPCR in four DM19 reporter lines carrying intact, single-copy integrations of which the genomic position was determined10. The variation in reporter gene expression levels between the lines was minor: the maximum difference in mTurquoise-eYFP RNA levels was 2-fold (Figure 8A).
Next, the ODM was carried out in these reporter lines. Three out of the four independent reporter lines showed rather similar ODM efficiencies (Figure 8B). However, one line, DM19[4]1, yielded a very low ODM efficiency compared to the other lines. These results indicate that the ODM is affected by the local genomic context. In what manner the local genomic context of the T-DNA integration in DM19[4]1 differs from that in the other lines remains to be identified. Analysis of available datasets on active and inactive chromatin marks at the genomic T-DNA integration sites in non-transgenic plants did not provide an answer10.
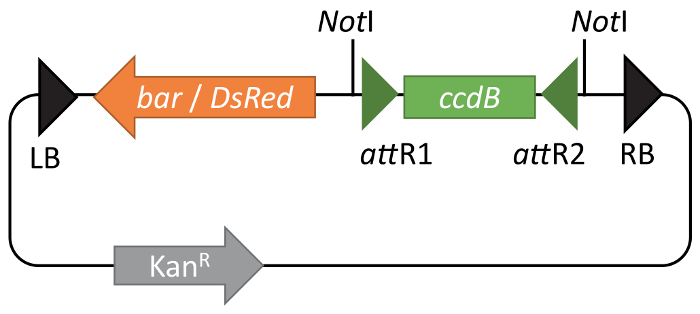
Figure 1: Functional maps of pBIBAC-GW vectors. pBIBAC-GW derivatives are available with either resistance to Glufosinate (bar) or DsRed fluorescence in seed coats (DsRed) as a selection marker in plants. For both vectors, a kanamycin resistance gene is the selection marker in bacteria. The Gateway ccdB cassette is shown between green arrowheads representing recombination sites attR1 and attR2. Please click here to view a larger version of this figure.

Figure 2: Mutagenesis reporter construct. The mTurquoise-eYFP reporter genes are driven by the 35S-CaMV promotor. The mTurquoise coding region is fused to an eYFP coding region carrying a C-A mutation at nucleotide position 120, resulting in a premature translational stop codon TAA, and premature termination of the translation of the fusion protein. The 3′ Nopaline Synthase (3'nos) polyadenylation signal is used to terminate the transcription of the construct22. Nuclear localization signal (NLS) is used to target the translated proteins to the nucleus. Please click here to view a larger version of this figure.
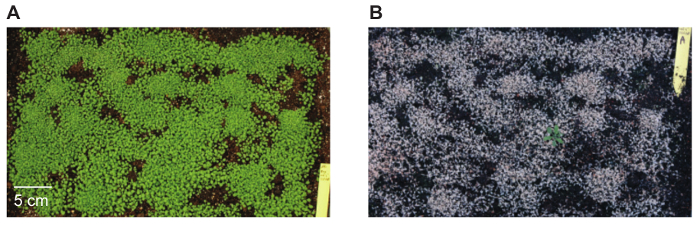
Figure 3: Tray filled with Arabidopsis seedlings before and after Glufosinate-ammonium treatment. Seedlings not expressing the bar gene that is present in the pBIBAC-BAR-GW T-DNA die after being sprayed with Glufosinate-ammonium solution. The photos show the same tray of seedlings (A) before spraying with Glufosinate-ammonium, 14 days after sowing, and (B) 10 days later, after being sprayed twice. Please click here to view a larger version of this figure.
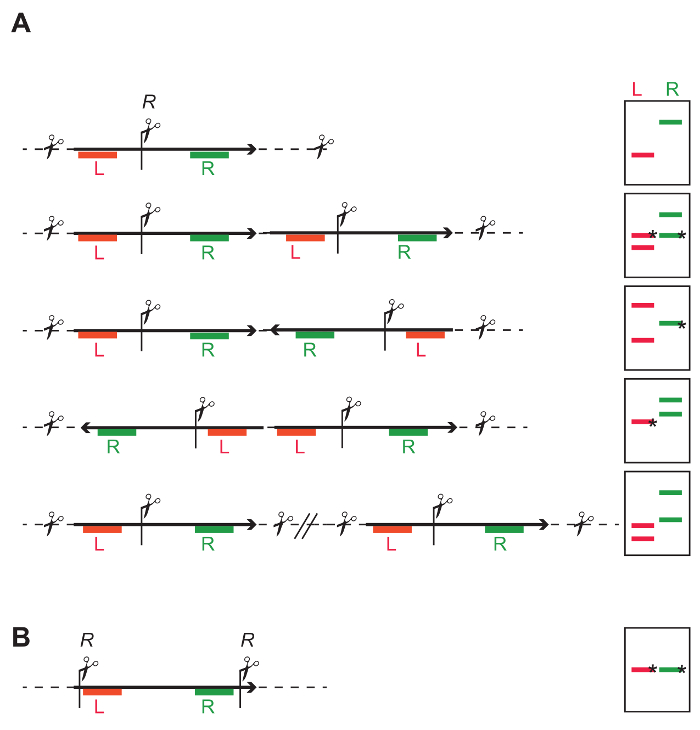
Figure 4: General DNA restriction strategy to identify the number and intactness of inserted T-DNAs. (A) One restriction site (R) in the middle of the T-DNA allows independent probing of the left (red L) and right part of the T-DNA (green R). The cartoons on the right show that depending on single- or multi-copy T-DNA integrations, different banding patterns are obtained with DNA blotting. Bands marked with an * have a defined length, while the length of other bands depends on the closest restriction site in the flanking genomic DNA. Single insert: The L and R probe both give one independent fragment. The expected average fragment size can be calculated based on the frequency of the restriction site in the genome. The minimal size is the distance from the restriction site to the Left Border (LB) or Right Border (RB), depending on which end of the integration is being probed, and if the T-DNA is intact. Tandem repeat: The probes for L and R give both two fragments; for each probe one of the fragments includes flanking genomic DNA, the second fragment has an expected size and is identified by both probes. Inverted repeat: Depending on the directionality of the integrated cassette, either one L and two R fragments, or two L and one R can be identified. Individual single insertions: The result is a number of independent fragments, and the number of fragments corresponds to the number of integrations. (B) Restriction sites at the extremities of the T-DNA allow determining the integrity of the fragment between the restriction sites. Please click here to view a larger version of this figure.
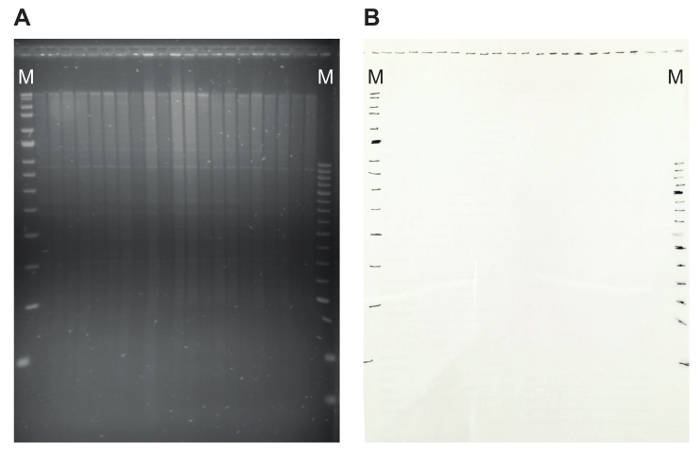
Figure 5: An agarose gel with restriction pattern and the matching transparency. (A) On the agarose gel, genomic DNA digested with EcoRI is shown. The proper digestion of the DNA is illustrated by the presence of discrete satellite bands. (B) Marking the position of the slots and marker bands on a transparency makes it possible to later easily calculate the size of hybridizing fragments. Here, MRC Holland markers (Blue and Red) are used, indicated by M. Please click here to view a larger version of this figure.
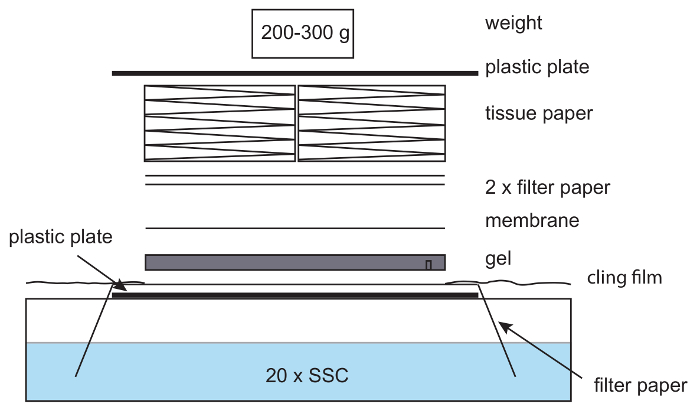
Figure 6: Setup for capillary blotting. In a capillary blotting setup, filter paper is placed on a plastic plate with the ends of the paper hanging in 20x SSC buffer. The paper is wetted with 20x SSC, and an agarose gel placed on top, followed by a nylon membrane, filter paper, and a stack of tissues. A light weight is placed on top. Care is taken to remove air bubbles between the gel, paper and membrane. Cling film is used to avoid drying out of the setup. Please click here to view a larger version of this figure.
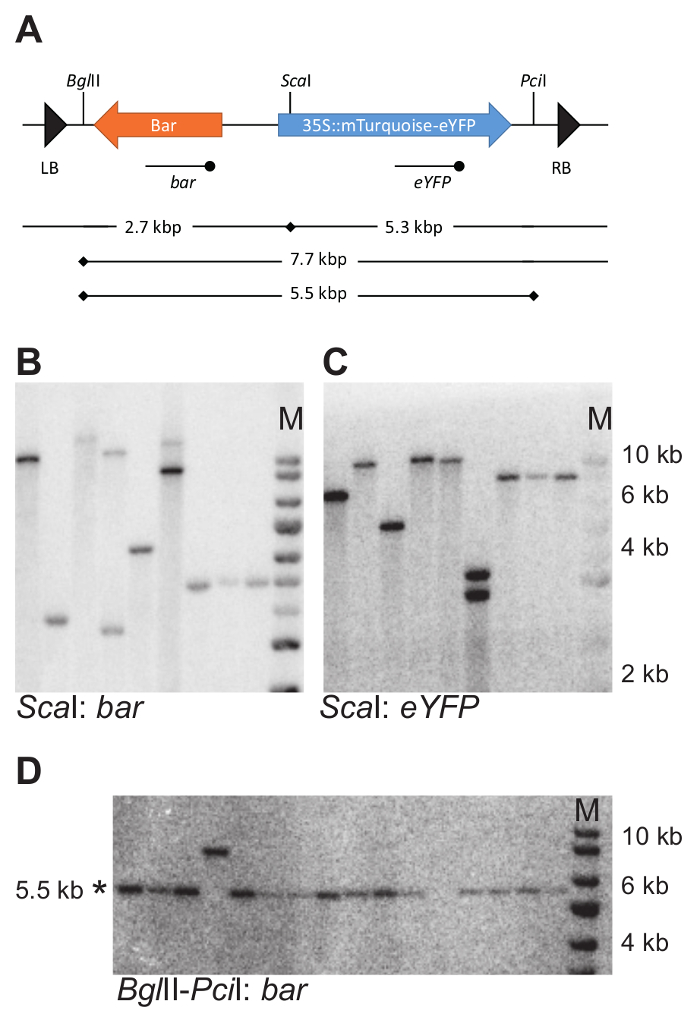
Figure 7: Example of DNA blotting strategy and experimental outcome. (A) DNA blotting strategy to determine the number and intactness of T-DNA integrations. Cutting locations of the selected restriction enzymes within the T-DNA are indicated with vertical bars. The eYFP and bar probes used for hybridization with digested genomic DNA are indicated using a line with the terminal dot below the T-DNA. (B–D) Example DNA blots. Genomic DNA was cut with ScaI and the blot was probed with both a bar and eYFP probe (B and C). Genomic DNA was cut with BglII and PciI and probed with a bar probe. Intact fragments are 5.5 kbp in size (D). Note that the set of samples in D differs from those shown in B and C. * indicates the expected fragment size; M, marker. In B, C, and D the same size marker is used. Please click here to view a larger version of this figure.
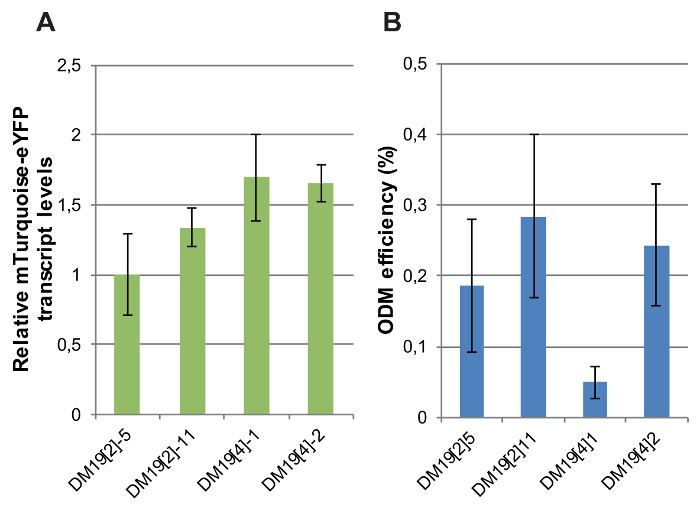
Figure 8: mTurquoise-eYFP expression levels and ODM efficiencies in independent mTurquoise-eYFP reporter lines. (A) Relative mTurquoise-eYFP transcript levels measured by RT-qPCR in DM19 reporter lines. For normalization transcript levels of Actin were used. (B) ODM efficiency measured in the DM19 reporter lines. For A and B, bars indicate the average of at least five biological replicates. Error bars indicate SEM. Please click here to view a larger version of this figure.
| Type of T-DNA locus | Nr of T-DNA integrations | Reporter line | Number of fragments detected | Integrity | |||
| ScaI | BglII | BglII/PciI | |||||
| bar | eYFP | bar | eYFP | eYFP | |||
| Single locus integrations | 1 | 19[2]-2 | 1 | 1 | 1 | 1 | + |
| 1 | 19[2]-5 | 1 | 1 | 1 | 1 | + | |
| 1 | 19[2]-9 | 1 | 1 | 1 | 1 | + | |
| 1 | 19[2]-11 | 1 | 1 | 1 | 1 | + | |
| 1 | 19[4]-1 | 1 | 1 | 1 | 1 | + | |
| 1 | 19[4]-2 | 1 | 1 | 1 | 1 | + | |
| 2, inverted repeat | 19[2]-10 | 2 | 1 | 1 | 1 | + | |
| 2, incomplete integration | 19[2]-3 | 1 | 2 | 1 | 1 | ND | |
| Multiple locus integrations | 2 | 19[2]-6 | 2 | 2 | 2 | ND | |
| 2 | 19[2]-7 | 2 | 2 | 2 | 2 | ND | |
| 3/4 | 19[2]-1 | 4 | 3 | 3 | 3 | ND | |
| ND – not determined. | |||||||
Table 1: Summary of the DNA blotting data for transgenics isolated after transformation with pDM19.
Discussion
Critical to generating transgenics with single, intact integrations of a transgene is the choice of the binary vector used. BIBAC family vectors have been used to deliver sequences of interests to many plant species23,24,25,26,27,28. BIBAC vectors, including BIBAC-GW, yield single-copy integrations with high efficiency: the average number of insertions per line is 1.5 to 2, compared to 3 or higher for most commonly used binary vectors5,9,29. As a major improvement compared to other BIBAC vectors, with the BIBAC-GW vectors, the sequences of interest can be easily inserted using Gateway recombination sites12. The modified vectors overcome the general problems of BIBAC vectors when used in conventional cloning strategies: i) a very limited number of unique restriction sites and ii) a low DNA yield. The Gateway recombination sites make BIBAC-GW vectors an attractive alternative to other binary vectors for generating transgenic plants.
Here a series of protocols, from generating BIBAC-GW derivatives containing sequences of interest, to plant transformation, and DNA blot analysis for number and intactness of the transgenic sequences is described. Several of the protocols reported in this paper, Gateway cloning, electroporation of bacteria, and plant transformation, are common practice in many laboratories and can also be carried out with slight modifications. It is important to know that BIBAC-GW is a single-copy vector in E. coli and in A. tumefaciens. Therefore, when isolating DNA, the yield is low; it is recommended to scale up the isolation procedure.
In transgenics carrying multiple T-DNA integrations, introduced transgenes are often subjected to gene silencing1,2,4,30, and should therefore for most applications be avoided. To identify transgenic plants with single, intact integrations, it is recommended to use DNA blot analysis. While methods other than DNA blotting can be used for determining T-DNA copy number and integrity of the T-DNAs in transgenic lines (segregation analysis, TAIL-PCR, quantitative PCR (qPCR), and digital droplet PCR), although labor intensive, DNA blotting is often the method of choice. Segregation analysis is not able to differentiate between multiple and single T-DNA integrations at single loci. TAIL-PCR often under-estimates the copy number, especially if more than one T-DNA integration is present31, and qPCR needs elaborate optimization for reliable results31,32. Digital droplet PCR is a rather precise method for copy number detection if the equipment required is available31. The added benefit of DNA blotting is facile detection of truncated T-DNAs, which is easily missed in all PCR-based techniques.
With DNA blot analysis, the hybridizing fragments on the blot need to be well identifiable in signal and size. Several factors are known to affect the outcome of DNA blotting. In addition to an appropriate selection of restriction enzymes (strategy indicated in Figure 4) and size markers, sufficient DNA of good quality is required. Less than 2 µg of genomic Arabidopsis DNA will not yield well-identifiable fragments. When dealing with bigger genomes, more DNA is required. To obtain sufficient amounts of Arabidopsis DNA, floral tissues or 1-week old seedlings can be used. A batch of seedlings grown on one Petri dish yields 2–8 µg of DNA. In order to avoid DNA degradation during isolation, care should be taken to process the plant material fast. Furthermore, genomic DNA should be resuspended in Tris-EDTA to reduce its degradation by nucleases, and stored at 4 °C rather than -20 °C to prevent DNA nicking due to repeated freezing-thawing cycles. If unsure that all DNA samples are digested completely, it is suggested to rehybridize the DNA blot with a probe recognizing an endogenous, unique genomic region. When selecting probe sequences for identifying transgenic or endogenous sequences, it is crucial to only select unique sequences. To be able to precisely determine the size of hybridized fragments, the positions of DNA gel slots and DNA marker bands should be marked on a transparency (Figure 5B) when visualizing an Ethidium Bromide-stained gel (Figure 5A) on a UV transilluminator. In case the marker sequences do not hybridize with the probe DNA, or hybridization is partial, it is the only way to track down the size of hybridizing fragments.
Once care is taken to achieve good hybridization signal and estimate of the fragment size, the interpretation of the blotting results is straightforward. When using only one restriction enzyme, and hybridizing with different probes detecting either the left or right part of the T-DNA, the number of detected fragments reflects the number of T-DNA insertions. For instance, Figure 7B, C shows the same DNA blot, hybridized with different probes, bar (Figure 7B) and enhanced Yellow Fluorescent Protein (eYFP) (Figure 7C), using the strategy shown in Figure 7A. All lanes, except 4, show an equal number of fragments on both blots: two fragments for line 6 and a single fragment for all other lines. This number of detected fragments is the number of T-DNA insertions.
When the number of fragments detected with probes binding either the left or right part of the T-DNA differs (as for Figure 7B, C, line 4), either incomplete insertions are present, or the T-DNAs have inserted in tandem arrangement. The tandem insertions display non-random fragment length for one of the T-DNA fragments (Figure 4A, right panel), and can be identified by comparing the hybridized fragment size with what is expected based on the restriction strategy. An additional blotting strategy may be needed to confirm the tandem arrangement of the T-DNA insertions. In the sample shown on line 4 (Figure 7B, C), two insertions are arranged in an inverted repeat orientation.
When estimating the intactness of a T-DNA, or part of it, the length of the hybridizing fragment can be calculated based on the restriction strategy. Any deviation from the expected size indicates the presence of an incomplete insertion. For instance, in Figure 7D, in lane 4, a hybridizing fragment migrates at 8 kbp, (instead of the expected 5.5 kbp) indicating an increased fragment size due to the lack of one of the restriction sites.
BIBAC-GW vectors are excellent tools for generating single-copy intact integrations in a number of plant species. The protocol reported here provides a reliable procedure to identify plants with single, intact integrations of a transgene of interest.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research is supported by the Dutch Technology Foundation STW (12385), which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs (OTP Grant 12385 to MS). We thank Carol M. Hamilton (Cornell University, United States) for providing pCH20, the backbone of the BIBAC-GW vectors.
Materials
| Kanamycin sulphate monohydrate | Duchefa | K0126 | |
| Gentamycin sulphate | Duchefa | G0124 | |
| Rifampicin | Duchefa | R0146 | |
| Tetracycline hydrochloride | Sigma | T-3383 | |
| DB3.1 competent cells | Thermo Scientific – Invitrogen | 11782-018 | One Shot ccdB Survival 2 T1R Competent Cells (A10460) by Invitrogen or any other ccdB resistant E. coli strain can be used instead |
| DH10B competent cells | Thermo Scientific – Invitrogen | 18290-015 | |
| Gateway LR clonase enzyme mix | Thermo Scientific – Invitrogen | 11791-019 | |
| tri-Sodium citrate dihydrate | Merck | 106432 | |
| Trizma base | Sigma-Aldrich | T1503 | |
| EDTA disodium dihydrate | Duchefa | E0511 | |
| Proteinase K | Thermo Scientific | EO0491 | |
| Bacto tryptone | BD | 211705 | |
| Yeast extract | BD | 212750 | |
| Sodium chloride | Honeywell Fluka | 13423 | |
| Potassium chloride | Merck | 104936 | |
| D(+)-Glucose monohydrate | Merck | 108346 | |
| Electroporation Cuvettes, 0.1 cm gap | Biorad | 1652089 | |
| Electroporator Gene Pulser | BioRad | ||
| Magnesium sulfate heptahydrate | Calbiochem | 442613 | |
| D(+)-Maltose monohydrate 90% | Acros Organics | 32991 | |
| Sucrose | Sigma-Aldrich | 84100 | |
| Silwet L-77 | Fisher Scientific | NC0138454 | |
| Murashige Skoog medium | Duchefa | M0221 | |
| Agar | BD | 214010 | |
| Glufosinate-ammonium (Basta) | Bayer | 79391781 | |
| Restriction enzymes | NEB | ||
| Ethidium Bromide | Bio-Rad | 1610433 | |
| Electrophoresis system | Bio-Rad | ||
| Sodium hydroxide | Merck | 106498 | |
| Hydrochloric acid | Merck | 100316 | |
| Blotting nylon membrane Hybond N+ | Sigma Aldrich | 15358 | or GE Healthcare Life Sciences (RPN203B) |
| Whatman 3MM Chr blotting paper | GE Healthcare Life Sciences | 3030-931 | |
| dNTP | Thermo Fisher | R0181 | |
| Acetylated BSA | Sigma-Aldrich | B2518 | |
| HEPES | Sigma-Aldrich | H4034 | |
| 2-Mercaptoethanol | Merck | 805740 | |
| Sephadex G-50 Coarse | GE Healthcare Life Sciences | 17004401 | or Sephadex G-50 Medium (17004301) |
| Dextran sulfate sodium salt | Sigma-Aldrich | D8906 | |
| Sodium Dodecyl Sulfate | US Biological | S5010 | |
| Salmon Sperm DNA | Sigma-Aldrich | D7656 | |
| Sodium dihydrogen phosphate monohydrate | Merck | 106346 | |
| Storage Phosphor screen and casette | GE Healthcare Life Sciences | 28-9564-74 | |
| Phosphor imager | GE Healthcare Life Sciences | Typhoon FLA 7000 | |
| UV Crosslinker | Stratagene | Stratalinker 1800 | |
| cling film (Saran wrap) | Omnilabo | 1090681 | |
| Agarose | Thermo Scientific – Invitrogen | 16500 | |
| Boric acid | Merck | 100165 | |
| DNA marker ‘Blauw’; DNA ladder. | MRC Holland | MCT8070 | |
| DNA marker ‘Rood’; DNA ladder | MRC Holland | MCT8080 | |
| Hexanucleotide Mix | Roche | 11277081001 | |
| Large-Construct Kit | Qiagen | 12462 | |
| Heat-sealable polyethylene tubing, clear | various providers | the width of the tubing should be wider than that of blotting membrane | |
| Heat sealer | |||
| Membrane filter disk | Merck | VSWP02500 | |
| Magnesium chloride | Merck | 105833 | |
| Hybridization mesh | GE Healthcare Life Sciences | RPN2519 |
References
- Jorgensen, R. A., Cluster, P. D., English, J., Que, Q., Napoli, C. A. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol. 31 (5), 957-973 (1996).
- Stam, M., et al. Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 12, 63-82 (1997).
- Stam, M., Viterbo, A., Mol, J. N., Kooter, J. M. Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol Cell Biol. 18 (11), 6165-6177 (1998).
- Jin, Y., Guo, H. S. Transgene-induced gene silencing in plants. Methods Mol Biol. 1287, 105-117 (2015).
- Oltmanns, H., et al. Generation of Backbone-Free, Low Transgene Copy Plants by Launching T-DNA from the Agrobacterium Chromosome 1[W][OA]. Plant Physiol. , (2010).
- Ye, X., et al. Enhanced production of single copy backbone-free transgenic plants in multiple crop species using binary vectors with a pRi replication origin in Agrobacterium tumefaciens. Transgenic Res. , (2011).
- Hamilton, C. M. A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene. 200 (1-2), 107-116 (1997).
- Frary, A., Hamilton, C. M. Efficiency and stability of high molecular weight DNA transformation: an analysis in tomato. Transgenic Res. 10 (2), 121-132 (2001).
- Vega, J. M., et al. Agrobacterium-mediated transformation of maize (Zea mays) with Cre-lox site specific recombination cassettes in BIBAC vectors. Plant Mol Biol. 66 (6), 587-598 (2008).
- Anggoro, D. T., Tark-Dame, M., Walmsley, A., Oka, R., de Sain, M., Stam, M. BIBAC-GW-based vectors for generating reporter lines for site-specific genome editing in planta. Plasmid. 89, 27-36 (2017).
- Hamilton, C. M., Frary, A., Lewis, C., Tanksley, S. D. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci U S A. 93 (18), 9975-9979 (1996).
- Belele, C. L., Sidorenko, L., Stam, M., Bader, R., Arteaga-Vazquez, M. A., Chandler, V. L. Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet. 9 (10), e1003773 (2013).
- Shizuya, H., et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A. 89 (18), 8794-8797 (1992).
- Shi, X., Zeng, H., Xue, Y., Luo, M. A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant Methods. 7, 33 (2011).
- Woodman, M. E., et al. Direct PCR of Intact Bacteria (Colony PCR). Curr Protoc Microbiol. , A.3D.1-A.3D.7 (2016).
- Ausubel, F. M., et al. Mol Biol. Current Protocols in Molecular Biology. 1, (2003).
- Edwards, K., Johnstone, C., Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 (6), 1349 (1991).
- Clarke, J. D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. (3), (2009).
- Lee, P. Y., Costumbrado, J., Hsu, C. Y., Kim, Y. H. Agarose Gel Electrophoresis for the Separation of DNA Fragments. J Vis Exp. (62), e3923 (2012).
- Green, M. R., Sambrook, J. . Molecular Cloning. , (2012).
- Sosa, J. M. Chromatography with Sephadex Gels. Anal Chem. 52 (6), 910-912 (1980).
- Depicker, A., Stachel, S., Dhaese, P., Zambryski, P., Goodman, H. M. Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet. 1 (6), 561-573 (1982).
- Feng, J., Vick, B. A., Lee, M. K., Zhang, H. B., Jan, C. C. Construction of BAC and BIBAC libraries from sunflower and identification of linkage group-specific clones by overgo hybridization. Theor Appl Genet. 113 (1), 23-32 (2006).
- Lee, M. K., et al. Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid Upland cotton (Gossypium hirsutum L). BMC Genomics. 14, 208 (2013).
- Wang, W., et al. A large insert Thellungiella halophila BIBAC library for genomics and identification of stress tolerance genes. Plant Mol Biol. 72 (1-2), 91-99 (2010).
- Wu, C., et al. A BAC- and BIBAC-based physical map of the soybean genome. Genome Res. 14 (2), 319-326 (2004).
- Xu, Z., et al. Genome physical mapping from large-insert clones by fingerprint analysis with capillary electrophoresis: a robust physical map of Penicillium chrysogenum. Nucleic Acids Res. 33 (5), e50 (2005).
- Zhang, M., et al. Genome physical mapping of polyploids: a BIBAC physical map of cultivated tetraploid cotton, Gossypium hirsutum L. PLoS One. 7 (3), e33644 (2012).
- Frary, A., Hamilton, C. M. Efficiency and stability of high molecular weight DNA transformation: An analysis in tomato. Transgenic Res. , (2001).
- Stam, M., Viterbo, A., Mol, J. N. M., Kooter, J. M. Position-Dependent Methylation and Transcriptional Silencing of Transgenes in Inverted T-DNA Repeats: Implications for Posttranscriptional Silencing of Homologous Host Genes in Plants. Cell Biol. 18 (11), 6165-6177 (1998).
- Głowacka, K., Kromdijk, J., Leonelli, L., Niyogi, K. K., Clemente, T. E., Long, S. P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 39 (4), 908-917 (2016).
- Stefano, B., Patrizia, B., Matteo, C., Massimo, G. Inverse PCR and Quantitative PCR as Alternative Methods to Southern Blotting Analysis to Assess Transgene Copy Number and Characterize the Integration Site in Transgenic Woody Plants. Biochem Genet. 54 (3), 291-305 (2016).

